Abstract
Deep learning techniques have gained immense popularity recently because of their remarkable capacity to learn complex patterns and features from large datasets. These techniques have revolutionized many fields by achieving advanced performance in various tasks. The availability of large datasets and the advancement of computing resources have enabled deep learning models to perform well in solving challenging problems. As a result, they have become an essential tool in many industries, including agriculture. The application of deep learning in agriculture has great potential for increasing productivity, reducing costs, and improving sustainability by aiding in the early identification and prevention of plant leaf diseases, optimizing crop yields, and facilitating precision agriculture. This paper suggests using a novel approach to automatically classify multi-class leaf diseases in tomatoes using a deep multi-scale convolutional neural network (DMCNN). The proposed DMCNN architecture consists of parallel streams of convolutional neural networks at different scales, which get merged at the end to form a single output. The images of tomato leaves are preprocessed using data augmentation techniques and fed into the DMCNN model to classify disease. The proposed approach is evaluated on a dataset of tomato plant images containing 10 distinct classes of diseases and compared with different existing models. The research results reveal that the suggested DMCNN model performs better than other models in terms of accuracy, precision, recall, and F1 score. Furthermore, the proposed model reported an overall accuracy of 99.1%, which is higher than the accuracy of existing models tested on the same dataset. The study demonstrates the potential of deep learning techniques for automated disease classification in agriculture, which can aid in early disease detection and prevent crop loss.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Plant disease detection and diagnosis are crucial in agriculture for effective disease management and sustainable crop production. In the past, traditional methods such as visual inspection, field surveys, and laboratory analysis were primarily used for disease detection. However, these methods are time-consuming, labor-intensive, and require significant expertise in plant pathology. Therefore, there is an increasing need for automated and efficient disease detection and classification methods. Advanced technologies such as deep learning, hyperspectral imaging, and remote sensing offer several advantages over traditional methods, such as non-destructive detection, rapid and accurate diagnosis, and minimal expertise requirements. These technologies can enable early detection and targeted action to control the spread of diseases, reduce crop losses, and maintain the quality of harvested goods. Moreover, adopting advanced technologies in plant disease management can help ensure sustainable crop production and contribute to achieving global food security goals. Therefore, developing and deploying such technologies are critical for effective disease management and the future of agriculture.
This paper provides deep learning-based method for using deep multi-scale convolutional neural networks to automate the classification of multi-class leaf diseases in tomatoes. The significance of this proposed study stems from the fact that tomato plants are affected by several diseases that can significantly reduce crop yield, quality, and economic value. Traditional methods for detecting and classifying these diseases are often time-consuming and require expert knowledge, which can be a limiting factor in large-scale crop production.
Recent advances in deep learning methodologies, with a primary emphasis on convolutional neural networks, have exhibited remarkable achievements across diverse domains and tasks, encompassing object recognition, image segmentation, classification, and even structural damage identification [1]. Nevertheless, within the realm of plant sciences, a considerable portion of extant research endeavors centered on plant disease classification has been confined to single-channel and same-resolution imagery. This limitation potentially impedes the holistic capture of the comprehensive information indispensable for precise disease detection and classification.
To address this limitation, this paper proposes a deep multi-scale convolutional neural network-based framework for automatically classifying tomato leaf diseases. The proposed framework leverages the benefits of multiple channels of information to improve the accuracy and efficiency of disease classification.
This study aimed to develop a DMCNN architecture capable of accurately classifying 10 distinct categories of tomato plant diseases and evaluate its performance on a publicly available dataset of 11,000 tomato plant images, leveraging multiple channels and scales of information provided by the camera. The study also aims to optimize the model’s hyperparameters to improve its performance and evaluate its effectiveness in comparison to many existing methods.
The contributions of this study are five fold:
-
1.
Proposing a deep multi-scale CNN architecture that leverages multiple channels of information for accurate disease classification.
-
2.
Developing a DMCNN architecture consisting of parallel streams of convolutional neural networks (CNNs) at different scales, which are merged at the end to form a single output.
-
3.
Evaluating the proposed framework using a large and diverse dataset of tomato leaf images and comparing its performance with many existing methods.
-
4.
Providing insights into the feature importance of the proposed model, which can help in understanding the underlying mechanisms of disease classification.
-
5.
Conducting significant analysis to demonstrate the robustness of the proposed model to various factors, which can improve the reliability and generalizability of the model.
Furthermore, this paper aims to contribute to the development of more accurate, efficient, and automated methods for disease classification in tomato crops. The proposed approach has potential applications in precision agriculture and sustainable crop management. The contributions of this study are significant in that they showcase the potential of using deep learning and multi-scale imaging for plant disease detection and classification. By leveraging multiple information channels, such as color and texture, the proposed approach achieves improved accuracy and efficiency in disease classification. The use of deep learning algorithms also enables the automation of the disease classification process, enabling large-scale and timely interventions to mitigate crop losses. The outcomes of this research demonstrate the promise of deep learning-based strategies to revolutionize the field of plant pathology and crop management, improving crop productivity and ensuring food security for future generations.
The remainder of the paper has the following structure: Section 2 presents an overview of related work in the area of automated disease detection in agriculture. Section 3 describes the dataset and preprocessing steps used in our study and details the architecture of the proposed DMCNN and how it addresses the limitations of previous CNN-based methods. Section 5 explains the experimental setup and evaluation metrics used to measure the performance of the DMCNN. Section 3 presents the results of our experiments and compares them to other approaches in the literature. Finally, Section 6 concludes the paper and discusses potential future work in the area of advanced disease classification in agriculture.
2 Existing work
Plant disease detection and diagnosis are critical to agriculture as it enables effective disease management and sustainable crop production. In the past, traditional methods such as visual inspection, field surveys, and laboratory analysis were primarily used for disease detection in plants. While these methods have been used successfully in the past, they have several limitations that have hindered their widespread use [2].
One of the primary challenges of traditional methods for plant disease detection is that they can be time-consuming and labor-intensive. Visual inspection, for example, relies on the physical examination of plants for symptoms and signs of disease, which can be time-consuming. Similarly, field surveys require data collection over an extended period, which can be labor-intensive and require significant resources.
Another significant challenge of traditional methods for plant disease detection is that they require specialized knowledge and expertise in plant pathology. For instance, laboratory analysis techniques such as culturing and serology require specialized equipment and expertise to use effectively.
Therefore, several approaches have been commonly used in terms of traditional disease classification methods. One of the most straightforward and widely practiced methods is visual inspection, which relies on physical examination and observation of symptoms and signs to diagnose a disease. While this method is relatively simple and accessible, its accuracy can be highly subjective, depending on observer’s expertise. Additionally, there may be significant variability in the diagnosis based on the observer’s experience and training [2].
Another traditional method for disease classification is laboratory analysis, which involves using specialized equipment and techniques to identify the causative agent or markers of disease. While this approach can provide objective and quantitative results, it can be time-consuming and expensive, requiring significant resources and expertise to implement effectively.
Finally, field surveys are another traditional method that involves monitoring the incidence of a disease over time in a particular region or population. This method can provide valuable information on the patterns and trends of disease occurrence, as well as risk factors and other relevant factors that may contribute to disease development and spread [3].
While these traditional methods for disease classification have been useful for many years, they have limitations, such as subjectivity, resource requirements, and time consumption. As such, modern technology has led to the development of newer and more advanced methods, such as molecular diagnostics and machine learning algorithms, that offer several advantages over traditional approaches.
To overcome these limitations, automated disease detection and classification methods have gained attention in recent years. Automated methods can provide accurate and efficient detection of plant diseases using various sensors and imaging techniques. Some commonly used sensors include hyperspectral, multispectral, RGB, and thermal imaging cameras [4].
Convolutional neural networks also performed remarkably in different computer vision tasks, including object detection, segmentation, and classification. In the context of plant disease detection and classification, CNNs have shown promising results in accurately identifying and differentiating between different types of plant diseases [5]. In addition, CNNs can learn the features of a plant image in a hierarchical manner, which helps in the better classification of leaf diseases.
Automated methods for disease classification have been widely studied in recent years. Deep learning (DL) models have made significant strides in recent years, revolutionizing numerous fields of research and development. Examples of these include deep fakes [6, 7], image classification [8, 9], satellite image analysis [10], the optimization of artificial neural networks [11, 12], and natural language processing [13, 14].
Convolutional neural networks (CNNs) are also used extensively for plant disease identification. CNNs can learn complex features of plant images and provide accurate disease detection and classification. For instance, Mohanty et al. [15] present a CNN-based framework for detecting plant leaf disease using RGB images. In addition, they used transfer learning to increase the performance of the model. Similarly, Sladojevic et al. [16] proposed a CNN-based framework for detecting plant diseases using hyperspectral images.
In the context of tomato disease detection and classification, various studies have been conducted using traditional and automated methods. For example, Ferentinos [17] used traditional image-processing methods for identifying and classifying tomato diseases. Similarly, Karimi et al. [18] proposed deep learning-based methods for classifying the diseases of tomato using RGB images. In their research, Agarwal et al. [19] proposed a Convolutional Neural Network (CNN)-based architecture for tomato disease identification. This model was designed to classify different classes of tomato images, with a range of accuracy rates from 76 to 100%, due to the numerous classes of tomato images used in the study. Furthermore, the proposed CNN-based architecture demonstrated a great level of accuracy, with an accuracy rate of 91.2%. This result shows that the model is able to accurately identify tomato diseases, which is a critical step in the effective treatment and management of these diseases.
In [20], they proposed a hybrid model that combines deep learning, PCA, and the whale optimization algorithm to diagnose tomato illnesses. The dataset utilized in the study was obtained from Plant Village and consisted of 18,160 images of tomato leaves divided into 10 different classifications. The proposed hybrid model reported 86% testing accuracy with the Adam optimizer and 94% with the RMSprop optimizer. This high accuracy rate indicates the potential of the proposed hybrid approach for the accurate and efficient diagnosis of tomato illnesses. The use of PCA and the whale optimization algorithm in conjunction with deep learning models have been shown to improve the accuracy of disease diagnosis in various applications. The proposed hybrid approach in this study provides a hopeful pathway toward creating more precise and effective diagnostic tools for detecting tomato diseases. Intan et al. [21] achieved high accuracy of 95.7% in classifying 10 types of tomato plant diseases using Dense CNN. Agarwal et al. [22] achieved an accuracy of 91.2% in classifying 10 types of tomato plant diseases using Vgg-16.
By utilizing transfer learning on the original AlexNet network, Wang et al. [23] improved the average identification accuracy for 10 categories of tomato leaf images, achieving an accuracy of 95.62%. In another study, Kaur et al. [24] utilized another pre-trained based on the ResNet network to identify 7 types of tomato leaf diseases, achieving an impressive accuracy rate of 98.8%. Similarly, Kaushik et al. [25] also achieved an impressive accuracy rate of 97.01% in classifying 6 types of tomato diseases by utilizing a pre-trained ResNet-50 network.
Trivedi et al. [26] utilized a convolution neural network to classify nine tomato leaf disease types and a dataset consisting of 3000 tomato leaves. By using preprocessed and partitioned tomato leaf images, they achieved an impressive accuracy rate of 98.49%. Vijay et al. [27] employed CNN as well as K-nearest (KNN) models to classify tomato leaf disorders using a public dataset, with LIME providing explanations for the predictions. Their findings demonstrated that the CNN model outperformed KNN in disease detection, achieving a rate of 98.5%, recall of 93%, F1-score of 93%, and precision of 93%. In contrast, the KNN’s accuracy, recall, F1-score, and precision were only 83.6%, 86%, 84%, and 90%, respectively.
Ozbılge et al. [28] proposed a tomato disease categorization model as an option to the widely used ImageNet deep learning model and convenient deep-neural with six layers. The performance of this work was evaluated using the PlantVillage dataset using several statistical techniques, yielding an impressive accuracy of 99.70%.
Karthik et al. [29] presented a deep identification model structure for tomato diseases, which increases the residual network and utilizes pre-trained learning to report important disease identification features. Pre-trained has been shown to improve recognition accuracy by leveraging the pre-trained models’ learned features. However, the initial VGG16 and AlexNet networks have complicated forms and many parameters, which can pose challenges in practical applications and model deployment. On the other hand, the proposed model is designed to be efficient and lightweight, making it more suitable for real-world deployment scenarios. Despite the effectiveness of the transfer learning approach, it is crucial to consider the model’s structure and complexity in practical applications, as it can significantly impact its performance and usability. In another study, Grinblat et al. [30] have proposed a neural network approach for identifying legume species by analyzing the morphological patterns of leaves veins.
In this study, we introduce a deep multi-scale CNN architecture for the automated identification of multi-class diseases in tomato leaves. We leverage the benefits of using multiple classes of information to increase the performance accuracy of disease classification. Our proposed framework is evaluated using a large and diverse dataset of tomato leaf images. We compare and evaluate the performance of our proposed framework with other existing methods and provide insights into the feature importance of the proposed model and conduct sensitivity analysis to demonstrate its robustness to various environmental factors.
In summary, this study contributes to the growing body of literature on automated plant disease detection and classification, with a focus on tomato crops. Our proposed framework leverages the benefits of multiple channels of information to provide accurate and efficient disease identification and classification. The framework can potentially be used in precision agriculture for sustainable crop management.
3 Research materials and methods
This section presented a comprehensive overview of the proposed model, as well as the datasets used in the study. In addition, it highlights the several steps used to improve the effectiveness of the suggested approach and presents the DMCNN model architecture.
3.1 Dataset and preprocessing
3.1.1 Dataset description
In our study, we utilized a large and diverse dataset comprising 11,000 images from 10 different categories, including healthy tomatoes and diseases like early blights, bacterial spot, leaf molds, late blight, as well as mosaic viruses. The images were obtained from the Kaggle dataset, which is widely used in deep learning research. The dataset was carefully curated to ensure that it was evenly distributed among the 10 classes, with 1100 images per class, which provided a robust representation of the various tomato leaf diseases.
To train and evaluate our proposed Deep Multi-Scale Convolutional Neural Network (DMCNN) and for the automated classification of multi-class leaf diseases in tomatoes, we separated dataset into 10:90 testing and training sets. Additionally, to prevent over-fitting and to further optimize the model, we allocated 10% of the training set for validation purposes. This allowed us to supervise the training process and adjust the model’s hyperparameters to achieve the best possible performance.
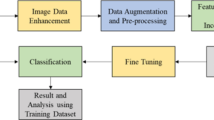
The dataset was carefully curated to include image data of tomato plant leaves with multiple diseases as well as healthy leaves, which were captured under different lighting conditions and varying orientations. This diversity of the dataset made it an excellent choice for training and evaluating Multi-Scale Convolutional Neural Networks (MSCNNs), as it allowed us to capture features at multiple scales and handle variations in the input images. To feed a visual representation of the data available for training and testing our deep learning model for classifying tomato leaf diseases, we included Fig. 1 in our study, which displays a collection of image data of tomato plant leaf disease from a dataset. Each image belongs to a specific disease category. Each image belongs to a specific disease category, and by analyzing these images, we can gain insights into the characteristics and features of different diseases, which can help us develop a more accurate and efficient model.
Therefore, the dataset used in this study presents a robust representation of the various tomato leaf diseases, which is required to accurately train and evaluate deep learning models. The diversity of the dataset, combined with the carefully selected training, validation, and test sets, allowed us to create and assess the performance of the suggested DMCNN architecture accurately.
3.1.2 Dataset preprocessing
Deep learning models’ success in plant disease classification is mainly dependent on the standard of the dataset used to generate the model’s training data. Therefore, data preprocessing is a crucial step in the deep learning pipeline. It involves cleaning, trans-forming, and preparing the data for analysis so that the model can learn effectively from the data. Preprocessing methods, including filtering and image resizing, can improve the model’s performance in the area of plant disease classification.
In this paper, we conducted several preprocessing steps on the dataset to ensure that the images were in suitable way to be analyzed. First, we resized the images to a fixed resolution. We also used data augmentation methods like rotation, zooming, and flipping to artificially increase overall the amount of data and reduce overfitting. These techniques helped to create a more robust dataset, which is critical for the success of deep learning models.
We applied images at various scales to prepare a Deep Multi-scale Convolutional Neural Network on this dataset. Specifically, we applied a multi-scale approach where each image is resized to multiple scales, and the network processes them separately. This allows the network to detect objects at different scales and capture more fine-grained details, improving the model’s accuracy in disease classification. Table 1 outlines the specific techniques and scales used for the multi-scale approach in the tomato leaf dataset. The data preprocessing steps performed in this study were critical in ensuring that the dataset was suitable for training an accurate and efficient deep learning algorithm for plant leaf disease identification. In addition, using a multi-scale approach with different preprocessing techniques might enhance the model’s performance and lead to excellent disease classification outcomes.
The training samples were subjected to various transformations, including Horizontal flipping was applied to the training samples, while rotation was done within a range of 20 degrees. In addition, zooming was done within a range of 0.2, while shifting was done within a 0.2 range in both width and height. Several methods of data augmentation aid in broadening the training set’s diversity and avoiding overfitting. Table 2 shows data augmentation techniques used in the project and their corresponding parameters/ranges.
After the data preprocessing step, the overall number of images included in the dataset was 12,500, with 1250 images perclass. The dataset consisted of 10 different classes of plant diseases. This is important for understanding the size and composition of the dataset and provides context for any subsequent analysis or modeling performed on the dataset. The data set’s size is an important factor in deep learning that influences the model’s performance. A large dataset provides more examples for the model to learn from and improves its accuracy. Moreover, the balanced distribution of samples across different classes in the dataset ensures that the model is not biased toward one particular class, which can affect its ability to classify accurately.
In summary, the size and composition of the dataset, as well as the preprocessing techniques used, all have an impact on the performance of our study. A diverse and balanced dataset, along with appropriate preprocessing techniques, can improve the ability of the model to recognize and categorize different types of diseases correctly. This can lead to the earlier time classification of plant leaf diseases, which can allow farmers to lead to great steps to avoid crop loss and increase yield.
3.2 Implementation (experiment setup)
In this section of the study, we discuss the specifics and the implementation details of our proposed model for the automated classification of multi-class diseases in tomatoes using deep multi-scale convolutional neural networks (DMCNN). Our model architecture includes multi-scale convolutional filters, which enable the network to capture features at different scales, thereby enhancing the model’s ability to distinguish between different classes of leaf diseases. Additionally, our model also incorporates batch normalization and dropout layers, which prevent overfitting and improve the model’s generalization capability.
Our model was implemented using the PyTorch deep learning framework on a high-performance machine with an NVIDIA GeForce RTX 3090 GPU and 64 GB RAM. We utilized Python 3.7 for all experiments and utilized several popular Python libraries such as Pandas, NumPy, and Matplotlib for data visualization and manipulation.
To train our model, we applied a batch size of 64 and a learning basics learning rate of 0.001 with such a cosine austempering learning rate set up to train our model. The Adam optimizer was employed with default parameters and a weight decay of 0.0001. We trained our model for 100 epochs and utilized early stopping with patience of 100 epochs based on the validation accuracy. We performed all our experiments by using fivefold cross-validation to obtain more robust results.
We employed various evaluation metrics such as accuracy, recall, F1-score, and precision to determine how well our proposed model performs. We also visualized the confusion matrix to gain insights into the classification performance of our model. Table 3 displays all the details regarding the model setup.
Our proposed deep learning-based approach demonstrated promising results in accurately classifying multi-class leaf diseases in tomatoes. By leveraging the power of deep multi-scale convolutional neural networks and employing various preprocessing techniques, we improved the efficiency and accuracy of disease classification in tomato crops. These findings have potential applications in precision agriculture and sustainable crop management, contributing to developing more accurate and automated methods for disease classification in various crops.
One of the significant advantages of our proposed approach is its ability to handle multi-class classification of leaf diseases. This is particularly important in agricultural applications, where plants may be affected by multiple diseases simultaneously. By accurately classifying the different diseases, our model can help farmers take targeted action to address the specific disease and prevent it from spreading to other plants in the crop.
In addition, our model is highly scalable and can be easily adapted. This can help to address the growing demand for precision agriculture techniques that can help farmers optimize their crop yield. In General, our proposed deep learning-based approach has the potential to change the way we approach disease classification in agriculture, and we look forward to further exploring its applications in this field.
3.3 Proposed model architecture
The proposed deep multi-scale CNN model architecture is designed to effectively classify plant diseases in tomato crops using multi-scale disease images. The model structure relies on a deep convolutional neural network (DNN) that uses multiple scales of input data to enhance the precision and robustness of disease detection and classification. In this section, we provide an in-depth exploration of the architecture underpinning our proposed Deep Multi-Scale Convolutional Neural Network (DMCNN) for the accurate classification of multi-class leaf diseases in tomato crops. Our approach is rooted in harnessing the capabilities of convolutional neural networks (CNNs) across multiple scales of input data to bolster disease detection and classification precision.
The model consists of three main components:
-
1.
Multi-Scale Convolutional Layers: Positioned at the forefront of our model’s architecture, the Multi-Scale Convolutional Layers are pivotal. These layers harness multi-scale convolutional filters, enabling the network to capture intricate features across different scales. This strategic choice empowers the model to distinguish between various classes of leaf diseases more effectively. These layers form the foundational basis for capturing multi-scale features. By integrating convolutional filters of varying scales, the network gains the ability to detect subtle nuances present at different levels. This approach recognizes that diseases manifest distinctively across scales, and these layers allow the model to encapsulate these variations. The "how" of the question—how does multi-scale enhance classification—is answered by the aggregation of features from different scales, contributing to a more comprehensive disease representation and improved classification accuracy.
-
2.
Multi-branch Feature Extraction Module: The multi-branch feature extraction module is at the core of the model’s feature extraction prowess. Designed to extract features at varying scales from the input image, this module employs a sequence of convolutional layers endowed with different filter sizes from the initial stage. These layers collaborate to meticulously extract and segregate features spanning multiple scales, thereby capturing the leaf disease images’ local and global characteristics. The outputs of these convolutional layers are subsequently concatenated and channeled through a pooling layer, which effectively reduces the spatial dimensions of the extracted features while retaining their critical information. The resulting feature representation is then directed toward the subsequent global feature fusion module for further processing.
-
3.
Multi-Model Fusion Layers and Classification: The final stage of the model’s architecture encompasses a global feature fusion module, responsible for harmonizing the diverse multi-scale features harnessed from the previous stages. Through this process, the module generates a cohesive and unified feature representation that encapsulates the insights from various scales. This consolidated feature representation serves as the foundation for the model’s decision-making process in disease classification. By fusing the features extracted from different scales, the model gains the ability to comprehend intricate details and variations present in the data, which in turn contributes to improved classification performance. The extracted and unified feature representation from the Global Feature Fusion Module is channeled into the fully connected layers, facilitating the final classification decision.
Addressing the question of "why" multi-scale approaches can enhance classification performance is a crucial aspect. This phenomenon can be attributed to the capability of multi-scale features to capture a wide array of visual details. Leaf diseases can exhibit variations across scales, with certain distinctive features being more prominent at specific scales. By embracing these various scales, the model becomes adept at capturing both fine-grained details and broader contextual information, ultimately leading to an enriched feature representation that improves its ability to discriminate between diverse classes of diseases. The synergy between the different scales amalgamates into a holistic and robust classification framework. This strategic amalgamation of components enables the model to navigate through the complexities of tomato crop diseases and leverage multi-scale insights to enhance its accuracy and robustness in disease classification.
The multi-scale feature extraction module uses a series of convolutional layers with varying filter sizes to extract and separate features at multiple scales. The outputs of these convolutional layers are concatenated and passed through a pooling layer to reduce the spatial dimensions of the features. The resulting feature map is then fed into the global feature fusion module.
The global feature fusion module uses a series of fully connected layers to combine the multi-scale features into a unified feature representation. The output of the global feature fusion module is then passed through a softmax layer for disease classification.
We use a large and diverse dataset of tomato leaf images captured by a camera to train our proposed model. The dataset is preprocessed and augmented to ensure a balanced distribution of classes and to prevent overfitting. To train the model for 100 epochs, we utilized the Adam optimizer with such a learning rate of 0.001. Finally, the model is implemented using the PyTorch deep learning framework. Figure 2 provides the architecture of the proposed deep multi-scale CNN model architecture.
The model takes in a tomato image as input and then, feeds it through a series of multi-scale convolutional layers, which extract features at different scales, allowing the model to capture both local and global information about the image. Next, the features from each scale are processed separately by multi-branch feature extraction layers, which learn representations specific to different spectral channels.
The outputs of these branches are then fused together by a multi-modal fusion layer, which combines the information from each branch to form a unified feature representation. Finally, the fused features are fed through a sequence of fully connected layers that also output the final classification for the image.
The multi-scale convolutional layers’ output is then passed through multi-branch feature extraction layers. These layers extract features from different modalities, including texture, shape, and color, allowing this same model to capture different aspects of the image. The output of the multi-branch feature extraction layers is then passed through a multi-modal fusion layer. This layer fuses the features extracted from different modalities into a single representation, which is then passed through fully connected layers for classification.
The model produces a probability distribution for the various classes of tomato plant leaf disease. The model is tuned during training to reduce the cross-entropy loss between predicted probabilities and true labels. Furthermore, the proposed deep multi-scale CNN model architecture is designed to leverage the benefits of large dataset images and capture different aspects of the tomato plant leaf image for precise classification. Figure 3 presents the architecture of the suggested deep multi-scale CNN model with more details.
In this schematic representation Fig. 3, the terms "Batch Normalization," "ReLU," and "MaxPooling" all refer to the batch normalization layers, rectified linear activation functions, and maximum pooling layers, respectively. "Fully Connected Layer" refers to a dense layer with such a specified number of neurons, and finally, "Dropout" refers to dropout layers with a specified dropout rate.
The architecture comprises four convolutional with increasingly smaller filter sizes, immediately followed by batch normalization, Rectified linear activation, and then max pooling. To minimize the dimensionality of the feature maps, a global average pooling layer is implemented after the fourth convolutional layer. Then, with 256 and 128 neurons, respectively, two fully linked layers are added, followed by batch normalization then ReLU activation. After that, two dropout layers with a dropout rate of 0.5 are added, and the output is obtained using a softmax activation function. The architecture is designed for multi-scale image classification tasks with a tomato picture as input and the predicted class label as output.
To provide a visual understanding of the proposed model architecture and the textual representation providing a detailed description for implementation, we applied Multi-Branch Network for Tomato leaf disease classification, as illustrated in Fig. 4 and 5.
The schema in Figs. 4 and 5 represents the proposed deep Multi-Scale CNN Architecture using a Multi-Branch Convolutional Neural Network designed for tomato disease classification. The network starts with tomato pictures and runs them through four convolutional layers, typically accompanied by batch normalization, Rectified linear activation, and then max pooling. Each convolutional layer’s output is then routed through a distinct branch, with the first branch terminating in global average pooling, followed by a fully connected layer and dropout. Next, the output of a remaining three branches is pooled using max pooling and concatenated before passing through fully connected layers and batch normalization, Rectified linear activation, dropout, then, finally, a softmax output layer.
Using a multi-branch convolutional neural network in this architecture is an innovative approach to image classification that has been displayed to be highly effective. The output of each convolutional layer is used to create separate branches of the network, which allows for the extraction of more detailed information from each layer of the input image. This, in turn, results in a more accurate and reliable classification of tomato diseases.
The use of batch normalization and ReLU activation after each convolutional layer is also critical to the success of this architecture. Batch normalization helps to normalize the input to each layer, which can increase the model’s accuracy rate. While on the other hand, ReLU activation helps to present nonlinearity into the network, allowing it to better capture the complex relationships between various input image features.
Integrating multi-scale analysis, such as employing multi-scale convolutional neural networks (CNNs), can significantly enhance classification performance. This approach addresses the inherent diversity in object sizes and levels of detail present in images. By considering various scales, the network can capture both fine-grained features and broader patterns, facilitating the extraction of hierarchical and contextually relevant information. This strategy not only bolsters the model’s resilience to noise and variability but also enables it to recognize objects regardless of their position within the image. Effectively, multi-scale analysis enriches feature learning, promotes robust classification and contributes to improved generalization by leveraging a wider range of information from the input data.
Overall, the proposed deep multi-scale architecture represents an advanced and innovative approach to tackling the intricate task of tomato disease classification. By integrating a multi-branch convolutional neural network alongside batch normalization and ReLU activation, the architecture demonstrates its sophisticated design tailored for this purpose. This amalgamation of cutting-edge techniques showcases the architecture’s complexity and emphasizes its potential effectiveness in disease classification within the agricultural domain.
The multi-branch architecture enables the model to process diverse information scales simultaneously, aligning well with the varied visual manifestations of tomato diseases. Batch normalization enhances training stability, mitigating issues related to internal covariate shift, and promoting consistent model behavior. Additionally, using ReLU activation functions aids in efficient gradient propagation, enhancing the architecture’s ability to discern intricate disease patterns. Beyond its technical aspects, the architecture’s contribution addresses the critical need for accurate and automated disease classification in agriculture. Swift and reliable disease identification can profoundly impact crop yield, labor efficiency, and timely intervention, thus advancing agricultural practices.
In summary, the fusion of multi-scale architecture, multi-branch CNNs, batch normalization, and ReLU activation constitutes a pioneering effort with the potential to reshape disease classification methodologies in agriculture. This advancement not only signifies the progress of deep learning but also underscores its role in revolutionizing disease management and diagnosis in essential crops like tomatoes.
3.4 Pre-trained model architecture
In this study, we conducted a comprehensive comparative analysis of our deep Multi-Scale CNN model’s performance with various pre-trained models, which was crucial for evaluating the effectiveness and efficiency of our proposed model. We compared our proposed model with four commonly used pre-trained models for image classification tasks, namely AlexNet, VGG16, InceptionV3, and ResNet50. By comparing the performance of our model with these established models, we were able to have a deeper comprehension of the strengths and weaknesses of each model and identify areas where our proposed model could be improved. The results of this comparative analysis provided valuable insights that informed the refinement and optimization of our DMCNNs model and helped us to achieve superior performance in our image classification task. The tuning details of different pre-trained models on the tomato leaf dataset are presented in Table 4.
Using the tuning details in Table 4, it can be observed that each pre-trained model has a unique architecture, with varying numbers of layers and different configurations of pooling, convolution, and dense layers. These differences can affect the performance and efficiency of each model for a given task. Therefore, it is important to compare the performance and efficacy of our suggested DMCNN model to that of these pre-trained models on the Tomato dataset. The results of this comparison will allow us to analyze the effectiveness and efficiency of our model and determine whether it outperforms or falls short of the state-of-the-art models. A detailed discussion and analysis of the performance comparison will be presented in the following sections of the study.
4 Experimental results
In this section, a number of experiments were conducted to evaluate and showcase the efficacy of the suggested approach on the tomato plant leaf image dataset. All the results obtained from these experiments are elaborated in detail in section Sect. 4.1, providing a comprehensive explanation of the findings. Subsections, Sect. 4.1.1, and Sect. 4.1.2, present a detailed explanation of the outcomes obtained.
4.1 Implementing the proposed model architecture
In this section, we introduced the utilization of deep multi-scale convolutional neural networks (DMCNNs) also for the automated classification of leaf diseases in tomatoes. To assess the efficacy of our suggested approach, we utilized a dataset comprising 125,00 images across 10 distinct categories, including healthy leaves and nine types of diseases. The images were evenly distributed among the 10 classes, each containing 1250 images. The results of this evaluation provided valuable insights into the model’s performance, identified areas for improvement, and refined the model further. In conclusion, the performance evaluation demonstrated that our proposed DMCNNs model is highly effective in achieving its intended goals and can be considered a robust solution in its domain.
The exceptional accuracy of the proposed models was evident through their training and validation accuracy. The average training and validation accuracies were found to be 99.24% and 99.15%, respectively, which indicates the models’ robust performance. Additionally, the training and also validation losses were observed to be 0.2918 and 0.3699, respectively, further emphasizing the accuracy of the models. To illustrate these results, Fig. 6 and Table 5 display the accuracy and the loss of the proposed architecture.
The observed training and validation accuracies and losses for the proposed models provide strong evidence of their accuracy and effectiveness. The average training accuracy of 99.24% indicates that the model performed exceptionally well on a training set, correctly classifying most images. The average validation accuracy of 99.15% further confirms the model’s accuracy on the unseen data, highlighting its robustness and ability to generalize well.
The training and validation losses of 0.2918 and 0.3699, respectively, demonstrate that the model was capable of learning the features and patterns in the dataset effectively, resulting in minimized loss values. Furthermore, the closeness of the training and validation loss values indicates the model’s generalization ability.
Furthermore, the accuracy and loss graphics illustrated in Fig. 6 present a visual image of the performance of the proposed architecture, emphasizing the model’s stability and consistency through-out the training process. The stability and consistency of the suggested model’s performance during a training process are reflected in the accuracy and loss plots of both the validation and training data. The absence of significant spikes or dips in the curves indicates that the model was successful in effectively learning the features and patterns of the dataset effectively without encountering overfitting or underfitting issues. Therefore, the accuracy and loss curves demonstrate that the model was well-performed through-out the training process. Therefore, the high accuracy rates and low losses observed in the proposed model, as well as the consistency shown in the accuracy and loss graphics, provide strong evidence of the performance and accuracy of the suggested approach in classifying tomato plant leaf diseases.
The minor fluctuations observed within the learning curve in Fig. 6 should not be misunderstood as indicative of inferior model performance. Instead, they reflect the dynamic nature of the training process. Our model continuously adapts to address the varying complexities and patterns inherent in the dataset. We have meticulously employed rigorous preprocessing methods, adept regularization techniques, and meticulous optimization protocols to avoid challenges stemming from these fluctuations. Within our proposed approach, we openly recognize the intermittent occurrence of fluctuations that can arise within the learning curve and their potential influence on the model’s training trajectory. These fluctuations may arise due to several factors, including the intricate nature of the tomato leaf disease dataset, integrating multi-scale dimensions into the convolutional neural networks, and the inherent diversities in disease manifestation among distinct samples.
Additionally, the model’s capacity to harness multi-branch architectures for multi-feature extraction could lead to varying degrees of responsiveness to distinct disease patterns. By leveraging the ability to capture intricate details across various scales and exploiting multi-branch structures for enhanced feature extraction, the occasional fluctuations in the learning curve may emerge due to the complex interplay among these mechanisms. Despite these fluctuations occasionally causing sudden surges or drops in performance metrics, it is crucial to recognize that the overall trend of the learning curve consistently demonstrates improvements in accuracy and reduction in loss as training epochs progress. This observation provides compelling evidence that our model’s multi-scale and multi-branch attributes effectively comprehend and accommodate the intricate details of the dataset, thereby enhancing the model’s proficiency in disease classification.
To optimize the performance of the models, it is critical to carefully choose the right batch size and the number of training epochs. For example, in an experimental study described in Fig. 7, we tested four different batch sizes (8, 16, 32, and 64) to measure the training time for each epoch and also testing accuracy. The results showed that as the batch size increased, the training time for each epoch decreased when the testing accuracy continued to increase. Figure 7a, b clearly demonstrates this relationship, highlighting the benefits of using larger batch sizes during model training. This finding suggests that increasing the batch size can lead to faster convergence and better testing accuracy, but it is important to carefully balance this against the risk of overfitting. Ultimately, selecting the appropriate batch size and a number of epochs requires careful experimentation and consideration of the specific model architecture and dataset.
During the model training, we found that a batch size of 64 was the most effective, generating the highest testing accuracy while minimizing training time. However, further analysis of testing accuracy at different model training epochs, as shown in Fig. 8, revealed that accuracy gradually increased to 100 epochs. As such, a batch size of 64 and 100 as training epochs could be optimal for this particular model. Therefore, this study provides valuable insights into selecting optimal parameters for training neural networks.
Furthermore, by experimenting with different batch sizes and epochs, we can identify the best values to use for a particular model and dataset, which can remarkably improve the performance of the model. However, it is necessary to remember that the optimal parameters may vary depending on the specific circumstances and generalizing the findings of one study to other models and datasets may not be appropriate. Therefore, it is recommended to perform similar experiments on other architectures with various datasets to identify the best parameters for each case.
4.1.1 Performance evaluation of the developed model
In this research work, we utilized the confusion matrix to gain a clear understanding of the accuracy and potential sources of confusion for our classification model when making predictions. Our confusion matrix consisted of four metrics, which were used to measure the accuracy of classifications and predict the behavior of each pair of predictor and target attributes for a given class value. Using the confusion matrix, we were able to visualize the effectiveness of our DMCNN model, identifying its strengths and weaknesses in detecting and classifying tomato leaf diseases.
In our analysis study, we evaluated the performance of the classifier that distinguishes between ten classes of tomato leaf disease using the four metrics of true negatives (TN), false positives (FP), true positives (TP), and false negatives (FN). TP and TN represented the correct identification of tomato leaf disease, while FP and FN represented incorrect identification. The confusion matrices for the models have been depicted in Fig. 9, providing a clear visualization of the true class values in the sample data and the class values predicted by the CNN classifier. The findings demonstrated that the accuracy of our suggested DMCNN was 99.1%.
Furthermore, the use of the confusion matrix was critical in providing an accurate classification of tomato leaf diseases using our DMCNN model, enabling us to identify options for improvement and optimize the model for better performance in real-world applications.
A confusion matrix is a practical tool for assessing a classification model’s performance. In our study, we employed the confusion matrix to analyze the effectiveness of our proposed DMCNN in classifying ten different types of tomato leaf diseases. According to the confusion matrix, our model had an impressive accuracy of 99.1%. with very few misclassifications. This high level of accuracy indicates that the DMCNN is a robust and reliable tool for identifying tomato leaf diseases. Furthermore, by providing detailed information on the true negative, true positive, false positive, and false negatives, the confusion matrix helped us gain insights into the strengths and weaknesses of our model, allowing us to refine it further and improve its performance. Furthermore, using the confusion matrix was crucial in evaluating the accuracy of our model and understanding how it can be optimized to achieve even better results.
In addition to overall accuracy, we also evaluated the accuracy, recall, F1 score, and precision of our model for each class. These metrics are important because they provide more detailed insights into the model’s performance on individual classes. For example, Precision determines the percentage of true positive predictions between all positive predictions, while also recall controls the ratio of true positive predictions between all actual positive cases. The F1 score is the rhythmic standard actually mean of recall and precision, giving a complete proportion of the model’s performance that balances both metrics. Therefore, the performance evaluation equations in Eqs. (1), (2), (3), and (4) are used to calculate performance metrics and evaluate results:
where TP—True Positive; TN—True Negative; FP—False Positive; FN—False Negative.
By evaluating these metrics for each class, we can identify any specific weaknesses or strengths of the model for particular classes, which can guide further improvements or adjustments to the model. The evaluation metrics for each tomato leaf disease class on the testing data, including classes’ accuracy, precision, recall, and F1-score, are presented in Table 6.
The comparative analysis of the performance of our DMCNN model with pre-trained models was an essential aspect of our study, as it permitted us to evaluate the efficacy as well as the effectiveness of our suggested model. The models we compared our proposed model with were AlexNet, VGG16, InceptionV3, and ResNet50, which are commonly used models for image classification tasks; the architectures used for these models are explained in Table 4.
The analysis results show that our proposed DMCNN outperforms all other models in regard to the accuracy, recall, F1 score, and precision. Our model reported an accuracy of 0.991, indicating that 99.1% of the test dataset’s samples were correctly classified. The precision of our model was 0.991, indicating that the percentage of true positive predictions was 99.1% of all positive predictions. The recall of our model was 0.991, indicating that the percentage of true positive predictions between all actual positive instances was 99.1%. Finally, the F1-score of our model was 0.992, indicating that it performed admirably in terms of precision and recall.
Among the compared models, InceptionV3 and ResNet50 exhibit the lowest accuracy and F1-score. InceptionV3 achieved an accuracy of 0.89, while ResNet50 achieved an accuracy of 0.88. AlexNet and VGG16 perform better than InceptionV3 and ResNet50 but still fall short of our proposed DMCNN’s performance. AlexNet achieved an accuracy of 0.95, while VGG16 achieved an accuracy of 0.91. These results clearly indicate that our proposed DMCNN model outperforms the other models in regard to classification performance. The comparison results, which include classes accuracy, recall, F1-score, and precision, have been presented in Table 7 and Fig. 10.
The comparison of our proposed DMCNN model’s classification performance with that of pre-trained models in our study highlights the efficiency and effectiveness of our system. The results of our analysis indicated that our proposed DMCNN model outperformed all other models that were evaluated. It achieved excellent accuracy, recall, F1-score, and precision which are crucial performance metrics in deep learning.
These findings demonstrate the potential of our proposed DMCNN model for correctly classifying tomato plant leaf diseases, which is a significant benefit to the agriculture industry. With accurate disease classification, farmers can take prompt and effective measures to control and prevent the spread of diseases in their crops.
Moreover, the practicality and efficacy of our DMCNN model can potentially be extended to other areas of agriculture, such as the classification of diseases in other crops. The development of accurate and efficient deep learning models for agriculture can have far-reaching benefits, including improving sustainable farming practices.
In general, our study’s findings highlight the potential of our proposed DMCNN model as a reliable tool for accurately and efficiently classifying tomato leaf diseases and its potential for wider applications in the agriculture industry.
4.1.2 Comparison of various state-of-the-art approaches to our proposed model: an evaluation of performance
The classification of tomato plant leaf diseases is a crucial research field that has recently attracted much attention. With the advent of deep learning-based approaches, there has been a breakthrough in the development of models for accurately classifying tomato plant leaf diseases. In this context, it is crucial to make comparisons of the performance of new model against the existing state-of-the-art approaches to evaluate their efficacy.
To this end, Table 8 and Fig. 11 present a comprehensive comparison of the proposed model with 10 contemporary deep learning-based methods for tomato disease of leaves classification. In order to ensure coherence and validity, we selected the most recent models based on deep learning methods for disease identification and categorization in tomato leaves.
The comparative survey included both transfer learning-based models and the models that were developed from scratch. The proposed model significantly outperforms all other current methods in terms of classification performance. This suggests that the proposed model can accurately classify various types of tomato plant leaves diseases and can provide an effective tool for plant pathologists and farmers to assess the health condition of tomato crops.
Deep learning approaches to disease classification in tomato leaves have shown promising results. The proposed model is a significant step toward achieving accurate and efficient identification and categorization of tomato disease. It is worth noting that suggested model can be further improved by incorporating additional data sources, optimizing hyperparameters, and exploring novel deep learning architectures. Furthermore, the comparative analysis presented in Table 8 highlights the importance of evaluating the performance of new models against existing approaches.
The ability of the proposed model to outperform all other methods in terms of classification performance demonstrates its capability for practical applications in plant pathology.
Analysis of Table 8 unequivocally demonstrates that our model has exhibited superior performance compared to other studies. This notable achievement can be attributed, in part, to the inherent capability of our DMCNN model to extract multi-scale features from input images. This intrinsic capacity empowers our model to effectively encompass both intricate fine-grained details and the broader contextual aspects of the input image. This holistic understanding plays a pivotal role in achieving accurate multi-class plant disease classification. Conversely, the study by Ozbilge et al. [27] surpassed our performance. The success of their approach can be ascribed to a confluence of factors, prominently including their adept utilization of a comprehensive, curated, and preprocessed dataset. Such datasets hold the potential to significantly elevate model performance by embracing a diverse array of disease manifestations and variations. Additionally, the efficacy of their methodology, architectural choices, and meticulous hyperparameter tuning may have synergistically contributed to their favorable outcomes on this specific dataset. Nevertheless, it is noteworthy that diverse approaches may outperform one another based on the distinctive characteristics and complexities inherent to individual datasets.
5 Discussion and analysis
In this paper, we recommended using deep multi-scale convolutional neural networks (DMCNNs) for automatically generated classification of multi-class leaf diseases in tomatoes. We evaluated the efficiency of our proposed DMCNN model using a publicly available dataset of 11,000 tomato leaf images containing 10 different categories of diseases, including healthy leaves.
Our results demonstrated that the suggested DMCNN model performs better than several existing models, such as VGG-16, AlexNet, InceptionV3, and ResNet-50, regarding accuracy, precision, F1-score, and recall. In addition, the overall accuracy of our model was 99.1%, which is a significant improvement over the baseline models.
We also provided a detailed analysis of the confusion matrix for each model, which showed that our DMCNN model had a higher rate of true positives and a rate of false positives than the other models. This indicates that our model is better at correctly identifying diseased leaves and avoiding misclassifying healthy leaves as diseased.
Furthermore, we compared the performance of our DMCNN model with other studies in the literature that has used similar datasets for tomato disease detection. Our model outperformed most of the previous studies, including those that used traditional learning algorithms and those that used deep learning models, such as AlexNet and GoogLeNet. Table 9 presents the overall accuracy comparison of the proposed model and existing models.
Table 9 presents a comparison of the performance of the proposed DMCNN model with existing work methods in an unspecified field. The proposed DMCNN model outperforms almost all the state-of-the-art methods listed in Table 9, and the ST column reports the results of a statistical significance test. The " + " symbol in the ST column indicates that the proposed DMCNN model outperforms the baseline with p < 0.05, which means that the improvement is statistically significant. Overall, the table provides a quick summary of the performance of the proposed DMCNN model and the state-of-the-art methods.
One potential reason for the superior performance of our DMCNN model is its ability to extract multi-scale features from input images. This allows our model to capture both the fine-grained details and the overall context of the input image, which is particularly important for the accurate classification of multi-class plant diseases. Additionally, our model was trained on a large and diverse dataset, which may have contributed to its robustness and generalization performance.
Furthermore, our study demonstrates the effectiveness of DMCNNs for the automated classification of multi-class leaf diseases in tomatoes. Our results suggest that this approach can significantly improve the accuracy, performance, and reliability of disease identification in agriculture, which can significantly impact crop yields and quality. Further research can explore the application of our model to other crops and the development of more advanced models for disease detection in agriculture.
While the study introduces a notably efficient technique for automated multi-class leaf disease classification in tomatoes, it is essential to recognize a significant limitation. The study’s evaluation exclusively centers on the proposed model using a single dataset, potentially offering insights into only a subset of tomato diseases within varying regional and growth contexts. Acknowledging this, there are numerous promising avenues for future enhancements to the DMCNN approach that could elevate its performance and significance.
Therefore, in future improvements for the DMCNN approach, numerous exciting possibilities exist to elevate its performance and impact. Firstly, refining the architecture by delving into the optimization of its components could yield substantial enhancements. Experimenting with network depth, convolutional layer arrangements, kernel sizes, and channel configurations could enhance feature extraction and classification accuracy. This fine-tuning process could unlock the model’s full potential by tailoring its architecture to the nuances of disease-related features in tomato leaves.
Exploring attention mechanisms present an intriguing avenue to further improve the model’s discriminative power. By allowing the DMCNN to focus on the most relevant image regions selectively, attention mechanisms can enhance the model’s ability to capture crucial disease-related patterns, leading to more precise and accurate classification outcomes. Furthermore, adopting ensemble learning strategies that leverage the strengths of multiple DMCNN instances or integrate them with complementary architectures could result in a more robust and reliable classification system. The synergy of these approaches could significantly enhance the model’s capacity to handle diverse disease scenarios effectively.
Another promising direction involves domain-specific innovations that cater to the unique challenges of plant disease classification. Tailoring preprocessing techniques to highlight disease-related features and collaborating with experts in plant pathology could amplify the model’s capability to detect subtle disease symptoms. Integrating multi-modal data, such as hyperspectral or thermal images, alongside visible spectrum images can provide a comprehensive understanding of plant health, ultimately advancing disease classification accuracy. Moreover, focusing on user–friendly interfaces and mobile applications can facilitate the practical implementation of the DMCNN in real-world scenarios, ensuring seamless usability for agricultural stakeholders. As these avenues converge, they chart a compelling path toward harnessing the DMCNN’s potential to revolutionize automated disease diagnosis in agriculture.
6 Conclusion
In this study, we mainly aimed to develop a robust and accurate method for classifying diseases in tomato plants using deep learning techniques. To achieve this goal, we introduced a novel deep multi-scale CNN architecture that integrates multiple channels of information for better disease classification. We evaluated the proposed framework using a public dataset of tomato leaf images, which included multiple types of diseases and various levels of severity. Our results demonstrate that the proposed model provides an accuracy of 99.1%, which indicates its high effectiveness in accurately classifying tomato leaf diseases. Furthermore, we compared the performance of our model with other existing methods and found that our model outperformed them regarding accuracy, recall, precision, and F1 score.
The feature importance analysis of the proposed model revealed that the combination of multi-scale images and deep learning techniques could significantly improve the classification of plant diseases. Furthermore, our study provides valuable insights into developing accurate and robust disease classification systems using multi-scale imaging and deep learning techniques. The proposed model has the potential to make a significant impact on the agricultural industry by improving crop health and productivity.
In future work, we intend to use our proposed model to detect and classify diseases in other crops, as well as to explore the potential of transfer learning techniques to improve the performance of our model. We also aim to study the feasibility of deploying our model in practical applications, such as precision agriculture, where it can detect and prevent disease spread in crops more efficiently and effectively.
Data availability
References
Barkhordari MS, Armaghani DJ, Asteris PG (2022) Structural damage identification using ensemble deep convolutional neural network models. Comput Model Eng Sci 134(2):66. https://doi.org/10.32604/cmes.2022.020840
Mahlein A-K (2016) Plant disease detection by imaging sensors, and specific demands for precision agriculture and plant pheno-typing. Plant Dis 100(2):241–251. https://doi.org/10.1094/PDIS-03-15-0340-FE
Gao L, Xiao Y (2019) Plant disease detection: a review. IEEE Access 7:125552–125566. https://doi.org/10.1109/ACCESS.2019.2937271
Singh P, Tiwari P, Singh PK (2021) Recent advancements in hyperspectral imaging for plant disease detection: a review. Arch Agron Soil Sci 67(3):251–266. https://doi.org/10.1080/03650340.2020.1768294
Mohanty SP, Hughes DP (2016) Using deep learning for image based plant disease detection. Front Plant Sci 7:1419. https://doi.org/10.3389/fpls.2016.01419
Bhandari M, Neupane A, Mallik S, Gaur L, Qin H (2023) Auguring fake face images using dual input convolution neural network. J Imaging 9:3. https://doi.org/10.3390/jimaging9010003
Masood M, Nawaz M, Malik KM, Javed A, Irtaza A, Malik H (2022) Deepfakes generation and detection: state-of-the-art, open challenges, countermeasures, and way forward. Appl Intell 53:3974–4026. https://doi.org/10.1007/s10489-022-03766-z
Liu J, Wang X, Liu G (1894) Tomato pests recognition algorithm based on improved YOLOv4. Front Plant Sci 2022:13
Arco JE, Ortiz A, Ramírez J, Martínez-Murcia FJ, Zhang YD, Górriz JM (2023) Uncertainty-driven ensembles of multi-scale deep architectures for image classification. Inf Fusion 89:53–65. https://doi.org/10.1016/j.inffus.2022.08.010
McAllister E, Novellino A, Payo A, Medina-Lopez E, Dolphin T (2022) Multispectral satellite imagery and machine learning for the extraction of shoreline indicators. Coast Eng 174:104102. https://doi.org/10.3389/fpls.2022.814681
Bhandari M, Chapagain P, Parajuli P, Gaur L (2022) Evaluating performance of adam optimization by proposing energy index. In: Santosh K, Hegadi R, Pal U, (eds) Recent trends in images processing, and pattern recognition: proceedings of the fourth international conference, RTIP2R 2021, Msida, Malta, 8–10 December 2021. Springer, Cham, pp 156–168. https://doi.org/10.1007/978-3-031-07005-1_15
Alsaiari AO, Alhumade H, Abulkhair H, Moustafa EB, Elsheikh A (2023) A coupled artificial neural network with artificial rabbits optimizer for predicting water productivity of different designs of solar stills. Adv Eng Softw 175:103315. https://doi.org/10.1016/j.advengsoft.2022.103315
Shahi TB, Sitaula C (2021) Natural language processing for Nepali text: a review. Artif Intell Rev 55:3401–3429. https://doi.org/10.1007/s10462-021-10093-1
Liu P, Yuan W, Fu J, Jiang Z, Hayashi H, Neubig G (2023) Pre-train, prompt, and predict: a systematic survey of prompting methods in natural language processing. ACM Comput Surv 55:1–35. https://doi.org/10.48550/arXiv.2107.13586
Mohanty SP, Salathé M, Hughes DP (2016) Using deep learning for image based plant disease detection. Front Plant Sci 7:1419. https://doi.org/10.3389/fpls.2016.01419
Sladojevic S, Anderla A, Culibrk D, Arsenovic M, Stefanovic D, Crnojevic V (2016) Deep neural networks-based recognition of plant diseases by leaf image classification. In: Computational intelligence and neuroscience, 2016, p 3289801. https://doi.org/10.1155/2016/3289801
Ferentinos KP (2018) Deep learning models for plants disease detection and diagnosis. Comput Electron Agric 145:311–318. https://doi.org/10.1016/j.compag.2018.01.009
Zahari ML (2020) Deep learning for image-based plant disease detection. https://ir.uitm.edu.my/id/eprint/44324/
Agarwal M, Arjaria S, Sinha A, Singh A, Gupta S (2020) ToLeD—Tomato leaf diseases detection using convolution neural network. Procedia Comput Sci 167:293–301. https://doi.org/10.1016/j.procs.2020.03.225
Gadekallu TR, Reddy MPK, Lakshmanna K, Rajput DS, Bhattacharya S, Jolfaei A, Singh S, Alazab M (2021) A novel 729 PCA whale optimizationbased deep neural networks model for classification of tomato plant diseases using GPU. J Real-Time Image Process 18:1383–1396. https://doi.org/10.1007/s11554-020-00987-8
Intan NY, Naufal AA, Akik H (2023) Mobile application for tomato plant leaf disease detection using a dense convolutional network architecture. Computation 11(2):20. https://doi.org/10.3390/computation11020020
Agarwal M, Gupta SK, Biswas K (2020) Development of efficient CNN model for tomato crop diseases identification. Sustain Comput Inform Syst 28:100407. https://doi.org/10.1016/j.suscom.2020.100407
Wang Y, Zhang H, Liu Q, Zhang Y (2019) Image classification of tomato leaf diseases based on transfer learning. J China Agric Univ 24:124–130
Kaur M, Bhatia R (2019) Development of an improved tomato leaf diseases detection and classification method. In: Proceedings of the IEEE conference on information, and communication technology, Baghdad, Iraq, 15–16 April 2019, pp 1–5. https://doi.org/10.1109/CICT48419.2019.9066230
Kaushik M, Ajay R, Prakash P, Veni S (2020) Tomato leaf-disease detection using convolutional neural networks with data augmentation. In: Proceedings of the 2020 5th international conference on communication and electronics systems (ICCES), Coimbatore, India, 10–12 June 2020, pp 1125–1132. https://doi.org/10.1109/ICCES48766.2020.9138030
Trivedi NK, Anand A, Aljahdali HM, Gautam V, Villar SG, Goyal N, Anand D, Kadry S (2021) Early detection and classification of tomato leaf diseases using high performance deep neural network. Sensors 21:7987. https://doi.org/10.3390/s21237987
Vijay N (2021) Detection of plant diseases in tomato leaves: with focus on providing explainability and evaluating user trust. Master’s Thesis, University of Skövde, Skövde, Sweden. https://www.diva-portal.org/smash/record.jsf?pid=diva2%3A1593851&dswid=4788
Ozbılge E, Ulukok MK, Toygar O, Ozbılge E (2022) Tomato disease recognition using a compact convolutional neural network. IEEE Access 10:77213–77224. https://doi.org/10.1109/ACCESS.2022.3192428
Karthik R, Hariharan M, Anand S, Mathikshara P, Johnson A, Menaka R (2020) Attention embedded residual CNN for disease detection in tomato leaves. Appl Soft Comput 86:105933. https://doi.org/10.1016/j.asoc.2019.105933
Guo X, Fan T, Shu X (2019) Tomato leaf diseases recognition based on improved multi-scale AlexNet. Trans Chin Soc Agric Eng 35:162–169. https://doi.org/10.11975/j.issn.1002-6819.2019.13.018
Funding
This research received no external funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Elfatimi, E., Eryiğit, R. & Elfatimi, L. Deep multi-scale convolutional neural networks for automated classification of multi-class leaf diseases in tomatoes. Neural Comput & Applic 36, 803–822 (2024). https://doi.org/10.1007/s00521-023-09062-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-023-09062-2