Abstract
The authors describe an ultrasensitive amperometric enzymatic assay for uranyl ion. It is based on the use of mesoporous silica nanoparticles (mesoSiNPs) loaded with Methylene Blue (MB) and functionalized with an UO2(II)-dependent DNAzyme. The electroactive label MB was sealed in the inner pores of the mesoSiNPs along with double stranded DNA (containing the DNAzyme and the substrate strand). In the presence of UO2(II), the DNAzyme is actived to cleave the substrate strands. This leads to the cleavage of the caps and the release of MB from the mesoSiNPs. The amount of released MB depends on the concentration of UO2(II) and can be determined amperometrically, best at a working voltage of −0.25 V (vs SCE), by using a chitosan coated carbon paste electrode. Response is linear in the 20 pM to 0.1 nM UO2(II) concentration range, and the detection limit is as low as 0.15 pM. Recoveries from spiked samples varied from 91.3 to 99.4%. The assay is highly specific, selective, and not interfered by other metal ions.

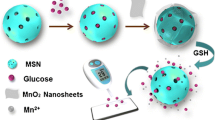
(a) Schematic representation of the synthesis, MB loading, and dsDNA Binding of the mesoSiNPs, as well as the release of MB from mesoSiNPs in the presence of UO2 2+. (b) Schematic illustration of the stepwise DNAzyme-based electrochemical sensor for uranium detection.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Uranium is a radioactive metal that is widely used in nuclear weapons, nuclear power plants, and so on [1]. Since uranium is one of the main sources in nuclear energy generation, it can be released into the environment and groundwater via uranium mining and improper disposal of nuclear waste [2]. Uranium exists in a variety of chemical forms in nature, while uranyl ion (UO2 2+) is the most stable chemical species in aqueous solution that represents the greatest risk to human beings and other living species because of its bioavailability [3]. Therefore, it’s imperative that the development of novel UO2 2+ detection methods that are efficient, sensitive and selective and applicable to aqueous systems. So far, various instrumental techniques, for example, spectroscopic analysis [4], mass spectrometry [5], atomic absorption spectroscopy [6], cold vapor atomic fluorescence spectrometry [7] were developed for the detection of UO2 2+ in aqueous solution. However, these techniques need expensive instruments, and moreover, the sample preparation is time-consuming and tedious.
To reduce instrumentation costs and time of sample preparation, the methods of detection of UO2 2+ have been reported. For example, Kumar et al. reported a simple analytical method for detection of UO2 2+ via photoluminescence quenching of amino-modified cadmium sulfide quantum dots [8]. In addition, Khashab et al. presented an efficient colorimetric peroxidase mimetic method to detect UO2 2+ by BSA-stabilized gold nanoclusters (BSA-AuNCs). In the absence of UO2 2+, the BSA-AuNCs showed an instrnsic peroxidase like activity [9]. In the presence of UO2 2+, this activity can be efficiently restrained. However, these methods have relatively poor selectivity, which do not meet the requirement of rapid detection of UO2 2+ in aqueous environment. Deoxyribozymes (DNAzymes) which are DNA molecules with high specificity for a number of metal ions have attracted more attention [10,11,12,13]. So far, a number of highly specific DNAzymes have been isolated and used for detecting metal ions such as Pb2+, Zn2+, Cu2+, Mn2+ and so on. Not long ago, a UO2 2+-specific DNAzyme was reported by Lu and his coworkers. Encouraged by the discovery of UO2 2+-specific DNAzyme, some DNAzyme based biosensors have been developed for the rapid and specific detection of UO2 2+. For example, Lu and his coworkers have prepared a efficient colorimetric sensor for detection of UO2 2+ based on gold nanoparticles (AuNPs) and UO2 2+-specific DNAzymes [14]. A fluorescent biosensor for detecting UO2 2+ was also developed based on UO2 2+-specific DNAzymes and the fluorescence quenching ability of molybdenum disulfide (MoS2) nanosheets [15]. Although some progress has been made over the past few years, it still is a significant challenge to design efficient and portable sensors for UO2 2+ detection with both high sensitivity and selectivity.
Among these techniques, electrochemical approaches have attracted more attention for good recognition capability, high sensitivity and selectivity, fast response and real time detection nature [16, 17]. Although our group prepared UO2 2+-specific DNAzymes electrochemical biosensors for detection of UO2 2+ [18, 19], these methods have an intrinsic limitation in sensitivity, as one UO2 2+ converts only one signal readout. To achieve signal amplification and highly sensitive detection of UO2 2+, mesoporous silica nanoparticles (mesoSiNPs) as carrier vehicles in this work, which has unique pore structure, biocompatibility, and ease of functionalization. More importantly, the large pore volume and surface area of mesoSiNPs make it possible to load large numbers of electroactive molecules.
In this system, we designed a novel electronic switch for detection of UO2 2+ by using MB-loaded and UO2 2+-specific DNAzyme induced mesoSiNPs. In the presence of UO2 2+, the release of more MB by the DNA substrate strand disrupting amplifies the electrochemical signal. The strategy establishes a efficient, high specificity and high sensitivity method for the detection of uranium, and provides a novel protocol for point-of-care testing of other metal ions.
Experimental
Chemicals and instruments
All DNA oligonucleotides used in this paper were designed according to articles and synthesized by Sangon Biotech Co., Ltd. (shanghai, China, http://www.sangon.com/), including the sequences (from 3′ to 5′) as described in Table S1 (in the Supporting Information) [14]. Cetyltrimethylammonium-bromide (CTAB), Sulfosuccinimidyl-4-(N-maleimidomethyl)-cyclohex -ane-1-carboxylate (Sulfo-SMCC), Tetraethylorthosilicate (TEOS), 3-aminopropyltriethoxysilane (APTES), Tris (hydroxymethyl) metyl aminomethane (Tris), Graphite powder, Paraffin oil, chitosan (chit) and Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) were purchased from Aladdin Reagent Corporation (Shanghai, China, http://www.aladdin-e.com/). All other chemicals were of analytical grade, and were used without further purification. All aqueous solutions were prepared with ultrapure water (18.2 MΩ·cm, Milli-Q, Millipore). DNA hybridization buffer (pH 7.4) contained 10 mM Tris-HCl, 1 mM ethylenediaminetetraacetic acid (EDTA). Phosphate -buffered saline (PBS, pH 7.4) contained 1.98 mM KH2PO4, 4.0 mM Na2HPO4, 2.6 mM KCl, 40 mM NaCl. Uranium nitrate hexahydrate was dissolved in water to make a 1 mM stock solution.
Electrochemical measurements were performed on a CHI660C electrochemical workstation (CH Instruments, Chenhua Co., Ltd. China, http://www.chinstr.com/). A conventional three-electrode cell assembly consisting of a modified carbon paste electrode (CPE) (3.5 mm in diameter) serving as the working electrode, a KCl saturated calomel electrode as the reference electrode and platinum (Pt) wire as the counter electrode. All measurements were carried out at room temperature. The scanning electron microscope (SEM) images were obtained with a JSM-6700F instrument (JEOL Ltd., Japan, http://www.jeol.co.jp/cn/). Zeta-potential analysis was performed on a Zetasizer (Nano-Z, Malvern, UK, http://www.chem17.com/st15315/product_ 139,196.html). Nitrogen absorption/desorptionmeasurement was obtained with a Autosorb IQ (Quantachrome Instruments U.S., http://www.quantachrome-china.com).
Preparation and functionalization of the MesoSiNPs
In this research, according to the related articles, mesoSiNPs was prepared by sol-gel methods [20]. Briefly, cetyltrimethylammonium bromide (CTAB, 1.8 g) was dissolved in double distilled water (864 mL), followed by addition of NaOH (2 M, 6.3 mL). The precipitation was stirred at 40 °C for 2 h and followed by adjusting the solution temperature to 80 °C. Next, the tetraethylorthosilicate (TEOS) (9 mL) was added dropwise to precipitate at a rate of 1 mL·min−1. After stirring 2 h, the white SiO2 precursor were washed and dried under vacuum at 100 °C for 12 h. Then it was transferred into a muffle furnace and slowly heated to 550 °C for 6 h. The obtained white powder was mesoSiNPs that removed the template phase.
Next, sample of the extracted mesoSiNPs (3 g) was refluxed in anhydrous ethanol (100 mL) with 3-aminopropyltriethoxysilane (APTES) (3 mL) inside a round bottom flask and stirred at 60 °C in water bath. After 6 h of reaction time, the reaction mixture was filtered, washed with ethanol and then placed in a 100 °C vacuum drying ovens to obtain amino-mesoSiNPs (mesoSiNPs-AM).
Then, mesoSiNPs-AM (40 mg) was added to MB (0.26 mM, 40 mL) solution and the mixture was shaken at 220 r·min−1 and 40 °C for 1 h. The solid product was filtered, rinsed with water and dried to obtain MB-loaded mesoporous silica nanoparticle (mesoSiNPs-AM@MB) [21, 22].
Then, sulfosuccinimidyl-4-(N-maleimidomethyl)- cyclohexane-1-carboxylate (Sulfo-SMCC) (120 μL) was dissolved in PBS (10 mM, 1 mL), and reacted with mesoSiNPs-AM@MB (12 mg·mL−1) for 30 min, and then precipitated by over desalting column and reacted with double-stranded DNA (dsDNA) for 30 min to immobilized the dsDNA onto the methylene blue-loaded silica nanomaterials and sealed the methylene blue in the pores of the silica (mesoSiNPs-AM@MB@dsDNA).
Fabrication of electrode
The bare CPE was prepared by mixing graphite powder (87 mg) and paraffin oil (21 μL) in an agate mortar. The paste was then tightly pressed into a polypropylene tube with an inner diameter of 3.5 mm. A copper wire was inserted into the carbon paste to provide the electrical contact. The surface of the bare CPE was smoothed on a weighing paper [23].
Chitosan (chit) (50 mg) was dissolved in acetic acid (1%) solution for 40 min, which was adjusted in six different pH varied from 5.0 to 7.5 with NaOH (2 M), respectively. The mixture of dropping the chit in different pH (10 μL) and mesoSiNPs-AM@MB@dsDNA (20 μL) were applied to CPE, natural dried. The coated process was carried out on a clean bench.
Electrochemical measurements
The modification process of the sensor was detected by cyclic voltammetry (CV), differential pulse voltammetry (DPV), and electrochemical impedance spectroscopy (EIS). The three electrode system, which KCl saturated calomel electrode as reference electrode, platinum electrode as auxiliary electrode and CPE as working electrode was used. CV and EIS measurements were performed in PBS containing [Fe(CN)6]3−/4- (5 mM). CV was conducted at the potential range of −0.1 V to 0.6 V under a scan rate of 0.1 V·s−1. EIS was performed at a frequency ranging from 0.1 Hz to 100 kHz with a amplitude of 5 mV. DPV was carried out in PBS (10 mM, pH 7.4) at the potential range of −0.6 V to 0.6 V under modulation amplitude of 50 mV and a pulse width of 0.2 s. All experiments were carried out at room temperature.
Results and discussion
Detection mechanism
The electrochemical strategy was mainly based on mesoSiNPs-AM@MB@dsDNA as the signal nanoprobe for UO2 2+ detection. As depicted in Scheme 1a, the specifically recognizing DNAzyme is immobilized on the aminated mesoporous silica and sealed MB in the mesopores. Sulfo-SMCC was heterobifunctional crosslinkers that contained N-hydroxysuccinimide (NHS) ester and maleimide groups. Its NHS esters reacted with primary mesoSiNPs-AM to form amide bonds, while maleimides reacted with the thiolated modified dsDNA to form stable thioether bonds, which made the dsDNA immobilized on the surface of MB-loaded amino-functionalization mesoSiNPs.
The general principle of UO2 2+-special DNAzyme as a stimuli-responsive cap for the controlled release of the MB loaded mesoSiNPs-based electrochemical biosensor for the detection of UO2 2+ (Scheme 1b). The chit and mesoSiNPs-AM@MB@dsDNA mixed dropp were applied to the CPE, when the presence of UO2 2+ induced the cleavage of the DNA substrate strand at the rA aposition to form two fragments, which led to the release of MB entrapped in the mesoSiNPs. Resulting in a larger electrochemical signal, by monitoring the change of the electrochemical signal that increased the peak current, the concentration of UO2 2+ can be indirectly determined with high sensitivity.
Choice of materials
According to the relevant articles, we found that the materials that can be applied to this sensor are graphene, polyaniline, polypyrrole (PPY) and mesoSiNPs and so on. But the irreversibility of graphene agglomeration leads to the poor capacitance performance; polyaniline has excellent conductivity, but its disorder of short fibrous and spherical morphology lead to its poor cycle stability; PPy lacks cycle stability and processing performance. Compared with other materials, mesoSiNPs as carrier vehicles in this work has many advantages of large specific surface area, unique pore structure, biocompatibility, and ease of functionalization, So that they have wide application in the drug release and enzyme immobilizatio. The enzyme immobilization process of mesoSiNPs can be applied in the field of enzyme immobilization, So the material was chosed to as electrode material.
Characterization of synthesized and MB-loaded mesoSiNPs
The morphology of mesoSiNPs was characterized by SEM (Fig. 1), which showed spherical particles of the uniform mesoSiNPs with a diameter around 100 nm.
The three mesoporous silica materials mesoSiNPs, mesoSiNPs-AM, mesoSiNPs-AM@MB and their precursors were analyzed by FT-IR, respectively (Fig. 2). The absorption peaks at 1228 cm−1 and 1093 cm−1 correspond to the asymmetric stretching vibration of Si-O-Si bond, and the absorption peaks at 787 cm−1 and 461 cm−1 belong to symmetry stretching of Si-O-Si bond Vibration and bending vibration, and the absorption peak at near 965 cm−1 attributes to Si-OH symmetrical stretching vibration. In Fig. 2a, the absorption peaks near 2930 cm−1 and 2847 cm−1 correspond to the stretching vibrations of CH3 and CH2 on the surfactant chain, respectively, and disappear completely in Fig. 2b. It is shown that by high temperature calcination, the template in the mesoporous material is effectively removed. As shown in Fig. 2c, a new vibration absorption peak at 1517 cm−1 is the deformation vibration of -NH2 group, which indicates that the amino group had been successfully grafted onto the surface of the silica mesoporous material. The absorption peak of CH3-N bond in MB is in the range of 2800–3500 cm−1, corresponding to the absorption peak at 3280 cm−1 and 2899 cm−1 in Fig. 2d, and the peak at 1592 cm−1 from (d), the absorption peak at 1488 cm−1 is due to the stretching vibration of the C = C bond, which proved that MB has been successfully supported on the mesoSiNPs-AM.
Zeta-potential analysis is used to characterize the preparation of mesoSiNPs (Fig. S1a, in the Supporting Information). After APTES modification, the mesoSiNPs nanoparticles become positively charged, indicating that the amino groups are successfully functionalized on the surface of mesoSiNPs nanoparticles (Fig. S1b) [24, 25]. The loaded MB slightly increases the chargeability of the mesoSiNPs (Fig. S1c). The phosphate backbone of DNA covalently immobilize on the surface of mesoSiNPs making the methylene blue sealed in the inner pores that results in a negative charge of the mesoSiNPs (Fig. S1d). The experimental results are in agreement with the theory.
The nitrogen adsorption-desorption isotherm of the mesoSiNPs-AM showed an average pore diameter of 3.9 nm (Fig. 3). The total pore volume and total specific surface of mesoSiNPs-AM were calculated to be 0.71 cm3·g−1 and 796 m2·g−1 by using the BJH and BET model on the adsorption branch of the isotherm, respectively.
Optimization of sensor construction conditions
Before quantity analysis of UO2 2+ using this method, we optimised the sensor construction condition, including the DNA hybridization time, DNA concentration, pH on the chit, the different dosages of chit and mesoSiNPs-AM@MB@dsDNA, and the reaction time of the sensor with UO2 2+ on the system. As shown in Fig. S2–S4, the optimum values were 12 h hybridization time (Fig. S2), 80 μM DNA concentration (Fig. S3), chit pH 6.7 (Fig. S4), the chit and the MSN-AM@MB@dsDNA were mixed in the ratio of 1:2, and 20 min of reactived time of the sensor with UO2 2+ (Fig. S5) on the system (See the Supporting Information).
Electrochemical characterization of different modified electrodes
The preparation process of the sensor was characterized by cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS). CV was performed in PBS containing [Fe(CN)6]3−/4- (5 mM) at a scan rate of 0.1 V·s−1 potential ranging from −0.1 V to 0.6 V, and the results were shown in Fig. 4.
As shown in Fig. 4, a pair of comparatively symmetrical redox peaks are observed at the bare carbon paste electrode (curve a), and the peak potential difference is 98 mV. After the chit is modified on the surface of the CPE, the peak current is increased due to the positive charge of chit, which makes the electrons easily reach the surface of the electrode (curve b). The increase of the peak current at the mesoSiNPs-AM @MB/chit/CPE (curve c) is due to the increase of the specific surface area and conductivity of the chit/CPE electrode, and with the addition of MB in the pores of the silica, the rate of surface diffusion of [Fe(CN)6]3−/4- to the electrode becomes faster. When dsDNA is modified on the surface of mesoSiNPs-AM@MB/chit/CPE, the peak potential is reduced by 159 mV and the peak current decreases correspondingly, indicating that dsDNA have fully been enclosed the MB in the pores of mesoSiNPs, and dsDNA themselves negative charge repulsion [Fe(CN)6]3−/4- leads to a slower electron transport rate (curve d).
EIS is also an effective method to characterize the surface properties of the modified electrode. The change of the semicircular diameter in the Nyquist spectrum can reflect the change of the surface property of the modified electrode.
As can be seen from Fig. 5, the semicircular diameter of the bare CPE is large, indicating that the electron transfer on the bare CPE is very slow. A dense membrane formed by modifying chit on the electrode surface increases the interface electron conduction rate and results in a lower impedance (Ret = 7.56 kΩ). After the mesoSiNPs-AM@MB is assembled on the chit-modified electrode, due to the nanomaterials increased the surface area of the electrode and the MB supported by the electroactive increased the conductivity of the electron, so the impedance is obviously decreased. When immobilized dsDNA onto the surface of the mesoSiNPs, the impedance increases from 7.56 kΩ to 11.4 kΩ, which proved that the dsDNA has sealed the MB in the pores of the mesoSiNPs successfully, the immobilized dsDNA prevented the diffusion of ferricyanide to the electrode surface. Based on the characterization of EIS and CV, it can be concluded that the modification process of the electrode reached the expected results.
Nyquist plots corresponding to (a) bare CPE, (b) chit/CPE, (c) mesoSiNPs-AM@MB/chit/CPE, (d) mesoSiNPs-AM@MB@dsDNA/chit/CPE electrodes in PBS (10 mmol/L, pH 7.4) containing 5 mM [Fe(CN)6]3−/4- as the redox probe. The impedance spectra were recorded within the range from 100 kHz to 0.1 Hz at the formal potential of [Fe(CN)6]3−/4−. The amplitude of the alternate voltage was 5 mV
Electrochemical detection of uranyl ion content
DPV detection was carried out in PBS containing different concentrations of UO2 2+ with mesoSiNPs-AM@MB@dsDNA/chit/CPE as the working electrode. As shown in Fig. 6a, the oxidation peak current of MB is increased with the increase of UO2 2+ concentrations.
The concentrations of UO2 2+ and the peak current at the concentration range of 20 pM to 100 pM show a good linear relationship (Fig. 6b). The linear equation is I/A = −10.398-0.2255 c/pM, (where I and c represent the peak current and UO2 2+ concentration, respectively.) The correlation coefficient is 0.9924 and the detection limit is estimated to be 0.15 pM based on 3 S/m, here S is the standard deviation of the intercept and m is the slope of the regression line [26]. This sensor is more effective than other reported uranium-based sensors (Table 1).
Specificity of biosensors
In practical applications, the composition of the measured solution is more complex, and the biosensor sensitivity and selectivity requirements are higher, so the sensor in a complex environment of the selectivity is necessary. UO2 2+, Th4+, Fe2+, Hg2+, Zn2+, Pb2+ and Mg2+ were added to the solution, respectively. The peak current I of the measuring sensor is compared by the relative intensity ∆I = (I-I0) (I0 represents the peak current of the biosensor when no metal ion is present in the test solution).
As shown in Fig. 7, DPV signals of the sensor slightly changed after adding the metal ions, namely, Th4+, Fe2+, Hg2+, Zn2+, Pb2+, and Mg2+ at 2 nM concentrations compared with UO2 2+ at 0.2 nM. The results confirmed that the response of the sensor to UO2 2+ was unaffected by the presence of other metal ions. Therefore, the sensor system has good selectivity for UO2 2+ in solution. The reproducibility and stability of the DNA sensor is investigated by the change of the peak current after the reaction on the modified electrode prepared by the same method, and precision and accuracy of the sensor, seeing the Supporting Information.
The actual sample detection
To investigate the validity of the procedure, the method was used to determine trace UO2 2+ in real sample by adding standard uranium solutions to lake water and tap water samples. The analytical results are listed in Table 2.
The results show that the sensor can detect trace UO2 2+ in aqueous solution, and the detection effect is excellent. From above results, it can be seen that the sensor can detect the trace amount of UO2 2+ in the aqueous solution, and the recovery rate is in the range of 91.3–99.4%.
Conclusions
In conclusion, an ultrasensitive electrochemical detection method for UO2 2+ based on specific DNAzyme and mesoSiNPs was designed. Results show that this strategy provides high selectivity and sensitivity for UO2 2+ detection. There exists a good linear correlation from 20 pM to 0.1 nM. The detection limit is 0.15 pM, which is lower than those of reported UO2 2+ detection methods. However, the analysis takes a few seconds than others due to the release of MB entrapped in the MSN process. Moreover, the assay was validated in real samples of spiked environmental water, and the results demonstrated its practical feasibility. Therefore, it is economical, effective, highly selective and practical. In view of these advantages, we deem that this assay will have some potential applications in environmental monitoring field.
References
Liu JW, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y (2007) A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. P Natl Acad Sci 104:2056–2061. doi:10.1073/pnas.0607875104
Gongalsky KB (2003) Impact of pollution caused by uranium production on soil macrofauna. Environ Monit Asses 89:197–219. doi:10.1023/A:1026031224658
Villa M, Manjon G, Hurtado S, García-Tenorio R (2011) Uranium pollution inan estuary affected by pyrite acid mine drainage and releases of naturally occurring radioactive materials. Mar Pollut Bull 62:1521–1529. doi:10.1016/j.marpolbul.2011.04.003
Alam MN, Rahman N, Azmi SNH (2008) Optimized and validated spectrophotometric method for the determination of uranium(VI) via complexation with meloxicam. J Hazard Mater 155:261–268. doi:10.1016/j.jhazmat.2007.11.055
Xiao G, Jones RL, Saunders D, Caldwell KL (2014) Determination of 234U/238U, 235U/238U and 236U/238U isotope ratios in urine using sector field inductively coupled plasma mass spectrometry. Radiat Prot Dosim 1:1–7. doi:10.1093/rpd/ncu023
Santos JS, Teixeira Leonardo SG, Dos Santos Walter NL, Lemos VA, Godoy JM, Ferreira Sérgio LC (2010) Uranium determination using atomic spectrometric techniques:an overview. Anal Chim Acta 674:143–156. doi:10.1016/j.aca.2010.06.010
Michon J, Frelon S, Garnier C, Coppin F (2010) Determinations of uranium(VI) binding properties with some metalloproteins (transferrin, albumin, metallothionein and ferritin) by flluorescence quenching. J Fluoresc 20:581–590. doi:10.1007/s10895-009-0587-3
Dutta RK, Kumar A (2016) Highly sensitive and selective method for detecting ultratrace levels of aqueous uranyl ions by strongly photoluminescent-responsive amine-modified cadmium sulfide quantum dots. Anal Chem 88:9071–9078. doi:10.1021/acs.analchem. 6b01943
Zhang D, Chen Z, Omar H, Deng L, Khashab NM (2015) Colorimetric peroxidase mimetic assay for uranyl detection in sea water. ACS Appl Mater Interfaces 7:4589–4594. doi:10.1021/am507361x
Breaker RR, Joyce GF (1994) A DNA enzyme that cleaves RNA. Chem Biol 1:223–229. doi:10.1016/1074-5521(94)90014-0
Hu KC, Lan DX, Li XM, Zhang SS (2008) Electrochemical DNA biosensor based on nanoporous gold electrode and multifunctional encoded DNA-Au bio bar codes. Anal Chem 80:9124–9130. doi:10.1021/ac8017197
Viswanathan S, Radecka H, Radecki J (2009) Electrochemical biosensor for pesticides based on acetylcholinesterase immobilized on polyaniline deposited on vertically assembled carbon nanotubes wrapped with ssDNA. Biosens Bioelectron 24:2772–2777. doi:10.1016/j.bios.2009.01.044
Zhang J, Shi PW, Yan PP, Wang MB, Tang QH, Cai FD, Deng AP, Li JG (2015) Quantum dots based electrochemiluminescent immunosensor for ultrasensitive and specific determination of mercury (II) ions using gold nanoparticles and a monoclonal antibody. J Electrochem Soc 162:B22–B26. doi:10.1149/2.0631501jes
Lee JH, Wang Z, Liu J, Lu Y (2008) Highly sensitive and selective colorimetric sensors for uranyl (UO2 2+):development and comparison of labeled and label-free DNAzyme -gold nanoparticle systems. J Am Chem Soc 130:14217–14226. doi:10.1021/ja803607z
Zhang HY, Ruan YJ, Lin L, Lin M, Zeng XX, Zeng X, Fu FF (2015) A turn-off fluorescent biosensor for the rapid and sensitive detection of uranyl ion based on molybdenum disulfide nanosheets and specific DNAzyme. Spectrochim Acta A 146:1–6. doi:10.1016/j.saa.2015.02.113
Baker SE, Cai W, Lasseter TL, Weidkamp KP, Hamers RJ (2002) Covalently bonded adducts of deoxyribonucleic acid (DNA) oligonucleotides with single-wall carbon nanotubes:synthesis and hybridization. Nano Lett 2:1413–1417. doi:10.1021/nl025729f
Zhang GY, Deng SY, Cai WR, Cosnier S, Zhang XJ, Shan D (2015) Magnetic zirconium hexacyanoferrate (II) nanoparticle as tracing tag for electrochemical DNA assay. Anal Chem 87:9093–9100. doi:10.1021/acs.analchem.5b02395
Ma DD, Yuan YL, Xiao XL, Gao YY, Li YH, Xu WH, Long W (2014) A label-free electrochemical biosensor for trace uranium based on DNAzymes and gold nanoparticles. J Radioanal Nucl Chem 299:1911–1919. doi:10.1007/s10967-013-2897-9
Tang Q, Yuan YL, Xiao XL, Guo P, Hu JB, Ma DD, Gao YY (2013) DNAzyme based electrochemical sensors for trace uranium. Microchim Acta 180:1059–1064. doi:10.1007/s00604-013-1021-8
Ren K, Wu J, Zhang Y, Yan F, Ju HX (2014) Proximity hybridization regulated DNA biogate for sensitive electrochemical immunoassay. Anal Chem 86:7494–7499. doi:10.1021/ac5012377
Qin QD, Ma J, Liu K (2009) Adsorption of anionic dyes on ammonium-functionalized MCM-41. J Hazard Mater 162:133–139. doi:10.1016/j.jhazmat.2008.05.016
Shao Y, Wang X, Kang Y, Shu Y, Sun Q, Li L (2014) Application of Mn/MCM-41 as an adsorbent to remove methyl blue from aqueous solution. J Colloid Interface Sci 429:25–33. doi:10.1016/j.jcis.2014.05.004
Xiao N, Deng J, Cheng JL, Ju SQ, Zhao HQ, Xie J, Qian D (2016) Carbon paste electrode modified with duplex molecularly imprinted polymer hybrid film for metronidazole detection. Biosens Bioelectron 81:54–60. doi:10.1016/j.bios.2016.02.041
van der Maaden K, Sliedregt K, Kros A, Jiskoot W, Bouwstra J (2012) Fluorescent nanoparticle adhesion assay:a novel method for surface pKa determination of self-assembled monolayers on silicon surfaces. Langmuir 28:3403–3411. doi:10.1021/la203560k
Vashist SK, Lam E, Hrapovic S, Male KB, Luong John HT (2014) Immobilization of antibodies and enzymes on 3-aminopropyltriethoxysilane-functionalized bioanalytical platforms for biosensors and diagnostics. Chem Rev 114:11083–11130. doi:10.1021/cr5000943
Gonza’lez AG, Herrador MA (2007) A practical guide to analytical method validation, including measurement uncertainty and accuracy profiles. Trac-Trend Anal Chem 26:227–238. doi:10.1016/j.trac.2007.01.009
Yun W, Cai D, Jiang J, Wang X, Liao JS, Zhang PC, Sang G (2016) An ultrasensitive electrochemical biosensor for uranyl detection based on DNAzyme and target-catalyzed hairpin assembly. Microchim Acta 183(4):1425–1432. doi:10.1007/s00604-016-1778-7
Zhou B, Wang YS, Yang HX, Xue JH, Wang JC, Liu SD, Zhao H (2014) A sensitive resonance light scattering assay for uranyl ion based on the conformational change of a nuclease-resistant aptamer and gold nanoparticles acting as signal reporters. Microchim Acta 181(11–12):1353–1360. doi:10.1007/s00604-014-1267-9
Acknowledgements
This research was supported by the National Natural Science Foundation of China (11405081), the Hunan Provincial Natural Science Foundation of China (2017JJ3276) and the Department of Education of Hunan Province (17B226).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author(s) declare that they have no competing interests.
Electronic supplementary material
ESM 1
(DOC 151 kb)
Rights and permissions
About this article
Cite this article
Wen, Y., Yuan, Y., Li, L. et al. Ultrasensitive DNAzyme based amperometric determination of uranyl ion using mesoporous silica nanoparticles loaded with Methylene Blue. Microchim Acta 184, 3909–3917 (2017). https://doi.org/10.1007/s00604-017-2397-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-017-2397-7