Abstract
Mycorrhizal symbiosis often displays low specificity, except for mycoheterotrophic plants that obtain carbon from their mycorrhizal fungi and often have higher specificity to certain fungal taxa. Partially mycoheterotrophic (or mixotrophic, MX) plant species tend to have a larger diversity of fungal partners, e.g., in the genus Pyrola (Monotropoideae, Ericaceae). Preliminary evidence however showed that the Japanese Pyrola japonica has preference for russulacean fungi based on direct sequencing of the fungal internal transcribed spacer (ITS) region from a single site. The present study challenges this conclusion using (1) sampling of P. japonica in different Japanese regions and forest types and (2) fungal identification by ITS cloning. Plants were sampled from eight sites in three regions, in one of which the fungal community on tree ectomycorrhizal (ECM) tips surrounding P. japonica was also analyzed. In all, 1512 clone sequences were obtained successfully from 35 P. japonica plants and 137 sequences from ECM communities. These sequences were collectively divided into 74 molecular operational taxonomic units (MOTUs) (51 and 33 MOTUs, respectively). MOTUs from P. japonica involved 36 ECM taxa (96 % of all clones), and 17 of these were Russula spp. (76.2 % of all clones), which colonized 33 of the 35 sampled plants. The MOTU composition significantly differed between P. japonica and ECM tips, although shared species represented 26.3 % of the ECM tips community in abundance. This suggests that P. japonica has a preference for russulacean fungi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mycorrhizal symbiosis is a mutualistic relationship in which carbohydrates from plants are exchanged against mineral nutrients from mycorrhizal fungi (Smith and Read 2008; van der Heijden et al. 2015). However, mycoheterotrophic (MH) plants, which are mostly achlorophyllous, represent exceptions to this general rule. Since they lost photosynthetic ability, both carbohydrates and minerals are obtained from associated mycorrhizal fungi (Merckx 2013). More recently, some green plants were suggested to obtain a significant proportion of fungal carbon (Selosse and Roy 2009; Selosse et al. 2016): the so-called mixotrophic (MX) plants are partially mycoheterotrophic but retain photosynthetic abilities (e.g., Gebauer and Meyer 2003; Gonneau et al. 2014).
In temperate regions, MH and MX plants evolved multiple times in the family Orchidaceae and in Monotropoideae in the family Ericaceae (Merckx et al. 2013). Molecular analyses revealed that mycorrhizal associations of MH and MX plants are often biased toward certain groups of ectomycorrhizal (ECM) fungi (Matsuda et al. 2011; Merckx et al. 2013) that receive carbon from surrounding autotrophic trees they simultaneously colonize (Björkman 1960). Thus, tripartite interactions within ECM networks facilitate the coexistence of trees and MH plants (Matsuda and Hijii 2004; Selosse et al. 2006; Simard et al. 2012; Nara 2015). Most MH and many MX species associate with a narrow range of fungi (e.g., Waterman et al. 2013). Such specificity becomes even higher at a local population scale, and populations inhabiting environmentally or geographically different sites may associate with different subsets of fungi (Dowie et al. 2012; Hynson et al. 2015).
Pyrola (Monotropoideae, Ericaceae), a plant genus of the northern hemisphere, comprises ca. 35 species (Liu et al. 2010) with a fully MH germination stage (Hashimoto et al. 2012; Hynson et al. 2013). It contains one MH and many MX species without mycorrhizal fungal specificity (Zimmer et al. 2007; Tedersoo et al. 2007; Hynson et al. 2009). These plants form arbutoid or pyroloid mycorrhizas that are characterized by the presence of intracellular hyphal coils, while fungal mantle formation ranges from apparent to absent (Robertson and Robertson 1985; Peterson et al. 2004; Massicotte et al. 2008; Vincenot et al. 2008). A root can be infected by multiple fungal species, especially when the mantle is absent (Tedersoo et al. 2007; Toftegaard et al. 2010). Molecular identifications of pyroloid mycorrhizal fungi have revealed associations with ECM fungi and a taxonomic range including several families such as Atheliaceae, Russulaceae, and Thelephoraceae per Pyrola species, which are therefore generalist at adulthood (Zimmer et al. 2007; Tedersoo et al. 2007; Vincenot et al. 2008; Hynson and Bruns 2009; Toftegaard et al. 2010; Hashimoto et al. 2012).
However, unlike the other Pyrola species, Asian Pyrola japonica was suggested to have a certain mycorrhizal preference. Although multiple ECM fungal taxa associate with P. japonica as with other Pyrola species, members of Russula account for 84 % of the analyzed sequences (Matsuda et al. 2012). This is currently the sole example of mycorrhizal preference in Pyrola, and it may have evolved secondarily from ancestors with generalist relationships, as suggested for some MX Orchidaceae (e.g., Girlanda et al. 2006). However, Matsuda et al. (2012) used direct sequencing of PCR products, and the rather low rate of successful sequencing might have caused a methodological error, neglecting admixture of co-infecting fungi.
The aim of this study was to identify more precisely the diversity of the mycorrhizal community associated with 39 individuals of P. japonica in three regions from central Japan (Fig. 1). A cloning method for PCR products refined fungal assemblages and occurrence frequencies in roots of the plant. We hypothesized that Russula is the most abundant taxon associated with P. japonica, irrespective of habitats. Moreover, we investigated whether the communities associated with P. japonica simply reflected those from surrounding ECM trees or a true preference for russulacean species. We hypothesized that Russula is not necessarily the most abundant taxon on surrounding ECM trees but that its abundance on P. japonica roots reflects a preferential association.
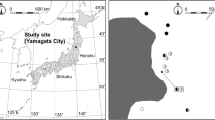
Location of sampling regions. Insets indicate study sites in each region. Aichi and Mie involved several sites at distances ranging from 1.8 to 15.9 km. M1 was the study site used in Matsuda et al. (2012)
Materials and methods
Study sites and sampling
Mature individuals of P. japonica were collected from eight sites in central Japan in 2012 and 2013 from three regions, namely sites A1–A3 in Aichi, M1–M4, in Mie, and K1 in Kyoto (Fig. 1). All sites harbor temperate deciduous or evergreen ECM forests that, despite roughly similar climatic conditions, differ by dominant ECM tree species (Table 1). No major understorey plants other than P. japonica were observed except the Poaceae Pleioblastus chino var. viridis at site M1 (Matsuda et al. 2012; Fig. S1). At each site, individuals were at least 20 m apart to secure independent sampling. At the site M4, we collected three plants with dried shoots because no intact healthy shoot was available at the sampling date. Plants were dug up with a 15 × 15 × 15-cm soil core to obtain the whole root system and kept at 5 °C until mycorrhizal observation. We collected 18 plants in 2012 and 21 plants in 2013 (Table 1). In the laboratory, excavated soil was washed out under running tap water on a 2-mm mesh sieve. Mycorrhizal roots were excised under a stereomicroscope (up to ×115 magnification; SZX16, Olympus, Tokyo). Since P. japonica mycorrhizas have no fungal mantle, we defined mycorrhizas as whitish to brownish zones characterizing the occurrence of intracellular hyphal coils (Matsuda et al. 2012; Fig. S1). Up to six 2–5-mm-long mycorrhizal root fragments were recovered per core, i.e., a total of 28 and 102 root fragments (accounting for a total 140 and 204 mm of root) in 2012 and 2013, respectively. They were stored at −80 °C until molecular analysis. In order to characterize ECM diversity on tree roots surrounding P. japonica, the surrounding ECM roots were also sampled around six plants from the Aichi sites (numbers in brackets in Table 1). From each soil core containing P. japonica root systems, 20 to 30 ECM tips were arbitrarily sampled and stored at −80 °C until molecular analysis.
Molecular analysis
Since the different epidermal cells of a single root fragment can associate with different mycorrhizal fungi, we cloned PCR products for barcoding. Genomic DNA was extracted from root samples using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). The fungal internal transcribed spacer (ITS) region of nuclear ribosomal DNA was amplified with the Ex Taq Kit (TaKaRa, Ohtsu, Japan) by PCR using TaKaRa PCR Thermal Cycler Dice (Model TP600, TaKaRa, Ohtsu, Japan). For each plant sample, primer pairs ITS1f and ITS4 or ITS1f and TW13 were used for amplification, as in Matsuda et al. (2012). Positive PCR products were cloned using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Twenty and 64 successfully inserted white colonies were picked up in 2012 and 2013, respectively. Selected colonies were further amplified with the Ex Taq Kit or EmeraldAmp MAX PCR Mix (TaKaRa, Ohtsu, Japan) according to the manufacturer’s instructions, using the previous primer sets. Thermal cycle conditions for PCR were as follows: preliminary denaturation at 94 °C for 3 min, 30 cycles at 94 °C for 30 s, 52 °C for 30 s, and 72 °C for 70 s, and final extension at 72 °C for 10 min. Positive clone products were submitted to restriction fragment length polymorphism (RFLP) analysis with Alu I and Hinf I (TaKaRa, Ohtsu, Japan). The digested fragments were separated on 4 % 3:1 Nusieve agarose gel in TAE buffer with a 100-bp DNA ladder (Bio Regenerations, Yokohama, Japan), and the length of fragments was estimated using a CS Analyzer 2.0 (Atto, Tokyo, Japan). Clones were grouped into RFLP types (but identical RFLP patterns detected from different plants were tentatively treated as different RFLP types in the first instance). One to four representative clones of each RFLP type were then sequenced. They were cleaned using Illustra GFX PCR DNA and the Gel Band Purification Kit (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) and sequenced using ABI 3730 (Applied Biosystems, Foster City, CA, USA) after reactions with the BigDye Terminator v3.1 Cycle Sequencing Kit (Invitrogen, Carlsbad, CA, USA) with primer ITS1f. When sequencing was not successful, the samples were sequenced bidirectionally using reverse primer ITS4 or TW13.
For the analysis of ECM tips, genomic DNA was extracted by homogenizing using a pestle and mortar and heated in buffer A (Thomson and Henry 1995) at 95 °C. KOD FX (TOYOBO, Tokyo, Japan) was used for PCR amplification with the following thermal cycle conditions: 94 °C for 2 min, 35 cycles of 98 °C for 10 s, 55 °C for 30 s, 68 °C for 70 s, and 72 °C for 10 min. Primers were ITSOF (Tedersoo et al. 2008) and TW13 in the first trial. When the amplification was not successful, the reverse primer was changed into LB-W (Tedersoo et al. 2007). Positive PCR products confirmed through electrophoresis were purified by Illustra GFX PCR DNA and the Gel Band Purification Kit or ExoSAP-IT (Affymetrix, Tokyo, Japan) and sequenced as above.
Data analysis
Obtained sequences were adjusted manually using the MEGA version 5.22 software (Tamura et al. 2011) and grouped into molecular operational taxonomic units (MOTUs) at a 97 % similarity cutoff using MOTHUR version 1.34.4 software (Schloss et al. 2009). The MOTU classification was validated by constructing a p distance tree. The representative sequences were selected by the abundance method for BLAST searches against international nucleotide sequence databases (INSD) for taxonomic placement (Altschul et al. 1997), and ≥97 % identity with known species were named as species. Putative chimera clones inferred from BLAST results were discarded manually from further analyses. Representative fungal ITS sequences were deposited in the DNA Data Bank of Japan (DDBJ) under accession numbers LC096270 to LC096953. In the following analyses, each MOTU and the occurrence frequency of MOTU were treated as taxon and abundance, respectively. For P. japonica MOTUs, the accumulation curves of taxa in the Aichi and Mie regions and all sites altogether were constructed using the EstimateS version 9.1 software (Colwell 2013). In addition, Shannon’s, Fisher’s alpha, and Simpson’s diversity indices of root-associated fungi were calculated to characterize the pattern of fungal assemblages of P. japonica. Non-metric multidimensional scaling (NMDS) and PERMANOVA were also performed to test if the community was structured in accordance with geographic regions. Clone abundance data of each site were used for ordination with the Horn dissimilarity index. We also used the function “envfit” implemented in the Vegan package (Oksanen et al. 2013) to identify variables that were significantly correlated with the community composition of ECM fungi. Eight variables relating to host trees, geographical location, climate, and soil conditions fitted as vectors with 999 permutations. The location was represented by the principal coordinates of neighborhood matrix (PCNM) calculated based on the distance matrix among study sites expressed as longitudinal and latitudinal coordinates (Dray et al. 2006). For comparison with fungal assemblages, MOTU compositions of P. japonica and ECM tips of pooled cores collected at Aichi were tested by means of the χ 2 test. In addition, the occurrence frequency of top five MOTUs on both P. japonica and ECM tips was examined using Fisher’s exact test. These analyses were accomplished by R 3.12 (R Core Team 2013) and the package vegan (Oksanen et al. 2013). Significance level was defined at p < 0.05 unless otherwise stated. Since Russulaceae were the dominant fungal family (see “Results”), phylogenetic clustering of recovered russulacean MOTUs was estimated by reconstructing a phylogenetic tree. Representative sequences of this study along with several russulacean sequences represented in the International Nucleotide Sequence Database Collaboration (INSDc) were aligned by MAFFT 7 with the L-ins-I strategy and 1 PAM scoring matrix or otherwise with default settings (Katoh and Standley 2013). A neighbor-joining (NJ) tree of the ITS region was constructed, using Lactarius sequences as outgroups, based on the Maximum Composite Likelihood model with 1000 bootstrap replicates using MEGA 5.22.
Results
Mycorrhizal fungi associated with P. japonica
PCR amplifications were successful for all P. japonica individual plants but one from A1, one from M3, and two from M4 (probably due to low colonization). In all, 1512 sequences from P. japonica were obtained out of 1706 clones, i.e., from 8 to 64 clones per individual plant. Based on a 97 % cutoff, these sequences clustered into 51 MOTUs that included 36 ECM taxa accounting for 96.0 % of all clones (Tables S1 and S2 and Fig. 2a; we included Helotiales whose status, ECM versus endophytic, is debated). Other taxa were minor in frequency (4.0 %) and matched with putatively saprotrophic or endophytic species. The accumulation curves for ECM taxa MOTUs did not saturate, when all the collected samples were pooled or when only samples in Aichi or Mie were examined (Fig. S2a). However, they saturated at the individual plant level (Fig. S2b), suggesting that new MOTUs in populations were due to differences between individuals rather than to insufficient sampling effort within individuals.
In all, 17 MOTUs belonged to the genus Russula, accounting for 76.2 % of all clones (Table S2 and Fig. 2a). MOTU 12 matched with Russula densifolia (AB291764) and MOTUs 15 and 30 with Russula violeipes (KF361779 and KF361796, respectively) with more than 97 % identity, whereas other Russula MOTUs did not match with any described taxon at the threshold (Table S1). The 11 most abundant MOTUs belonged to Russula, except for the fourth, which matched with Laccaria amethystina (KF692083; 99 %) and accounted for 113 (7.5 %) of all clones. Russula MOTUs were detected from 33 plants (out of 35 for which barcoding succeeded) and dominated in 28 of these. Although half of the MOTUs were shared among plants within sites or a region, only two MOTUs (Russula spp. 2 and 7) were found in different regions. L. amethystina occurred in 12 plants from 5 sites and 2 regions and dominated in 3 plants where it accounted for 47 to 100 % of clones. Finally, three individual plants were respectively dominated by Amanita ceciliae, Rhizopogon sp. 1, and Helotiales sp. 1 that accounted for 72, 69, and 43 % of their respective clones. Other MOTUs, especially saprotrophic or endophytic, were less abundant in all or individual plants (<2 % of all clones) and were mostly detected from a single site or individual (Table S2). Shannon’s, Fisher’s alpha, and Simpson’s diversity indices for each site ranged from 0.26 to 2.26, from 0.51 to 5.29, and from 0.10 to 0.87, respectively, and were higher at a regional scale for Aichi and Mie (Table 2). Since this was also observed for Fisher’s alpha, a diversity index that corrects for sampling intensity, this means that different sites reveal different partners. Sites with multiple ECM tree species (M1 and A1) tended to show higher values (Table 2), but rarefaction analyses in Aichi and Mie at n = 11 showed similar estimated numbers of 21.7 and 17.2 MOTUs, respectively (Table 2 and Fig. S2a). In an NMDS scattering plot, although MOTU assemblages in Aichi and Mie tended to be separated on the first axis, the separation according to regions was not significant (PERMANOVA, p = 0.1, Fig. S3a). Of the eight variables fitted as vectors to the NMDS plot of the mycorrhizal community of P. japonica, PCNM was significantly correlated (Fig. S3b).
ECM fungal communities around P. japonica at Aichi
ECM communities surrounding P. japonica were assessed around 6 plants in the Aichi region (four in A1, one in A2 and A3), and 137 sequences of ECM tips were recovered that fell into 33 MOTUs (Table S1) belonging mostly to Russula (42.3 %), Helotiales (15.3 %), Cortinarius (10.9 %), and Corticiaceae (5.8 %) (Fig. 2b). This is more than the 20 MOTUs found on corresponding P. japonica, and indeed diversity indices tended to be higher for the ECM community (Table 2). Although the sampling effort of ECM tips was lower than for P. japonica clones, rarefaction in six soil cores of ECM communities at Aichi revealed more MOTUs (15) than did six P. japonica plants (five MOTUs; Fig. S4). In addition, the slightly different barcoding method between ECM tips and P. japonica clones was applied, but the primer LB-W used for the former samples is considered to target basidiomycetes (Tedersoo et al. 2008) and thus valid for the comparison of russulacean fungal occurrences between them. Six out of 47 MOTUs detected in Aichi were shared between P. japonica and ECM tips in the same core (Fig. S5): these MOTUs accounted for 35.8 % (132/369 clones) of the P. japonica community versus 26.3 % (36/137 sequences) of the ECM tip community (Table S3). Three were Russula spp. (spp. 5, 6, and 10), which accounted for 35.6 % (47/132 clones) of the shared community in P. japonica; the others were Rhizopogon sp. 1, Strobilomyces confusus, and Helotiales sp. 1, which together accounted for 64.4 % (85/132) of the shared community in P. japonica (Tables S2 and S3). Moreover, four taxa found on ECMs were present on P. japonica roots from other sites, namely Russula spp. 1 and 11, Cenococcum sp. 1, and Helotiales sp. 4 (Tables S2 and S3). As expected, Russula spp. were frequent in the shared community, but strikingly Russula sp. 1, which dominated in the Mie region on P. japonica (48 % of plants), was absent from P. japonica in the Aichi region: this is unexpected because it frequently occurred on ECM tips in Aichi (19 % of ECM tips, in five out of six cores). Yet, total MOTU compositions were significantly different between P. japonica and ECM tips at Aichi (Fig. 3; χ 2 = 381.07, df = 46, p < 0.001).
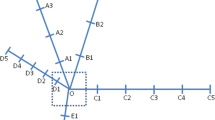
Frequencies of mycorrhizal taxa detected from Pyrola japonica roots (white square) and surrounding ECM roots (black square) at Aichi. Data were calculated as percentages of the focus taxa within a whole sample (n = 369 clones for P. japonica and n = 137 root tips for ECM roots). Asterisks indicate for each MOTU significant differences at p < 0.05 (*) and p < 0.001 (***) of the occurrence frequency in the two communities according to Fisher’s exact test
Phylogenetic positions of russulacean MOTUs
Phylogenetic positions of russulacean MOTUs obtained from P. japonica and ECM tips were analyzed using the NJ method (Figs. 4 and S5). Representative sequences of the MOTUs shared between P. japonica roots and ECM tips were supported by 100 % of bootstrap values. MOTUs 12 and 15 were confirmed to be R. densifolia and R. violeipes, respectively. No clear clustering of MOTUs was observed, although some were grouped into close positions, such as Russula spp. 4, 5, and 9 and Russula spp. 1, 2, and 15.
Neighbor-joining phylogeny with positions of russulacean MOTUs from both Pyrola japonica roots and ECM tips recovered in this study (in bold). Bootstrap tests (1000 replicates) larger than 90 % are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method. MOTUs derived from ECM tips are followed by an “e.” Some interior branches were compressed into solid elongated triangles whose thickness was proportional to the number of sequences (an uncompressed tree is to be found in Fig. S5)
Discussion
We analyzed fungal mycorrhizal associates of P. japonica grown under various ECM forests of three sites. Most mycorrhizal fungi belonged to ECM taxa, and 76.2 % of the clones sequenced were affiliated to Russula spp., which colonized 94 % of sampled P. japonica individuals. In the ECM community studied on tree roots surrounding P. japonica at Aichi, Russula spp. also dominated. The mycorrhizal community on ECM tips differed significantly from that on P. japonica roots, but the different primer sets may cause PCR biases.
A preference of P. japonica for Russula spp.
The predominance of Russula spp. was congruent with our previous report (Matsuda et al. 2012) from a single P. japonica population (under Quercus serrata and Quercus acutissima at Tsu City, central Japan; Fig. 1), where 84 % of identified fungi belonged to Russula. However, in this previous study, the exclusive use of direct sequencing of fungal ITS discarded many root samples with multiple colonizations: the success rate in Matsuda et al. (2012) was lower (44 % of root fragments) than the current one (85 %). Population origins in central Japan and methodological differences thus had limited impacts on the conclusion that Russula spp. dominate among fungi associating with P. japonica. Similarly, nearby ECM tree taxa, including Pinus, Castanopsis, or Quercus, did not affect the predominance of russulacean fungi on P. japonica. This predominance thus reflects a mycorrhizal preference of this plant for Russula. Russula spp. from the delica group are known to associate specifically with the MX orchid Limodorum abortivum (Girlanda et al. 2006; Bellino et al. 2014), and Russula spp. are partners to many MH orchids (Merckx 2013). The predominance of russulacean fungi was also reported in the MH Monotropoideae (Ericaceae), a sister group of Pyrola plants (Merckx et al. 2013; Matsuda et al. 2011). Although circumstantial evidence has accumulated, it remains unclear why Russula spp. support MX and MH plants.
Another unexpected observation is the dominance in some individuals from Aichi and Mie of L. amethystina. This Euro-Asiatic taxon (Vincenot et al. 2012) has already been found in European Pyrola chlorantha (Tedersoo et al. 2007), as have other Laccaria spp. in European P. minor (Zimmer et al. 2007) and Japanese Pyrola asarifolia (Hashimoto et al. 2012). Yet, Laccaria spp. were always a minor component of the diverse communities found on the Pyrola host plants. We speculate that the local abundance of L. amethystina in our study is relatively boosted by the depauperate fungal community of P. japonica, but we cannot exclude a marginal preference for Laccaria. Thus, it may be that Laccaria acts as a mycorrhizal substitute when the usual dominant fungal partner is missing or no longer available.
Based on our data, P. japonica thus displays distinctive features compared with other investigated Pyrola spp. that harbor diverse ECM fungal taxa not only in green, MX, or autotrophic species (Tedersoo et al. 2007; Vincenot et al. 2008; Hynson and Bruns 2009; Toftegaard et al. 2010; Hashimoto et al. 2012) but also in achlorophyllous MH species (Hynson and Bruns 2009). Thus, the genus Pyrola is often considered as a generalist lineage in the evolution of mycoheterotrophy (Hynson et al. 2013). Although achlorophyllous MH plant species often show higher mycorrhizal specificity (Merckx 2013), the fully MH P. aphylla has a low specificity, comparable to that of the MX P yrola picta (Hynson and Bruns 2009). In the case for P. japonica, mycorrhizal fungal richness was far lower than for other Pyrola species (consider Fisher’s alpha values that are independent of the sampling effort in Table 2), although taxa also reported for other Pyrola plants (e.g., Amanita, Laccaria, Thelephoraceae, and Russulaceae) were marginally involved. Shannon and Simpson diversities were also lower at most sites because evenness was distorted by the dominance of Russula.
Habitat characteristics could affect the mycorrhizal community of P. japonica. In MH Monotropoideae and MH orchids, surrounding tree species or geography can affect the mycorrhizal community. In the former plants that are phylogenetically close to the genus Pyrola, Monotropa uniflora and Pterospora andromedea associated with different Russula (Bidartondo and Bruns 2001) and Rhizopogon species (Dowie et al. 2012) depending on the locality, respectively. Similarly, Taylor and Bruns (1999) reported that MH orchids in the genus Corallorhiza, growing in either oak- or conifer-dominated forests, associate with different Russula communities. In our study, the P. japonica mycorrhizal community seems to differ depending on the sites inferred from an NMDS plot, and indeed diversity values are higher at the regional scale than the site scale, suggesting a spatial variation of the fungal associates within Aichi and Mie. Yet, tree taxa were not a significant factor; however, the statistical power of our sampling for this issue was limited (Fig. S3) and the closely located A2 and A3 sites (1.8 km), which differed by tree taxa (Fagaceae versus Pinaceae), had quite different communities. Remarkably, Russula sp. 1 was one of the most frequent MOTUs on P. japonica at the Mie sites, and among ECM tips at the Aichi sites, but it was never detected from P. japonica in the Aichi region (Tables S2 and S3; the proportion of individuals with Russula sp. 1 significantly differed between regions χ 2 = 5.56, df = 1, p < 0.05). Thus, tree taxa as well as geographical locations may influence community patterns of mycorrhizal fungi of P. japonica, although this deserves closer investigation. Considering our limited sampling efforts as well as the wide geographical distribution of P. japonica in East Asia (Takahashi 1991), further extensive sampling could allow robust investigations of regional and host differences.
To describe all fungal associates, the species accumulation curve should ideally be saturated, but identifying major ECM fungal associates needs only at most several tens of clones or even direct sequencing (Tedersoo et al. 2007; Kennedy et al. 2011). In this study, we collected up to 64 clones from each plant and that was an effort comparable to the most intense analyses of mycorrhizal fungi associated with MX orchids (Abadie et al. 2006; Girlanda et al. 2006). Although observed MOTU richness does not saturate with 64 clones in most individuals, the diversity index was mostly saturated by the first 20 to 30 clones (Fig. S2b). The Simpson index takes evenness into account so that it rapidly reaches a plateau when one of community member is far more abundant than the others. Thus, even without description of all fungi present in the sample, 20–30 clones efficiently characterized the major partners associated with P. japonica and its preferential association with Russula spp. In the future, “next-generation” sequencing technologies may provide a more comprehensive picture of P. japonica mycorrhizal associations.
A community filtered from the surrounding ECM community
The ECM communities of trees surrounding six Aichi P. japonica plants were also analyzed, and five individuals shared at least one MOTU with neighboring ECM tips from the same core (Table S3). Although shared MOTUs do not necessarily mean a direct mycelial connection with ECM roots, these results are congruent with the presence of common mycorrhizal networks connecting MX plants with surrounding canopy trees (Selosse and Roy 2009; Simard et al. 2012). The existence of a few shared species (12.8 % of MOTUs) that are very abundant (colonizing ca. one third of sampled roots on each partner) is congruent with a previous study of P. asarifolia and surrounding trees (Hashimoto et al. 2012) and is also a common trait of many ECM communities in that multihost species are often abundant (e.g., Henry et al. 2015). Moreover, such abundant root colonization may favor abundant mycelial links and thus efficient carbon transfer to the C-receiving MX plant.
Although our analyses of 20–30 ECM tips are likely too limited to provide a comprehensive view of the ECM community at each study site (Miyamoto et al. 2014), the fungal composition significantly differed between P. japonica and surrounding trees. For the five most frequent MOTUs on P. japonica roots, the relative frequency of russulacean MOTUs was significantly higher than that on ECM tips (Fig. 3), e.g., R. violeipes 1 (p < 0.001) or Russula sp. 10 (p < 0.05). Similarly, among the five most frequent MOTUs on ECM tips, the frequencies of Russula spp. 1 and 6 were significantly higher (p < 0.001) on ECM tips than on P. japonica. PCR biases derived from different primer sets used for either Pyrola roots or ECM tips may mask the actual differences in ECM fungal assemblages between them. However, the LB-W primer for ECM tips is known to more specifically amplify basidiomycetes (Tedersoo et al. 2008), and thus any bias might have favored the detection of russulacean groups on ECM tips. The primer change and the use of cloning for P. japonica (but not for ECM tips) may have introduced a bias (e.g., cloning could have led to overrepresentation of Russula spp. in P. japonica), so our comparison is only valid provided that this bias is limited. Under the assumption of a limited bias, the ECM communities were rich in Russula spp. (six species representing 42.3 % of the sampled tips), suggesting, as expected, that Russula-rich spots may be favorable for P. japonica settlement (a similar conclusion was made for the MH Sarcodes sanguinea (Ericaceae) and its specific associates Rhizopogon ellenae; Bidartondo et al. 2000). On the other hand, the absence of some fungi abundant in the surrounding ECM community is significant, and the much lower diversity on P. japonica than in the surrounding ECM community suggests that this plant filters the surrounding ECM community. Such filtering, although with much less diversity reduction, may help to explain why the Japanese populations of the related P. asarifolia display fungal associates that somehow differ from ECM communities of surrounding trees (Hashimoto et al. 2012).
Conclusions
The mycorrhizal fungi associated with P. japonica confirm our preliminary conclusion (Matsuda et al. 2012) that they are dominated by russulacean fungi, in spite of a diverse local ECM community. This mycorrhizal preference, although not unusual among MX plants, is unique compared with other Pyrola spp. Since all study sites were in central Japan, and since we suspect a potential for local variation as discussed above, wider sampling of P. japonica covering the natural host distribution would provide more insights into its pattern of fungal preferences.
References
Abadie JC, Puttsepp U, Gebauer G, Faccio A, Bonfante P, Selosse MA (2006) Cephalanthera longifolia (Neottieae, Orchidaceae) is mixotrophic: a comparative study between green and nonphotosynthetic individuals. Can J Bot 84:1462–1477
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bellino A, Alfani A, Selosse MA, Guerrieri R, Borghetti M, Baldantoni D (2014) Nutritional regulation in mixotrophic plants: new insights from Limodorum abortivum. Oecol 175:875–885
Bidartondo M, Kretzer AM, Bruns TD (2000) High root concentration and uneven ectomycorrhizal diversity near Sarcodes sanguinea (Ericaceae): a cheater that stimulates its victims? Am J Bot 87:1783–1788
Bidartondo MI, Bruns TD (2001) Extreme specificity in epiparasitic Monotropoideae (Ericaceae): widespread phylogenetic and geographical structure. Mol Ecol 10:2285–2295
Björkman E (1960) Monotropa hypopitys L.—an epiparasite on tree roots. Physiol Plantarum 13:308–327
Colwell RK (2013) EstimateS: statistical estimation of species richness and shared species from samples, ver. 9.1. http://purl.oclc.org/estimates. Accessed 15th January 2016
Dowie NJ, Hemenway JJ, Miller SL (2012) Identity, genetic lineages and putative hybrids of an obligate mycobiont associated with the mycoheterotrophic plant Pterospora andromedea in the south-central Rocky mountains. Fungal Ecol 5:137–146
Dray S, Legendre P, Peres-Neto PR (2006) Spatial modelling: a comprehensive framework for principal coordinate analysis of neighbour matrices (PCNM). Ecol Model 196:483–493
Gebauer G, Meyer M (2003) 15N and 13C natural abundance of autotrophic and myco-heterotrophic orchids provides insight into nitrogen and carbon gain from fungal association. New Phytol 160:209–223
Girlanda M, Selosse MA, Cafasso D, Brilli F, Delfine S, Fabbian R, Ghignone S, Pinelli P, Segreto R, Loreto F, Cozzolino S, Perotto S (2006) Inefficient photosynthesis in the Mediterranean orchid Limodorum abortivum is mirrored by specific association to ectomycorrhizal Russulaceae. Mol Ecol 15:491–504
Gonneau C, Jersáková J, Tredern E, Till-Bottraud I, Sauve M, Saarinen K, Roy M, Hájek T, Selosse MA (2014) Photosynthesis in perennial mixotrophic Epipactis spp. (Orchidaceae) contributes more to shoot and fruit biomass than to hypogeous survival. J Ecol 102:1183–1194
Hashimoto Y, Fukukawa S, Kunishi A, Suga H, Richard F, Sauve M, Selosse MA (2012) Mycoheterotrophic germination of Pyrola asarifolia dust seeds reveals convergences with germination in orchids. New Phytol 195:620–630
van der Heijden MGA, Martin F, Selosse M-A, Sanders IR (2015) Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol 205:1406–1423
Henry C, Raivoarisoa JF, Razafimamonjy A, Ramanankierana H, Andrianaivomahefa P, Selosse MA, Ducousso M (2015) Asteropeia mcphersonii, a potential mycorrhizal facilitator for ecological restoration in Madagascar wet tropical rainforests. For Ecol Manag 358:202–211
Hynson NA, Bruns TD (2009) Evidence of a myco-heterotroph in the plant family Ericaceae that lacks mycorrhizal specificity. Proc Roy Soc Lond B 276:4053–4059
Hynson NA, Preiss K, Gebauer G, Bruns TD (2009) Isotopic evidence of full and partial myco-heterotrophy in the plant tribe Pyroleae (Ericaceae). New Phytol 182:719–726
Hynson NA, Madsen TP, Selosse MA, Adam IKU, Ogura-Tsujita Y, Roy M, Gebauer G (2013) The physiological ecology of mycoheterotrophy. In: Mycohetertrophy. Merckx VSFT, Springer, New York, pp 297–342
Hynson NA, Bidartondo MI, Read DJ (2015) Are there geographic mosaics of mycorrhizal specificity and partial mycoheterotrophy? A case study in Moneses uniflora (Ericaceae). New Phytol 208:1003–1007
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kennedy AH, Taylor DL, Watson LE (2011) Mycorrhizal specificity in the fully mycoheterotrophic Hexalectris Raf. (Orchidaceae: Epidendroideae). Mol Ecol 20:1303–1316
Liu ZW, Zhou J, Liu ED, Peng H (2010) A molecular phylogeny and a new classification of Pyrola (Pyroleae, Ericaceae). Taxon 59:1690–1700
Massicotte HB, Melville LH, Tackaberry LE, Peterson RL (2008) A comparative study of mycorrhizas in several genera of Pyroleae (Ericaceae) from western Canada. Botany 86:610–622
Matsuda Y, Hijii N (2004) Ectomycorrhizal fungal communities in an Abies firma forest, with special reference to ectomycorrhizal associations between seedlings and mature trees. Can J Bot 82:822–829
Matsuda Y, Okochi S, Katayama T, Yamada A, Ito SI (2011) Mycorrhizal fungi associated with Monotropastrum humile (Ericaceae) in central Japan. Mycorrhiza 21:569–576
Matsuda Y, Shimizu S, Mori M, Ito SI, Selosse MA (2012) Seasonal and environmental changes of mycorrhizal associations and heterotrophy levels in mixotrophic Pyrola japonica (Ericaceae) growing under different light environments. Am J Bot 99:1177–1188
Miyamoto Y, Nakano T, Hattori M, Nara K (2014) The mid-domain effect in ectomycorrhizal fungi: range overlap along an elevation gradient on Mount Fuji, Japan. ISME J 8:1739–1746
VSFT Merckx (2013) Mycoheterotrophy: an introduction. In: VSFT Merckx (ed) Mycoheterotrophy. Springer, New York, pp 1–17
VSFT Merckx, Freudenstein JV, Kissling J, Christenhusz MJM, Stotler RE, Crandall-Stotler B, Wickett N, Rudall PJ, Maas-van de Kamer H, Maas PJM (2013) Taxonomy and classification. In: VSFT Merckx (ed) Mycoheterotrophy. Springer, New York, pp 19–101
Nara K (2015) The role of ectomycorrhizal networks in seedling establishment and primary succession. In: Horton TR (ed) Mycorrhizal networks. Springer, Berlin Heidelberg, pp 177–201
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2013) Vegan: community ecology package. R package version 2.0-10. http://CRAN.R-project.org/package=vegan
Peterson RL, Massicotte HB, Melville LH (2004) Mycorrhizas: anatomy and cell biology. NRC Research Press, Ottawa
R Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, http://www.R-project.org/
Robertson DC, Robertson JA (1985) Ultrastructural aspects of Pyrola mycorrhizae. Can J Bot 63:1089–1098
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Selosse MA, Richard F, He X, Simard S (2006) Mycorrhizal networks: les liaisons dangereuses. Trends Ecol Evol 11:621–628
Selosse MA, Roy M (2009) Green plants that feed on fungi: facts and questions about mixotrophy. Trends Plant Sci 14:64–70
Selosse MA, Bocayuva MF, Kasuya MCM, Courty PE (2016) Mixotrophy in mycorrhizal plants: extracting C from mycorrhizal networks. In Martin F (ed) Mycorrhizal ecology. Springer, Berlin-Heidelberg.
Simard SW, Beiler KJ, Bingham MA, Deslippe JR, Philip LJ, Teste FP (2012) Mycorrhizal networks: mechanisms, ecology and modelling. Fungal Biol Rev 26:39–60
Smith SE, Read DJ (2008) Mycorrhizal symbiosis, 3rd edn. Academic, London
Takahashi H (1991) Distribution of Japanese Pyrola and Orthilia, and a distribution pattern with a gap in the central to southern Tohoku district. Acta Phytotax Geobot 42:23–43
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Taylor LD, Bruns TD (1999) Population, habitat and genetic correlates of mycorrhizal specialization in the ‘cheating’orchids Corallorhiza maculata and C. mertensiana. Mol Ecol 8:1719–1732
Tedersoo L, Pellet P, Kõljalg U, Selosse MA (2007) Parallel evolutionary paths to mycoheterotrophy in understorey Ericaceae and Orchidaceae: ecological evidence for mixotrophy in Pyroleae. Oecol 151:206–217
Tedersoo L, Jairus T, Horton BM, Abarenkov K, Suvi T, Saar I, Kõljalg U (2008) Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytol 180:479–490
Thomson D, Henry R (1995) Single-step protocol for preparation of plant tissue for analysis by PCR. Biotechniques 19:394–397
Toftegaard T, Iason GR, Alexander IJ, Rosendahl S, Taylor AFS (2010) The threatened plant intermediate wintergreen (Pyrola media) associates with a wide range of biotrophic fungi in native Scottish pine woods. Biodivers Conserv 19:3963–3971
Vincenot L, Tedersoo L, Richard F, Horcine H, Kõljalg U, Selosse MA (2008) Fungal associates of Pyrola rotundifolia, a mixotrophic Ericaceae, from two Estonian boreal forests. Mycorrhiza 19:15–25
Vincenot L, Nara K, Sthultz C, Labbe J, Dubois M-P, Tedersoo L, Martin F, Selosse MA (2012) Extensive gene flow over Europe and possible speciation over Eurasia in the ectomycorrhizal basidiomycete Laccaria amethystina complex. Mol Ecol 21:281–299
Waterman RJ, Klooster MR, Hentrich H, Bidartondo MI (2013) Species interactions of mycoheterotrophic plants: specialization and its potential consequences. In: VSFT Merckx (ed) Mycoheterotrophy. Springer, New York, pp 267–296
Zimmer K, Hynson NA, Gebauer G, Allen EB, Allen MF, Read DJ (2007) Wide geographical and ecological distribution of nitrogen and carbon gains from fungi in pyroloids and monotropoids (Ericaceae) and in orchids. New Phytol 175:166–175
Acknowledgments
We thank Mr. S Iio for habitat information regarding the Hashirida shrine, Iyama shrine, and Makiba Mizunosaka West Hill and the Aichi prefecture government office for permission to access the sites. We also thank T. Chikada, Life Science Research Center, Center for Molecular Biology and Genetics, Mie University, for helping out with DNA analyses, the members of the Laboratory of Forest Pathology and Mycology, Graduate School of Bioresources, Mie University, for their help in field sampling, and D. Marsh for correcting our English. This study was partly supported by KAKENHI (22688011, 23658124, 25304026) to YM and the Fondation de France (fond Ars Cuttoli & Appel) to M-AS.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
BLAST results for ITS consensus sequences of each MOTU. (DOC 92 kb)
Table S2
Fungal taxa mycorrhizal on Pyrola japonica. Putative trophic status: ECM, ectomycorrhizal; ERM, ericoid; END, endophytic; SAP, saprotrophic; NEM, nematophagous. Asterisk indicates MOTUs that were detected from Pyrola japonica and tree roots but not identical soil cores. (DOC 130 kb)
Table S3
Fungal taxa detected from Pyrola japonica and tree roots in the same cores. MOTUs shared between Pyrola japonica and ECM tips in one core are enclosed. Hosts, Py: Pyrola japonica, Q: Quercus glauca, P: Pinus densiflora, C: Castanopsis cuspidata. (DOC 130 kb)
Fig. S1
Images of each study site: (a) Aichi, (b) Mie and (c) Kyoto, (d) flowering Pyrola japonica, (e) root fragment, (f) light and (g) fluorescent microscopic views of P. japonica roots made by hand cross section. Arrows in (e), (f) and (g) indicate hyphal coils, and no fungal mantles were formed on epidermal cells. Double arrowheads in (e) and (f) indicate plant cells that contained no hyphal coils. Bars 1 mm for (e) and (f) and 100 μm for (g). (DOC 728 kb)
Fig. S2
Rarefaction curves of (a) MOTUs in Pyrola japonica collected at all study sites (white circle), Aichi (black square) and Mie (black up-pointing triangle) and (b) Simpson’s diversity of clones derived from each plant collected. Plants collected at Kyoto were not analyzed owing to the lack of replicates (n = 3). Plant ID is shown with the combination of site codes, overstory trees (P for Pinus and Q for Quercus) and replicates. (DOC 81 kb)
Fig. S3
A non-metric multidimensional scaling plot of the mycorrhizal community of Pyrola japonica. Clone abundance data of each site were used for ordination with Horn dissimilarity index (a). White up-pointing triangle, Aichi sites A1 to A3; black square, Mie site M1 to M4; white circle, Kyoto unique site K1. Grouping according to region was not significant (PERMANOVA, p = 0.1). Eight variables were tested for significant correlation with mycorrhizal community dissimilarities (b). Significantly correlated variables are shown in the plot as arrows. The length of the arrow is proportional to the strength of the correlation with the ordination (*p < 0.05). PCNM is the principal coordinates of neighborhood matrix and see for “Materials and methods” in details. (DOC 61 kb)
Fig. S4
Rarefaction curves of MOTUs in Pyrola japonica (white circle) and its surrounding ectomycorrhizal roots (black circle) collected in Aichi. (DOC 43 kb)
Fig. S5
Neighbor-joining phylogeny of russulacean MOTUs recovered in this study. Bootstrap tests (1000 replicates) larger than 90 % are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method. MOTUs detected in this study are in bold, and when they were derived from ECM tips denoted by an ‘e’ following the MOTU names. (DOC 103 kb)
Rights and permissions
About this article
Cite this article
Uesugi, T., Nakano, M., Selosse, MA. et al. Pyrola japonica, a partially mycoheterotrophic Ericaceae, has mycorrhizal preference for russulacean fungi in central Japan. Mycorrhiza 26, 819–829 (2016). https://doi.org/10.1007/s00572-016-0715-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-016-0715-2