Abstract
Purpose
The classification of three-dimensional (3D) spinal deformities remains an open question in adolescent idiopathic scoliosis. Recent studies have investigated pattern classification based on explicit clinical parameters. An emerging trend however seeks to simplify complex spine geometries and capture the predominant modes of variability of the deformation. The objective of this study is to perform a 3D characterization and morphology analysis of the thoracic and thoraco/lumbar scoliotic spines (cross-sectional study). The presence of subgroups within all Lenke types will be investigated by analyzing a simplified representation of the geometric 3D reconstruction of a patient’s spine, and to establish the basis for a new classification approach based on a machine learning algorithm.
Methods
Three-dimensional reconstructions of coronal and sagittal standing radiographs of 663 patients, for a total of 915 visits, covering all types of deformities in adolescent idiopathic scoliosis (single, double and triple curves) and reviewed by the 3D Classification Committee of the Scoliosis Research Society, were analyzed using a machine learning algorithm based on stacked auto-encoders. The codes produced for each 3D reconstruction would be then grouped together using an unsupervised clustering method. For each identified cluster, Cobb angle and orientation of the plane of maximum curvature in the thoracic and lumbar curves, axial rotation of the apical vertebrae, kyphosis (T4–T12), lordosis (L1–S1) and pelvic incidence were obtained. No assumptions were made regarding grouping tendencies in the data nor were the number of clusters predefined.
Results
Eleven groups were revealed from the 915 visits, wherein the location of the main curve, kyphosis and lordosis were the three major discriminating factors with slight overlap between groups. Two main groups emerge among the eleven different clusters of patients: a first with small thoracic deformities and large lumbar deformities, while the other with large thoracic deformities and small lumbar curvature. The main factor that allowed identifying eleven distinct subgroups within the surgical patients (major curves) from Lenke type-1 to type-6 curves, was the location of the apical vertebra as identified by the planes of maximum curvature obtained in both thoracic and thoraco/lumbar segments. Both hypokyphotic and hyperkypothic clusters were primarily composed of Lenke 1–4 curve type patients, while a hyperlordotic cluster was composed of Lenke 5 and 6 curve type patients.
Conclusion
The stacked auto-encoder analysis technique helped to simplify the complex nature of 3D spine models, while preserving the intrinsic properties that are typically measured with explicit parameters derived from the 3D reconstruction.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Adolescent idiopathic scoliosis (AIS) refers to a complex deformation of the spine in three-dimensional (3D) Cartesian space with unknown aetiopathogenesis. Standardized comparisons between treatment strategies or long-term management plans involve a classification system of spinal deformities in order to establish the optimal surgical strategy for example. Ponseti et al. [1] made a first endeavor by categorizing spinal curves according to the location and visual patterns of the curve. King et al. [2] proposed to consider the configuration (as observed in the coronal plane), magnitude and degree of flexibility of the scoliosis deformity. Five different curve types were described for spinal arthrodesis recommendations. Their classification system excludes the lumbar segment and the sagittal profile and yields poor validity, reliability and reproducibility. Currently, AIS characterization and treatment recommendations rely mostly on the more comprehensive Lenke classification system [3]. A specific curve type, a lumbar spine modifier and a sagittal thoracic modifier define distinctive spine curves. Nevertheless, Lenke classification is based on the conventional measurement of two-dimensional (2D) geometric indices such as Cobb angle or central sacral vertebral line. Describing spine deformities with only 2D parameters is insufficient to capture the intricate 3D variability of scoliosis [4].
Classification systems need to improve upon the 2D assessment of scoliosis, which is tied to radiographs in the coronal and sagittal planes. Similar 2D profiles on both coronal and sagittal planes may actually come from different 3D spine geometries [4], where vertebral anatomical landmarks are expressed in Cartesian coordinates (x, y, z). A better understanding and characterization of deformation mechanisms should lead to more appropriate treatments and accurate evaluations. The Scoliosis Research Society agreed on a rationalized 3D terminology to describe spinal deformity [5] and a task force was instructed to assess the clinical relevance and impact of 3D analysis for AIS. Recent efforts have been made to use 3D reconstructions of scoliotic deformities in order to propose accurate and reproducible classification systems, which take into account the 3D nature of the deformity. Numerical methods create new alternatives to current classification systems. First, advanced 3D indices of scoliosis were investigated to discriminate between different types of deformation. Poncet et al. [6] introduced a 3D classification method of scoliotic deformities, based on the geometric torsion of the vertebral body line categorized several curve patterns. Kadoury et al. [7] extended the local geometric torsion measure to regional curves with a parametric curve fitting that was less prone to inaccuracies in the 3D reconstruction. A fuzzy c-means classifier further created subgroups based on the regional geometric torsion indices. Secondly, regional measures were also explored to provide discriminant indices. Sangole et al. [8] included the axial rotation of the apical vertebrae and the orientation of the plane of maximum curvature (PMC) in the main thoracic (MT) region. Thoracic curve types (Lenke 1) were further subdivided in three different groups with the ISOData algorithm. Duong et al. [9] also considered the orientation of the best-fit plane in the set of 3D parameters. Two different subgroups were found in their small dataset of Lenke 1 curve types. Overall, these studies [7–9] share a similar framework. Their classification systems are derived from the clustering of hand-engineered parameters, which were calculated from 3D spine reconstructions. However, relying on geometric indices sets out on a quest in search of the best characteristics to describe the 3D nature of scoliotic spines.
Numerical methods should be able to capture within a simplified space, the high resolution and complex nature of a fully geometric 3D reconstruction of the spine, both on a regional (spinal) and local (vertebra) levels. This implies directly analyzing the 3D spine models instead of expert-based features as it has been experimented previously. Duong et al. [10] proposed a wavelet-based compression technique of the spinal curves. Kadoury and Labelle [11] investigated a manifold learning algorithm based on locally linear embedding for dimensionality reduction of 3D spine models of the Lenke 1 curve types. However, these local techniques for dimensionality reduction tend to suffer from the curse of dimensionality and to be sensitive to data models which tend further away from the general trend of the normal distribution [12]. Hence, increasing the number of landmarks to describe the 3D spine models or including other Lenke types will lead to miss-classification of an important number of samples. Global nonlinear techniques for dimensionality reduction could overcome these drawbacks [12] by preserving the global properties of the 3D spine models.
In this study, we propose to use recent advances in artificial intelligence to simplify the high-resolution (238 vertebral anatomical landmarks expressed in Cartesian coordinates) and complex nature of geometric 3D spine reconstructions for classification purposes. This highly non-linear transformation discriminates between AIS scoliotic curves by learning the intrinsic properties of 3D spine reconstructions by preserving the global properties. Once a low-resolution representation has been learned from a cohort of 3D spine models, new classes can be derived from their simplified description.
Materials and methods
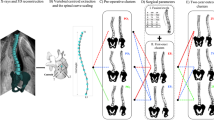
We evaluate the relevance of a machine learning algorithm, namely the stacked auto-encoders, on a large database that comprises 915 reconstructions of all Lenke types (i.e. from Lenke 1 to Lenke 6). The proposed framework is illustrated in Fig. 1 and consists of four main steps: (1) reconstruction of a 3D spine model from biplanar X-rays for each patient; (2) encoding each 3D spine in a low-resolution space; (3) clustering of the encoded spines models; (4) validation of the sub-groups with clinical data.
Flowchart of the method. The system sequentially: 1 reconstructs a 3D spine model, x of D dimensions, from biplanar X-rays for each patient; 2 maps the high-resolution spine reconstruction to a low-resolution space, called a code, with stacked auto-encoders of symmetric layer sizes which continuously compresses to a d dimension, called a code; 3 clusters the low-resolution spines into k sub-groups; 4 validates the cluster relevance with the clinical data. In our experiments, each 3D spine models consisted of a vector of size 714 (i.e. the high-resolution space) that are compressed to a code vector of size 25 (i.e. the low-resolution space) by using stacked auto-encoders
Patient data
In this retrospective study, data of 663 preoperative AIS patients from nine scoliosis centers (New York City, Baltimore, Philadelphia, CHOP, Miami, San Diego, Wilmington, Montreal, Vancouver) during an 18-year period (1994–2012) were selected. From this group, 151 patients had repeat measurements from multiple clinic visits (mean = 2.7 visits), yielding a total of 915 visits. All patients were diagnosed with an adolescent idiopathic scoliosis in the thoracic and/or lumbar spine. The mean of the major Cobb angle was 58.8° ± 15.2° (range = 21.3°–113.6°). Note that a major Cobb angle corresponds to the maximum value between the main thoracic (MT) Cobb angle and the thoracolumbar/lumbar (TLL) Cobb angle, both measured in the plane of maximum curvature (PMC). Using coronal, sagittal and bending radiographs, members of the 3D Classification of the SRS assigned a Lenke type to all visits, which are divided in: 312 Lenke 1, 118 Lenke 2, 152 Lenke 3, 122 Lenke 4, 113 Lenke 5, and 98 Lenke 6 curve types.
3D reconstruction of the spine
The spine was reconstructed in 3D from calibrated coronal and sagittal radiographs of the patient in a standing position [13, 14]. A statistical model from a database of scoliotic patients was used to reconstruct an initial spine model in 3D. Anatomical landmarks on each vertebra were further refined with an iterative process based on several features extracted from the radiographs. Finally, an experienced user at our institution corrected and validated the anatomical landmark positions on each vertebra to generate a personalized 3D reconstruction of the spine for each patient. This reconstruction process from biplanar radiographs produced high shape accuracy and high vertebral position precision for asymptomatic subjects and idiopathic scoliosis patients when compared with a reconstruction performed from CT scans [14]. Concretely, a 3D reconstruction of the spine consists of fourteen anatomical landmarks per vertebra (12 thoracic, five lumbar): center, left, right, anterior and posterior of both superior and inferior vertebral endplates (ten landmarks); and tips of both pedicles (four landmarks). All 3D spine models were normalized with regards to their height and rigidly translated to a common referential at the L5 vertebra. Hence, each 3D spine model consists of a vector of 714, which corresponds to the concatenation of the 14 landmarks with 3D coordinates in the x, y and z axis, identified on each of the 17 vertebrae. Radiographs were acquired from both the low-dose EOS low dose imaging device (EOS imaging, Paris, France), which simultaneously acquires biplanar radiographs, and conventional radiographic imaging systems. 437 reconstructions of the spine (from 290 patients) came from radiographs acquired with the EOS imaging system while 478 reconstructions of the spine (from 373 patients) came from conventional radiography. Both methods showed to yield similar levels of accuracy for the 3D reconstruction of the spine, in comparison to CT models and expert annotations [13, 14].
Encoding of 3D spine models
The geometric 3D spine models were then simplified into a low-resolution encoding in order to capture the main factors of variation in the shape of the spine from the given cohort. In other words, the purpose is to compress geometric 3D spine models that are represented in a high-resolution vector (size of 714), into a low-resolution vector (compressed code). Encoding 3D spine models will help discover patterns relevant for adolescent idiopathic scoliosis. To perform this step, a stacked auto-encoder (SAE) was used to simplify the representation of the 3D spine models. A SAE consists of a specific artificial neural network architecture. Artificial neural networks can be thought of as complex and highly non-linear mathematical functions which parameters are learnt to transfer the input observed variables to the output variables.
A SAE is composed of two stages. First, an encoding stage learns the parameters to map the input vector to a latent representation. Second, a decoding stage regenerates the input vector from the latent representation. More specifically, the algorithm for encoding 3D spine models is performed as follows. Each spine is represented as an input vector that consists of the 3D coordinates of all the anatomical landmarks (denoted as x in Fig. 1). The SAE first attempts to compress each spine into a low-resolution latent representation using an encoder function. The lower-resolution representation is considered as a compressed version of the input, called a code. Once a code is obtained, the algorithm then attempts to regenerate an output vector that consists of a 3D spine model (denoted as z in Fig. 1) from the code using a decoder function. However, this reconstruction process is not flawless and will generate some errors (denoted as ε in Fig. 1). The objective would then be to learn parameters that yield a low reconstruction error. Hence, the encoding–decoding procedure is optimized by minimizing in an iterative fashion the difference between the inputs x and outputs z. Stacking several auto-encoders helps the artificial neural network to become invariant to most local changes and disentangle the main factors of variation in the dataset [15, 16].
Clustering
Once a large database of 3D reconstructed spine models were encoded into low-resolution codes, the k-means++ clustering algorithm [17] partitioned the spine dataset into k separate sub-groups. This clustering algorithm is a variant of the traditional k-means clustering algorithm that integrates a probabilistic seeding initialization method. The selection of the right number of clusters k is based on the validity ratio [18], which minimizes the intra-cluster distance and maximizes the inter-cluster distance.
Statistical analysis
We validated the clustered data points with standard geometrical indices in the main thoracic (MT) and thoracolumbar/lumbar (TLL) regions. For each spine, the Cobb angles and the orientations of the PMC were computed in both regional curves. The kyphotic angle was measured between T2 and T12 on the sagittal plane. The lumbar lordosis angle was defined between L1 and S1 on the sagittal plane. The axial rotation of the apical vertebra in the MT region was computed by the Stokes method [19]. Finally, the pelvic incidence (PI) was measured between the line perpendicular to the sacral plate at its midpoint and the line connecting this point to the axis of the femoral heads [20]. One-way ANOVA tested differences between the cluster groups with a significance level α = 0.05. The p values were adjusted with the Bonferroni correction. Moreover, an experienced surgeon at our institution performed a clinical assessment of the ten closest 3D spine reconstructions near the centroid of each cluster.
Results
The cohort of 915 visits from 663 patients was randomly divided into a training set (645 visits), a validation set (135 visits) and a testing set (135 visits) for unbiased evaluation. In order to determine the hyper-parameters of the neural network, an exhaustive grid search was performed on the validation set by minimizing the mean squared error. The architecture yielding the lowest error is presented in Fig. 1. We used an encoder with four latent layers of size (layer 1: 1000 nodes; layer 2: 500 nodes; layer 3: 250 nodes; code: 25 nodes) and a symmetric decoder with tied weights to map the high-resolution patient’s spine models into low-resolution codes. Weight parameters were initialized by a denoising auto-encoder to capture the statistical dependencies between the inputs. The final model was trained by using the entire dataset of 915 visits. Note that five visits were further excluded because the pelvis radiograph was not available.
The k-mean++ clustering detected eleven different groups from the low-resolution encoding of 3D geometrical models based on the validity ratio. Table 1 presents the clinical statistical data analysis for these eleven groups. The mean values of all geometric parameters are listed for all eleven groups and the differences between all groups were found to be statistically significant (α = 0.05) for each parameter. Table 2 presents the Lenke curve type distribution across the eleven clusters, while Table 3 offers a summary description of each cluster based on the observed parameters. Figure 2a, b presents samples for all these eleven clusters detected by the analysis framework. In order to visualize the distribution of samples in this low- resolution space, a principal component analysis was performed on the encoded samples of size 25, in order to project the encoded spine reconstructions to 3D and 2D views. Figure 3 depicts the visualization of the first three principal components (PC) from this analysis. The first PC explains 46 % of the variance in the encoded geometric spines, representing the location of the major curve. High values in the first PC tend to increase angulation of TLL plane and the axial rotation angle of the apical vertebra while decreasing the Cobb angle and the angulation of MT plane. The second PC explains 26 % of the variance and is related to the lordotic angle. The third PC explains 11 % of the variance and is related to the kyphotic angle.
a Samples for clusters I–VI found by the clustering algorithm. For each cluster sample, coronal/sagittal radiographs, Da Vinci views [4], coronal and top views of the 3D reconstruction model are presented. b Samples for clusters VII–XI found by the clustering algorithm. For each cluster sample, coronal/sagittal radiographs, Da Vinci views [4], coronal and top views of the 3D reconstruction model are presented
Visualization of the eleven clusters found by the k-Means++ algorithm from the low-resolution encoding of 3D geometrical models. Each colored point represents a single 3D spine reconstruction in a low-resolution space. a 3D scatter plot of all 915 visits in the low-resolution space using principal component analysis. The 3D view is projected onto 2D views with (b) first and second principal components, and (c) second and third principal components
Discussion
In this 3D analysis of spinal deformities, a novel method simplifying the representation of the geometric 3D reconstruction of a patient’s spine was presented to study 3D morphology as a stepping stone for a new classification system based on a machine learning algorithm. Previous systems based on 2D radiographic images covered all types of curve patterns and provided a reliable set of measures which take under account the deformity in the sagittal plane, along with specific modifiers [2, 3]. Still, relying on 2D projections of a complex 3D curve as encountered in AIS represents a considerable limitation to these standard approaches. On the other hand, evaluating the deformity based on discrete local 3D measurements, such as axial rotation or geometric torsion [7], is inevitably linked to the quality of the 3D reconstruction and to the inter-rater variability of these pre-defined measurements. In this paper, we attempt to analyze 3D patterns based on the global representation of the spine without using explicit parameters derived from the 3D reconstruction of the spinal shape. The approach was able to detect eleven sub-groups based on their low-resolution representation. The differences in clinical measurements (Cobb angles and orientation of PMC, kyphosis, lordosis, pelvic incidence) between all these new 3D sub-groups were found to be statistically significant.
Two clinically relevant groups emerge among the eleven different clusters of patients. In the first, clusters VII, XI, and III (illustrated as shades of blue in Fig. 3) represent the clusters with small thoracic deformities and large lumbar deformities. An increase of the TLL Cobb angle in the PMC and a decrease of the axial rotation in the MT region from cluster VII to cluster III are clearly apparent from Table 1. On the other hand, the MT Cobb angles in the PMC remain low. A high distribution of Lenke 5 and 6 curve types in these three clusters confirm these patterns. In the second group, clusters II, V, I and X (illustrated in shades of red/orange in Fig. 3) represent the clusters with large thoracic deformities and small lumbar deformities. An increase of the MT Cobb angle and the MT orientation in the PMC, from cluster II to cluster X, is observable. A similar behavior for the axial rotation of the apical vertebra in the MT region is observed. The geometrical parameters obtained in the lumbar segment remain low. An absence—or a very small presence—of Lenke 5 and 6 curve types in these five clusters confirms these patterns. These observations reveal the fact that the location of the major curve (thoracic, lumbar, thoraco-lumbar/lumbar) is the most discriminant clinical factor in distinguishing different classes of deformity. Within these two groups, there exists an important range of kyphotic and lordotic profiles, as well as a spectrum of varying curve severity that is observable, thus suggesting that there exists variability with single or double major curves, either in the thoracic and lumbar regions.
Clusters can also be stratified based on their kyphotic and lordotic profiles. Clusters II, III and VIII represent the clusters with hyper-kyphotic and hyper-lordotic profiles. However, cluster II and cluster III have completely different deformities in their respective MT and TLL regions. Cluster VIII denotes the cluster with high deformities in both MT and TLL regions. This is confirmed by the highest percentage of Lenke type-4 from all clusters (29 %). Clusters I, VI and IX represent the clusters with hypo-kyphotic and hypo-lordosis profiles. Cluster VI has the highest thoracic deformities and relatively high lumbar deformities. This behavior is similar to the spine reconstructions included in cluster VIII. Cluster IX differs from cluster VI with lower Cobb angles in both MT and TLL regions in the PMC. Finally, cluster IV represents the cluster with hypo-kyphotic and hyper-lordotic profiles with large thoracic deformities. These findings confirm also the existence of hypo-kyphotic profiles (clusters IV and VI) within groups exhibiting high thoracic deformities. The difference between these two clusters is the major difference in angulation and Cobb angle of the plane of maximal deformity in the thoracic region, thereby suggesting that regional angulation is still an important factor in assessing the deformation.
The ultimate application of the method proposed in this study is the classification of spinal deformities from personalized 3D models of the patient’s spine geometry. In AIS, classification is not only useful for understanding the progression of the disease, but more importantly, for selecting the optimal surgical strategy before the patient goes for surgery based on specific curvature profiles. Hence by better assessing the type of deformation and risk of progression for a specific pattern, this increases the surgeon’s chance of choosing the proper instrumentation tools and surgical plan, and ultimately the patient’s outcome.
We propose in this paper to use artificial intelligence to simplify high-resolution and complex 3D spine reconstructions for classification purposes. An automated classification method, called stacked auto-encoders, discovers sub-groups within a large pool of patients with both thoracic and lumbar deformations. The code layer of the auto-encoder learns a distributed low-resolution representation that aims to capture the main factors of variation in the clinical dataset. However, different examples from the distribution of the training dataset may potentially yield to high reconstruction errors. Therefore, having a large and representative training dataset of AIS is critical. This will also prevent the model from overfitting.
Clinically relevant clusters can then be discovered to explain the variation within AIS patients. This demonstrates the feasibility of an optimization approach to increase an orthopaedic surgeon’s ability to personalize a treatment plan of idiopathic scoliosis based on the individual 3D spinal and rib cage geometry. Experts very well know the various appearances of scoliotic deformities. Indeed, two markedly different curves can have the same coronal angle but have dramatically different sagittal profiles as well as axial deformities. Therefore, a classification system is needed; it should be able to group subjects, by summing up this huge number of parameters. The traditional classification systems are based on arbitrary choices made by experts in the field of interest who decide to focus on specific clinical aspects. To make the best choices, physicians will need clinically validated parameters and comparisons between different classification systems. This is where the work proposed here can help clinicians to better appreciate subtleties not easily detected with current imaging protocols. Some top view parameters seem to represents the optimal parameter able to always define the characteristics of different scoliosis pattern. Moving from a 2D to a 3D classification will help mainly for two crucial aspects: first of all, it will help to perform a more precise prognosis for each patient. Secondly, it will help clinicians in applying a more personalized surgical treatment based on the specific changes observed in 3D.
A number clinical parameters such has the spinous process angle and apical vertebral rotation, which have both shown to strongly correlate with the Cobb angle and improve the prediction of the main curve’s deformation [21], are important in determining the appropriate deformation class aimed for surgery planning. In fact, apical vertebral rotation was shown to strongly correlate with surgical outcomes [4] and a few alternatives for assessing vertebral rotation in non-invasive methods have also been proposed to reduce the need of X-ray images [22]. In an attempt to propose a new classification based on three-dimensional modeling of the spine, quantifying the structural nature of a curve is pivotal to integrate flexibility characteristics within the classification paradigm, such as in the Lenke classification. Cobb angle deviations or methods estimating fulcrum flexibility can be considered a potential alternative measure of spinal flexibility [23]. A limitation of the present study is that curve flexibility was not integrated into the auto-encoding process. Future work will attempt to include 3D geometrical models obtained from supine or bending radiographs in order to estimate spine flexibly. Finally, methods based on the concept of reflection and a process termed rotoinversion, which is a combination of reflection and rotation, were proposed to assess asymmetry in adolescent idiopathic scoliosis with geometric models of the torso [24]. In this study, spine asymmetry is intrinsically incorporated into the morphology descriptors found by the encoded models, which help to differentiate different patterns of symmetry within the study population.
The current study evaluated the 3D sub-groups of all Lenke types for thoracic and lumbar scoliotic curves, suggesting that shape variability is present within an existing 2D group used in clinical practice. However, these types of approaches include complex synthetization tasks, which require sizeable datasets to improve the data representation within the code layer. Therefore, a larger multicentric dataset may help to significantly increase the number of patients from various sites and obtain a more reproducible and robust model. Furthermore, the development of computational methods will ultimately lead to more reliable classification paradigms, helping to identify possible patients who might progress with time. Future work will use longitudinal data for surgical treatment planning, whereas each case is considered independently in the current framework. Finally, a reliability study will be undertaken to evaluate the relevance of classification systems in terms of surgical strategy.
References
Ponseti IV, Friedman B (1950) Prognosis in idiopathic scoliosis. J Bone Joint Surg Am 32(2):381–395
King HA, Moe JH, Bradford DS, Winter RB (1983) The selection of fusion levels in thoracic idiopathic scoliosis. J Bone Joint Surg 65(9):1302–1313
Lenke LG, Betz RR, Harms J, Bridwell KH, Clements DH, Lowe TG, Blanke K (2001) Adolescent idiopathic scoliosis. J Bone Joint Surg 83(8):1169–1181
Labelle H, Aubin CE, Jackson R, Lenke L, Newton P, Parent S (2011) Seeing the spine in 3D: how will it change what we do? J Pediatr Orthopaed 31:S37–S45
Stokes IA (1994) Three-dimensional terminology of spinal deformity: a report presented to the scoliosis research society by the scoliosis research society working group on 3-D terminology of spinal deformity. Spine 19(2):236–248
Poncet P, Dansereau J, Labelle H (2001) Geometric torsion in idiopathic scoliosis: three-dimensional analysis and proposal for a new classification. Spine 26(20):2235–2243
Kadoury S, Shen J, Parent S (2014) Global geometric torsion estimation in adolescent idiopathic scoliosis. Med Biol Eng Comput 52(4):309–319
Sangole AP, Aubin CE, Labelle H, Stokes IA, Lenke LG, Jackson R, Newton P (2009) Three-dimensional classification of thoracic scoliotic curves. Spine 34(1):91–99
Duong L, Mac-Thiong JM, Cheriet F, Labelle H (2009) Three-dimensional subclassification of Lenke type 1 scoliotic curves. J Spinal Disord Tech 22(2):135–143
Duong L, Cheriet F, Labelle H (2006) Three-dimensional classification of spinal deformities using fuzzy clustering. Spine 31(8):923–930
Kadoury S, Labelle H (2012) Classification of three-dimensional thoracic deformities in adolescent idiopathic scoliosis from a multivariate analysis. Eur Spine J 21(1):40–49
van der Maaten LJ, Postma EO, van den Herik HJ (2009) Dimensionality reduction: a comparative review. J Mach Learn Res 10(1–41):66–71
Kadoury S, Cheriet F, Labelle H (2009) Personalized X-ray 3-D reconstruction of the scoliotic spine from hybrid statistical and image-based models. IEEE Trans Med Imaging 28(9):1422–1435. doi:10.1109/TMI.2009.2016756
Humbert L, De Guise JA, Aubert B, Godbout B, Skalli W (2009) 3D reconstruction of the spine from biplanar X-rays using parametric models based on transversal and longitudinal inferences. Med Eng Phys 31(6):681–687
Vincent P, Larochelle H, Lajoie I, Bengio Y, Manzagol PA (2010) Stacked denoising autoencoders: learning useful representations in a deep network with a local denoising criterion. J Mach Learn Res 11:3371–3408
Vincent P, Larochelle H, Bengio Y, Manzagol PA (2008) Extracting and composing robust features with denoising autoencoders. In: Proceedings of the 25th international conference on machine learning. ACM, pp 1096–1103
Arthur D, Vassilvitskii S (2007) k-means++: the advantages of careful seeding. In: Proceedings of the eighteenth annual ACM-SIAM symposium on discrete algorithms. Society for Industrial and Applied Mathematics, pp 1027–1035
Ray S, Turi RH (1999) Determination of number of clusters in k-means clustering and application in colour image segmentation. In: Proceedings of the 4th international conference on advances in pattern recognition and digital techniques, pp 137–143
Stokes IA, Bigalow LC, Moreland MS (1986) Measurement of axial rotation of vertebrae in scoliosis. Spine 11(3):213–218
Legaye J, Duval-Beaupere G, Hecquet J, Marty C (1998) Pelvic incidence: a fundamental pelvic parameter for three-dimensional regulation of spinal sagittal curves. Eur Spine J 7(2):99–103
Morrison DG, Chan A, Hill D et al (2015) Correlation between Cobb angle, spinous process angle (SPA) and apical vertebrae rotation (AVR) on posteroanterior radiographs in adolescent idiopathic scoliosis (AIS). Eur Spine J 24(2):306–312
Mangone M, Raimondi P, Paoloni M et al (2013) Vertebral rotation in adolescent idiopathic scoliosis calculated by radiograph and back surface analysis-based methods: correlation between the Raimondi method and rasterstereography. Eur Spine J 22(2):367–371
Keenan BE, Izatt MT, Askin GN, Labrom RD, Pearcy MJ, Adam CJ (2014) Supine to standing Cobb angle change in idiopathic scoliosis: the effect of endplate pre-selection. Scoliosis. doi:10.1186/1748-7161-9-16
Hill S, Franco-Sepulveda E, Komeili A, Trovato A, Parent E, Hill D, Lou E, Adeeb S (2014) Assessing asymmetry using reflection and rotoinversion in biomedical engineering applications. Proc Inst Mech Eng H 228(5):523–529
Acknowledgments
This paper was supported in part by the CHU Sainte-Justine Academic Research Chair in Spinal Deformities, the Canada Research Chair in Medical Imaging and Assisted Interventions, the 3D committee of the Scoliosis Research Society, the Natural Sciences and Engineering Research Council of Canada and the MEDITIS training program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no potential conflict of interest.
Rights and permissions
About this article
Cite this article
Thong, W., Parent, S., Wu, J. et al. Three-dimensional morphology study of surgical adolescent idiopathic scoliosis patient from encoded geometric models. Eur Spine J 25, 3104–3113 (2016). https://doi.org/10.1007/s00586-016-4426-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00586-016-4426-3