Abstract
BackgroundAlzheimer’s disease (AD) is a degenerated condition of the brain where memory loss is fully depleted for elderly individual. Efficient machine learning methods are accessible, producing low classification accuracy since single modality features are being evaluated. In this paper, the multimodal approach is developed and execution of comprehensive validation for structural atrophy through Magnetic Resonance Imaging decreases metabolism through Fluorodeoxyglucose Positron Emission Tomography (FDG-PET), and accumulation of amyloid plaques through Pittsburgh compound B (PiB-PET), as well as cognitive assessment for identifying the early onset of AD. It has been stated that additional information from multiple image modalities would ameliorate the classification accuracy while diagnosing early AD. The novel classifier, Adaptive Hyperparameter Tuning Random Forest Ensemble Classifier (HPT-RFE), is proposed for three binary classifications. In this classifier, the tunning of hyperparameters is automated for computing the best features while constructing the optimum size of Random Forest. The advantage of using the classifier is computationally much faster when compared with Support Vector Machine, Naïve Bayes, K-Nearest Neighbour and Artificial Neural Network. Simulation results show that the performance of the Adaptive HPT-RFE classifier has been regarded as best among all binary classifications in the ADNI dataset. For AD versus Normal Control (NC) binary classification, 100% accuracy, sensitivity, and specificity have been achieved, whereas the accuracy of 91% and 100% specificity for NC versus Mild Cognitive Impairment (MCI) classification and 95% accuracy, 100% specificity, 80% sensitivity for AD versus MCI classification are compared with four state-of-the-art techniques.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
One of the global challenges in the health care industry is AD, affecting more than 5 million American people [1]. Due to adverse complications in AD, it has been predicted that more than 7 million people died aging more than 65 years [2]. The cases of AD patients would be double every five years after attaining the age of 65 years. In addition to that, AD is almost one-third of the total population aging more than 85 years [3]. For memory disability and other cognitive problems, the characterization of AD is exhibited by neurofibrillary entanglement [4, 5]. For an association in the memory losses in AD patients, molecular mechanisms are still unknown. Some healthy individuals do not have memory loss but possessing plaques and deposits [6]. Risk factors such as obesity, age, diabetics, and inflammation increment in the brain occurred in AD [7]. APOE is the most vital supplementary risk factor among CR1, FERMT2, and COMT genotypes [8]. According to the National Institute of Aging and the Alzheimer’s Disease Association, preclinical AD, MCI, and dementia are the three stages of AD [9]. Encephalopathy or cognitive impairment occurs at the first stage (preclinical) AD. The primary cause of AD is dementia which encompasses mild, moderate, and severe phases. Amyloid positivity, Asymptomatic cerebral amyloidosis [10], synaptic dysfunction are the divisions of Preclinical AD. Declining of cognitive functions and neurodegeneration are shreds of evidence found in all three divisions. Clinical diagnosis, the background of the disease, psychological tests with additional information are some of the methods in diagnosing AD. To assist AD, non-protruding eye tests are being conducted. In addition to that, an association of beta-amyloid protein present in the eye has significant roles in the levels of the brain [11]. Neuroimaging method poses a crucial part of evaluating suspected AD patients. The two other modalities for diagnosing AD are PET and MRI. Brain’s structural and functional information is acquired through MRI using the detailed characterization of tissues with a difference in soft tissues. This technique can easily differentiate white matter and grey matter where brain tissues are displayed. Molecular and metabolic information of the brain is obtained by PET imaging which is not restrained to Aβ and glucose. High sensitivity is achieved for the distribution of tangles and plaque lesions in AD patients. As a result, this modality helps to diagnose normal and different stages of AD qualitatively and quantitively [12]. Hence, combined imaging techniques of MRI and PET would facilitate diagnosing AD other than any other technique. In the last few decades, the application of hybrid imaging models is gaining importance in the clinical fields. Some of the models include PET/Computed Tomography (CT), single-photon emission computed tomography (SPECT), and fluorescence molecular tomography (FMT). In radiation areas, the PET/MR hybrid imaging model is applied to AD patients for better performance [13]. For correction in attenuation of soft tissues, Dixon MR produces satisfactory results and is applicable in FDG uptakes to identify lesions in brain tissues. Hence, PET/MR imaging model finds its potential application in the areas such as classification, different stages of the disease, diagnostic evaluation, and comprehension of pathomechanisms [14]. In addition to that, a single imaging session produces all imaging and biomarkers information that assists patients and referred physicians [12].
Previous studies show that there has been a remarkable increase in retention of PiB Biomarker, which is observed as elevated plaques levels comparing with HC subjects. PiB patterns produce retention in parietal and frontal cortices of brain regions, which is entirely different for AD patients [15]. In cortical areas of AD patients, PiB retention has been considered most prominent, whereas lowest in areas of white region processing the studies related to a post-mortem of Aβ plaques [16]. In AD’s frontal, temporal, parietal cortex of the brain, retention of PiB has been observed. Significantly affected areas in the brain’s occipital and lateral temporal cortex have prudently been observed in mesial temporal regions. In the previous studies relating extensive Aβ deposition, retention of striatal PiB has been significantly observed in AD striatum patients [17]. Hence, the description of Aβ deposition is mentioned and confirmed by using PiB in AD and CN subjects [18]. The main highlights of the paper are as follows:
-
1.
In this study, multimodal approach is developed for comprehensive validation in structural atrophy through MRI, decrease metabolism through FDG-PET and accumulation of amyloid plaques through PiB-PET as well as cognitive assessment for studying early patterns for diagnosing AD.
-
2.
FreeSurfer 6.0.1 has been applied in MRI, FDG-PET and PiB-PET images for extracting different features, such as morphometric features, Region of Interest (ROI), Surface features, Standard Uptake Values (SUV) and cognitive assessment in High Performance Computing (HPC).
-
3.
Adaptive HPT-RFE classifier is based on one versus one classification which leads to three binary classification, namely AD versus NC, MCI versus NC and AD versus MCI and compared with SVM, NB, KNN, and ANN classifiers which makes model a robust and stable one.
-
4.
The performance of Adaptive HPT-RFE classifier has been applied on 102 subjects which has been taken from Alzheimer’s disease Neuroimaging Initiative (ADNI) database and compared with other four state-of-the-art techniques in each binary classification tasks where simulation results indicate that model has a good potential for generalisation.
The paper’s organization is as follows: Sect. 2 describes the related works. Sect. 3 describes methods. Section 4 illustrates Image Processing. Classification methods are discussed in Sect. 5. Simulation results and discussions are presented in Sect. 6. Finally, the conclusion of the proposed study is explained in Sect. 7.
2 Related works
There has been no concrete treatment for AD where modern therapies are modified to degrade the progression of the disease. The main criteria of conducting trials are to measure cognitive functions by applying ADAS-cog [19] and CDR Scale [20]. It has been observed that clinical symptoms of patients vary according to the progression of AD. This disease does not closely correlate with beta-amyloid (Aβ) proteins and tau deposition with losses [21]. Applying ADAS-Cog psychometric tests [22] and brain cognitive reserve capacity [23], modification of clinical symptoms is well presented. Therefore, biomarkers are used for assessing the effectiveness of the disease in convenient histopathological features of AD [24]. Biomarkers, such as 11C- PiB, are applied in cerebrospinal fluid (CSF) for predicting accumulation of Aβ1–42 [21]. CSF p-tau and tau PET are the two biomarkers used in tau pathology. Biomarkers, such as PET, take uptake values for specific regions in FDG, MRI for neuronal loss atrophy, and CSF in elevated t-tau protein [21]. Most biomarkers are investigated for clinical symptoms by calculating the following parameters: specificity, validity, transition, and correlation with other biomarkers [25]. For the same patient, rare studies are conducted at many time intervals for more than two biomarkers, and all biomarkers are not readily available for all subjects.
Amyloid or tau PET are popular imaging methods for treating patients having dementia about complaints in cognitive functions induced by changes in neurodegenerative. Interdependence between aspects of pathology and physiology are investigated. Different studies are conducted, which shows that experimental analysis of imaging methods is becoming potential for evaluating pathophysiologic changes. For observing the dynamic changes in the brain, conventional imaging methods such as CT and MRI are applied for cognitive impairment. In the neuropsychiatric field, PET/MRI hybrid brain imaging system has been developed, which is beneficent to assess the changes in the cerebral pathophysiological [26]. Hence, this hybrid brain imaging system helps diagnose cerebrovascular diseases and AD [27]. Several research works illustrate the relationships between functional or morphological MRI information and change inflow of blood or metabolism. But limited studies are conducted on integrated analysis of functional MRI and amyloid/tau imaging.
3 Methods
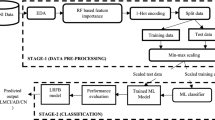
In the proposed framework, there are two processing steps: (1) Pre-processing of images: From the grey matter segmentation of the brain, different sizes of patches are obtained where each feature is extracted from MRI, FDG-PET, and PiB-PET images (2) Classification: Train an HPT-RFE classifier to learn the patterns and discriminate AD, MCI, and NC individuals and applied for three binary classifications.
3.1 Materials
3.1.1 Preparation of dataset
In 2003, the ADNI dataset [28] was created by M.W. Weiner as a public–private partnership for researchers worldwide. The critical ingredient of ADNI is to measure the progression of MCI to early AD by biomarkers such as MRI, PET, neuropsychological and clinical assessment.
In validating our proposed method, T1WI MRI scans, PiB PET, FDG PET images, and cognitive assessments of 102 ADNI subjects have been used in the study. These subjects have been divided into three cohorts at the time of preparing the manuscript. (1) NC: 11 Male and 8 Female subjects are diagnosed for all points (2) MCI: 44 Male and 21 Female subjects are evaluated for more than two years. (3) AD: 18 subjects are considered with a clinical diagnosis. Here, cognitive assessments of all subjects such as MMSE, GDSCALE, Global CDR, and FAQ scores are displayed in Table 1. In the second row, the numbers in the bracket mention the total number of male and female subjects. In contrast, the rest five rows indicate the two numbers, i.e., minimum and maximum values of parameters used in cognitive assessment. In addition to that, over 4100 MRI images, more than 500 FDG-PET images, and 223 PiB-PET images are applied in categorizing into three cohorts. Procedures of image processing protocols, post-acquisition pre-processing, and comprehensive ADNI cohort are described [29].
3.1.2 MRI protocol
All T1 Weighted 3D MRI (T1WI) Protocol are acquired from SAGITTAL Acquisition Plane, 3D Acquisition Type, 8HRBRAIN Coil, 1.5 Tesla Magnetic Strength, 8˚ Flip Angle, GE MEDICAL SYSTEMS Manufacturer, 256 pixels of Matrix X and Matrix Y, 166 Pixels of Matrix Z, SIGNA HDx Manufacturer Model, 0.9 mm of Pixel Spacing X and Y, RM Pulse Sequence, 1.2 mm of Slice thickness, 3.8 TE ms, 1000 TI ms, 8.6 TR ms [30].
3.1.3 FDG-PET protocol
All glucose metabolism 3D FDG-PET are acquired from 0.096000 cm−1 Attenuation, Rectangle: 4.300000 mm Ax, rectangle 6.500000 mm Rad Convolution Kernel, EMISSION Counts Source, START Decay Correction, 6 Frames, GE MEDICAL SYSTEMS Manufacturer, Discovery RX Mfg Model, 128.0 pixels of number of rows and columns, 47 Slices, 2 mm of X and Y Pixel Spacing, F-18 Radioisotope, 18F-FDG Radiopharmaceutical, RTSUB Randoms Correction, 3D Kinahan–Rogers Reconstruction, Model-Based Scatter Correction, 3.3 mm Thickness [31].
3.1.4 PiB-PET protocol
All amyloid 3D 11C PiB-PET are acquired from ramp Convolution Kernel, and Dynamic EMISSION Counts Source, START Decay Correction, 27 frames, Siemens/ CTI Manufacturer, HR + Manufacturer Model, 128.0 pixels of the number of rows and columns, 63 Slices, 2.1 mm of X and Y Pixel Spacing, C-11 Radioisotope, 11C PiB PET Radiopharmaceutical, CPU iterative Reconstruction, Simulated 3D Scatter Correction, 2.4 mm Thickness [32].
3.1.5 FreeSurfer
Nowadays, the primarily used software in VolBM is FreeSurfer (surfer.nmr.mgh.harvard.edu), where complicated image processing operations are implemented [33]. Freesurfer calculates the corresponding volumes and a massive number of anatomical structures from the segmentation of incoming scans. Here, FreeSurfer version 6.0.1 has been applied in the hippocampus, total GM, temporal GM, and ventricular volume output as an imaging biomarker for the AD brain. Computational complexity is a significant hindrance for FreeSurfer as compared to HPC for limited application in clinical routines. Time taken for the FreeSurfer pipeline for an up-to-date Single processor PC takes 6 to 24 h to complete each scan. A clustering network is used in HPC, composed of 2 Master and Computer Node, 1 GPU Computer Node, and 1 Cloud Node as explained in Fig. 2.
3.1.6 HPC
The HPC configurations [34] are as follows: 2 numbers of Master node and Computer Node of E5-2630 v3 Intel Xeon 2.4 GHz processors with 8-core, Hard Disk Capacity of 500 GB; 1 GPU Compute Node of 2 numbers of Nvidia K20 GPU and 64 GB memory; 1 Cloud node of E5-2620 V3@2.4, 6 core processors E5-2620 and 64 GB Memory, Hard Disk Drive of 1 TB. The overview of cluster network is shown in Fig. 1.
4 Image processing
Due to parameters, such as similar pose, scale, and less heterogeneity along with FDG-PET, PiB-PET, and T1WI MRI images, a small database of images has been acquired from the ADNI dataset. By applying machine learning techniques, high classification accuracy has been achieved. Hence, Voxel-Based Morphometry (VBM), Region of Interest (ROI), Positron Emission Tomography Partial Volume Correction (PET PVC), FDG and PiB features and Standard Uptake Values (SUV) are shown in Fig. 2.
4.1 VBM
It is a neuroimaging analysis method for investigating differences in the brain’s focal anatomy by applying statistical tools. To identify the difference in the brain structure and perform quantitative measurements, morphometry analysis has to become a vital tool for research. The main advantage of using the VBM method is to produce an unbiased score that can represent differences created by the brain’s anatomical structure [35]. For VBM, 3D volumetric T1WI MRI images are provided. Using various statistical tools, VBM tests all the voxel-based images in the brain. For instance, t tests are performed on each image’s voxel for recognizing the pattern differences between two subject groups. In addition to that, measurements of MRI in brain atrophy marked the tracking of AD’s progression. A large number of literature reviews related to VBM are studied [36] VBM is applied for studying the volumetric atrophy of grey matter located in the brain’s neocortex. The criteria for selecting brain structures are to classify volumes during AD’s early or advanced stage [37]. In total, 22 volume features are extracted from the 3D T1WI Global MRI measure of Volume (called subject/stats/ aseg. stats). Four best features are selected by normalization of brain features and ventricular volumes. Figure 3 shows the volume and intensity structure-wise FS segmentation. Also, characteristics of Volume are mentioned in Table 3.
4.2 Region of interest (ROI)
3D grey level volume for various brain structures is extracted from 102 subjects [38, 39]. 1 mm3 resolution is required in high-quality T1WI MRI images for pre-processing using FreeSurfer Software. By entering the command “recon-all–I file-name. nii–all”, FreeSurfer Software is executed without the intervention of anyone. This software is used as a process mode for pipelining the preprogramed processing of the patient’s data. By applying the default parameters, a total of 110 features of Cortical and subcortical volume (Fig. 3) are computed. During the process, Bilateral ROIs are obtained. Between WM, cortical WM, and pial surfaces, FreeSurfer develops a model in the cortical surface stream (Fig. 3). An array of anatomical features such as CTH, curvature, folding, surface normal and area calculations are computed for every cortex part. Desikan-Killiany atlas is adopted for our study, which is applied to 68 labelled regions of the cortex area. In segmenting the automated subcortical regions of the brain, 40 labels are assigned for representing 40 subcortical regions. For each subject, approximately 12 h are required for completing the segmentation of subcortical volume. Table 2 lists the features extracted from T1WI MRI images.
4.3 PET partial volume correction (PVC)
A comprehensive automatic pipelining PET data analysis is executed on the brain’s cortical surface in this study. For PVC, PET Surfer software provides a required number of tools. Applying the spmregister tool from FreeSurfer 6.0.1, the registration of PET images to T1 images is the primary step of executing the pipeline. The intensities of PET images are normalized for comparing inter subjects. The MNI space pick atlas generated values of SUVs by dividing PET images considering mean uptake values from the reference region [40]. Here, the reference region is taken to be Pons for FDG-PET images [41]. By applying SPM 12, MNI space registration is performed. At first, estimating the transformation to MNI template from T1 and applying inverse deformation in the reference region. 6-mm sphere is eroded for masking of pons so that voxels are considered only within the pons while computing mean uptake. For restricting the outside cortex’s spill-out activity, PVC is performed. In calculating the better computational efficiency, the iterative Yang algorithm [42] has been applied to PET-PVC [43]. In this algorithm, the VOI method is used in which uniform activity within the region is assumed, and parcellated T1 image is obtained. From Freesurfer, the gtmseg tool has been applied to parcellated the T1 image of the subject into 112 areas. Suggestions made by the developer of PET-PVC, the fusion of certain regions, are fused to decrease the number to 50. Applying the mris_preproc tool, the standard template is constructed for each subjects’ cortical surface, where spherical registration is performed [44]. SUVR values are measured, and images are normalized using a template and atlas.
SUVR is defined as the ratio of the concentration of radioactivity CPET(T) (kBq/mL) to injection dose administered (MBq) at the same time by the weight of the body (Kg), which is given by formula
The Pipeline process of PET-PVC is shown in Fig. 4.
4.4 Feature selection
In this paper, 109 ROIs have been extracted from FDG-PET, PiB-PET, and MRI images to apply automatic feature extraction. Here, some features do not have any correlation with the output. These features take considerable computational time and space, making the performance degrades. The problems caused by extra features are referred to as the “curse of dimensionality”. Hence, overfitting occurs as a challenging task since the computation of features takes longer. By selecting the right parts, the dimensionality of features can be reduced. As a result, the classification process is faster, which decreases the computational time in training and testing datasets, thereby facilitating classification accuracy. Firstly, the normalization of features is executed by Standard Scalar Function from 0.23.1 version of the Scikit-learn library [45]. After that, this dataset is transformed in such a way that data redundancy and dependency are reduced. Next, random tree embedding [46] is employed for high-dimensional data acquired from the Scikit-learn library (0.23.1) [45]. By using SelectKBest features from the Scikit learn library, we have selected the best features among them.
5 Classification methods
In this work, five different popular classifiers are used for evaluating performance based on single as well as combined features.
5.1 Adaptive HPT-RFE classifier
Random Forest is regarded as an ensemble machine learning algorithm where many decision trees are agglomerated for classification and regression tasks [47]. To overcome overfitting problems, different subsets of data are trained individual and then averaged value is taken.
In this work, Adaptive HPT-RFE classifier has been proposed and applied to the three binary classifications. The parameters adjusted in this method are n_estimators and max_features. The former parameter is defined as the number of trees. The time of computation is more for complicated trees. Beyond the critical trees, the results obtained are not significantly changing. The latter parameter is defined as the generation of random subsets when every node is being splatted. The default values of max_features = ‘Sqrt’ for the problems regarding classification. For achieving good results, max_depth = None along with min_samples_split = 2. Moreover, another parameter, bootstrap = True, is set as default, and bootstrap = False means extra trees are added. By setting oob_score = True, the generalization accuracy can be achieved by applying bootstrap sampling. For deciding the number of nodes in a particular branch of a decision tree, Gini Index plays important role for performance in Random forests. To determine the value of Gini for a particular branch, a formula is framed which uses each class and probability that is most likely to occur.
here in this equation, N represents the number of classes in the dataset, \(p_{j}\) indicates the relative frequency of a particular class in the dataset. Importance of nodes is calculated for two child nodes which is given in equation.
here Nk = node importance at k, wsk = weighted samples at node k, Ik = value of impurity at node k, L(k) = left split at node k, R(k) = right split at node k.
The calculation of each feature in random forest is given by:
here FI = Feature Importance, NK = Node Importance at k.
Then, features are normalised to value 0 and 1 by dividing all the features importance are
The importance of all final features, at a level of Random Forest with Hyperparameter Tunning, is the sum total of average of decision trees. The resultant feature importance is given by the ratio of the sum of all feature importance to the total number of trees (T).

5.2 SVM
For separating the two data groups, SVM is commonly used as a supervised machine learning algorithm for multivariate classification [48]. A hyperplane is required for classifying the data into various classes. Mathematically, it can be represented as
where W represents as weight vectors, namely W = {P1, P2, P3,…, PN}; N indicates the number of features and b as the bias. As the bias term is influenced by the hyperplane, additional weight P0 has been taken into consideration. Therefore, the above equation can be written as
If any point lies above the hyperplane, the equation is rewritten as
If any point lies below the hyperplane, the equation is rewritten as
For the above two equations, it can be understood that the “sides” of the SVM margin are written as
Combining the two inequalities of Eq. (11) and (12), we get
5.3 NBC
NBC [49] is a vital integrant in machine learning algorithm while considering calculation of the different set of probabilities with frequency counts. An assumption is made where all variables are mutually independent in classification problem [9]. The main idea behind NBC is on the basis of theorem of Bayes and total probabilities which is given by (11).
where P(hp1|xi) is posterior probability for hypothesis hp1. Prior probability for hypothesis hp1 and hp2 is given by \(P\left( {hp1} \right)\) and \(P\left( {hp2} \right)\), whereas \(P \left( {xi{|}hp1} \right)\) and \(P \left( {xi{|}hp2} \right) \) represent the likelihood of hp1 and hp2 hypothesis.
5.4 KNN
KNN is one of the nonparametric algorithms for machine learning. Here, instance data of training are stored by this classifier as Generalised Internal model is difficult to construct. At every point, classification values are computed by using majority vote. For every data class, assigned query point represents the most valued within k-NN [50]. A linear transformation matrix of size n_features × n_components maximizes the sum over the samples s with a probability \(P_{S}\) that are correctly classified as given in Eq. (15).
In the learned embedded space, probability \(P_{S}\) is given as
where \(P_{sk}\) is defined as the softmax over Euclidean Distance and Ds are the set of points that belong to same class for each sample.
5.5 ANN
ANN is a computational scheme which as a representation of biological neural networks existed in human brains which is expressed through connected nodes [51]. In the architecture of ANN, nonlinear activation function is used in a feedforward structure. Biases are added in each layer for finding outputs in the layer (\(Z_{k}\)) using mathematical expressions.

6 Results and discussion
6.1 Experimental evaluation
For addressing the K-best features, i.e., one versus one binary classification tasks such as AD versus NC, MCI versus NC, AD versus MCI, five classifiers have been applied on sagittal, axial and coronal orientations of T1WI MRI images. Here, 102 subjects are acquired from the ADNI database which has been considered in our study. The samples are randomized so that 70% of the samples are trained, and 30% are used for testing. Random forest tree is generated by calculating Gini Index and tunning hyperparameters such as Random state and number of estimators trained by five Classifiers. Hence, Accuracy, Precision, Sensitivity, and Specificity are calculated from the Confusion Matrix of testing samples. By using the majority voting of the classifier, classification labels are identified. The detailed procedure of experimentation is given in Fig. 5.
6.2 Performance evaluation metrics
For evaluating performance metrics, diagonal elements of the Confusion matrix represent rightly predicted by the classifier. These elements are further divided into True Positive (TP) and True Negative (TN) correctly labelled. False Negative (FN) and False Positive (FP) are non-diagonal elements for incorrectly labelled classes. Accuracy (ACC), Sensitivity (SEN), Specificity (SPEC), F1 Score (F1SC), and Precision (PREC) are defined as follows [52]:
Features are extracted from different modalities and converted into a vector consisting of a single feature where classifiers are trained on a particular feature vector. Functions are called for importing libraries from Sckit Learn 0.23.1 in tunning the hyperparameters executed on Python IDE 3.6 environment. Hence, each optimized hyperparameter is trained in five different classifiers for the training dataset and then evaluating the model’s performance using the test dataset. The given experiments are run on the system having the configuration of OS Windows 10, Intel® Core™ i7-9750U CPU @ 2.60 GHz processor, six-core, 12 Logical Processors, and 16 GB RAM with Ubunutu 20.04.2. Summarisation of classification report of three binary classification is shown in Table 3.
For evaluating various diagnostic tests in biomedical research for testing the performance in classification problems and different prediction models, Accuracy of ROC Curve (ROC-AUC) is used as a fundamental graph. Therefore, the ROC-AUC plot has True positive rate (TPR) and False Positive Rate (FPR) parameters which are performance measure in positive and negative part of the dataset. ROC curves have been drawn for each classifier model for three binary classifications such as AD versus NC, AD versus MCI and MCI versus NC which is given in Fig. 6.
6.3 Comparison with other state-of-the-art techniques
Detecting AD right its onset has been a significant problem in the process of AD diagnosis. Most of the past work has achieved sufficient accuracy in classifying whether a subject has AD or normal [51]. However, the binary classification of AD versus MCI and MCI versus NC is still an open problem for the community of researchers. The result obtained from the proposed work for three binary classifications of AD versus MCI, MCI versus NC, and NC versus AD is reported in Tables 4, 5 and 6. The modalities of the proposed work given in Tables 4, 5 and 6 only mention the image-based biomarkers however the work also includes the quantitative values of cognitive assessments. It is instinctive that differentiating between NC and AD should be more accessible, which is also evident in our results as the binary classification of AD versus NC has been achieved 100% accuracy. The accuracy of the other two binary classifications: AD versus MCI and NC versus MCI, has achieved 95% and 91%, respectively. Compared to the classification of AD versus NC, this reduction inaccuracy is because the features obtained after segmentation of cortical and subcortical regions and the SUVR are not as distinct for MCI and AD subjects.
The results obtained also justify the use of the quadruple biomarkers. The accuracy, specificity and sensitivity values generated by the proposed work have been compared with the state-of-the-art techniques and given in Tables 4, 5 and 6. Table 4 shows a comparison with previous researches which have used a single biomarker, Table 5 shows a comparison of previous works using a combination of two biomarkers and Table 6 shows a comparison with techniques using triple biomarkers. Most of the works mentioned in Tables 4, 5 and 6 have worked on ADNI or a dataset that is similar to the ADNI dataset. It can be seen that the accuracies reported by previous works which have used either single, double, and triple biomarkers are less compared to the proposed work. Therefore, it can infer that the quadruple biomarkers are more efficient in early diagnosing of AD. Another observation from the obtained result is that the system’s sensitivity is lesser than the specificity for all three binary classifications. This would mean that the system can prevent a more significant number of misdiagnoses which is also seen in most previous works. The author would analyse the effect of considering multiple biomarkers on specificity and improve it in future work.
7 Conclusion and future work
In this study, Adaptive HPT-RFE Classifier has been proposed for one versus one three binary classifications using single modality features (MRI, FDG-PET, PiB-PET) or multi-modality features (MRI and FDG-PET, FDG-PET and PiB-PET, MRI, FDG-PET and PiB-PET). Here, FreeSurfer Software is used in HPC to extract features of MRI, FDG-PET, PiB-PET Images from 102 subjects. Best classification accuracy, sensitivity, and specificity results are produced while comparing four different classifiers for each binary classification problem. Also, this technique performs best for comparison with other state-of-the-art methods. Therefore, these results indicate that it may be beneficial as a potential tool for diagnosing the different Alzheimer’s disease classes.
Meanwhile, there are other learning techniques such as deep learning algorithms such as hybrid architectures of Convolutional Neural Networks (CNNs) [70], Long Short-Term Memory (LSTM), Deep Autoencoders (DAE), and more variants of Artificial Neural Networks (ANN) could be used in future works for getting much-improved classification accuracy for all various classes of AD. Moreover, an amalgamation of feature selection techniques and deep learning models would be applied on single modality and multi modalities, which can be addressed as a future problem mainly focussing on the interpretability of clinical diagnosis. Also, researchers are primarily concentrating on the methods based on feature selection but not in the classification of AD from NC.
In the future, the colossal size of the dataset can be applied for classification purposes. In addition to that, other machine learning tools can be used. Hence, these models can also be learned to diagnose other diseases such as Parkinson’s disease, Brain Cancer, Eplisey, asthma, and blood cancer. Based on this idea, different ensemble models can be generated for increasing the performance in the classification of other datasets, such as text mining, text classification, which is written in various languages and formats.
References
Thies W, Bleiler L (2013) 2013 Alzheimer’s disease facts and figures. Alzheimers Dement 9(2):208–245
2016 Alzheimer's disease facts and figures. (2016). Alzheimer's Dementia, 12(4), 459–509
Alzheimer's Disease. (2010). New England J Med 362(19), 1844–1845
Jack C, Knopman D, Jagust W, Shaw L, Aisen P, Weiner M et al (2010) Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol 9(1):119–128
Li X, Li T, Andreasen N, Wiberg M, Westman E, Wahlund L (2013) The association between biomarkers in cerebrospinal fluid and structural changes in the brain in patients with Alzheimer’s Disease. J Intern Med 275(4):418–427
Perez-Nievas B, Stein T, Tai H, Dols-Icardo O, Scotton T, Barroeta-Espar I et al (2013) Dissecting phenotypic traits linked to human resilience to Alzheimer’s pathology. Brain 136(8):2510–2526
Birch A, Katsouri L, Sastre M (2014) Modulation of inflammation in transgenic models of Alzheimer’s disease. J Neuroinflamm. https://doi.org/10.1186/1742-2094-11-25
Karch C, Goate A (2015) Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiat 77(1):43–51
Brayne C (2014) A population perspective on the IWG-2 research diagnostic criteria for Alzheimer’s disease. Lancet Neurol 13(6):532–534
Park S, Kim J, Kim H, Kim T, Kim Y, Lee D et al (2013) Preliminary study for a multicenter study of Alzheimer’s disease cerebrospinal fluid biomarkers. Dementia Neurocognit Disorder 12(1):1
Bandelow S, Clifford A, Wardt V, Hogervorst E, Madden M, Lindesay J, Gale A (2011) P1–139: Accurate non-invasive diagnoses of Alzheimer’s Disease using eye scanning. Alzheimer’s Dementia. https://doi.org/10.1016/j.jalz.2011.05.419
Barthel H, Schroeter M, Hoffmann K, Sabri O (2015) PET/MR in dementia and other neurodegenerative diseases. Semin Nucl Med 45(3):224–233
Kumari R, Pushkar S (2020) Analysis of biomedical image for Alzheimers disease detection. In: Examining Fractal Image Processing and Analysis Advances in Computational Intelligence and Robotics, 224–251
Jadvar H, Colletti P (2014) Competitive advantage of PET/MRI. Eur J Radiol 83(1):84–94
Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, Långström B (2004) Imaging brain amyloid in Alzheimers disease with Pittsburgh compound-B. Ann Neurol 55(3):306–319
Thal DR, Rüb U, Orantes M, Braak H (2002) Phases of Aβ-deposition in the human brain and its relevance for the development of AD. Neurology 58(12):1791–1800
Braak H, Braak E (1990) Alzheimerʼs disease. J Neuropathol Exp Neurol 49(3):215–224
Edison P, Archer HA, Hinz R, Hammers A, Pavese N, Tai YF, Brooks DJ (2006) Amyloid, hypometabolism, and cognition in Alzheimer disease: An [11C] PIB and [18F] FDG PET study. Neurology 68(7):501–508
Bråne G, Gottfries CG (1986) The GBS scale: a new rating scale for dementia syndromes. Nord Psykiatr Tidsskr 40(2):125–134
Morris JC, Ernesto C, Schafer K, Coats M, Leon S, Sano M, Woodbury P (1997) Clinical dementia rating training and reliability in multicenter studies. Neurology 48(6):1508–1510
Mckhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Kawas CH, Phelps CH (2011) The diagnosis of dementia due to Alzheimers disease: recommendations from the National Institute on Aging-Alzheimers Association workgroups on diagnostic guidelines for Alzheimers disease. Alzheimers Dementia 7(3):263–269
Podhorna J, Krahnke T, Shear M, Harrison JE (2016) Alzheimer’s disease assessment scale-cognitive subscale variants in mild cognitive impairment and mild Alzheimer’s Disease: change over time and the effect of enrichment strategies. Alzheimers Res Therapy. https://doi.org/10.1186/s13195-016-0170-5
Stern Y (2012) Cognitive reserve in ageing and Alzheimers disease. Lancet Neurol 11(11):1006–1012
Morris JC, Selkoe DJ (2011) Recommendations for the incorporation of biomarkers into Alzheimer clinical trials: an overview. Neurobiol Aging 32:S1
Grimmer T, Riemenschneider M, Förstl H, Henriksen G, Klunk WE, Mathis CA, Drzezga A (2009) Beta amyloid in Alzheimers disease: increased deposition in brain is reflected in reduced concentration in cerebrospinal fluid. Biol Psychiat 65(11):927–934
Drzezga A, Barthel H, Minoshima S, Sabri O (2014) Potential clinical applications of PET/MR imaging in neurodegenerative diseases. J Nuclear Med 55(Supplement 2):47S-55S. https://doi.org/10.2967/jnumed.113.129254
Zhang XY, Yang ZL, Lu GM, Yang GF, Zhang LJ (2017) PET/MR imaging: new frontier in Alzheimers disease and other dementias. Front Mol Neurosci. https://doi.org/10.3389/fnmol.2017.00343
http://adni.loni.usc.edu/pibpet-analysis-method/pibpet-analysis
Fischl B (2012) FreeSurfer. Neuroimage 62(2):774–781
https://www.bitmesra.ac.in/Visit_Department_Page?cid=1&deptid=70&pid=112
Ashburner J, Friston KJ (2001) Why voxel-based morphometry should be used. Neuroimage 14(6):1238–1243
Whitwell JL (2009) Voxel-based morphometry: an automated technique for assessing structural changes in the brain. J Neurosci 29(31):9661–9664
Schmitter D, Roche A, Maréchal B, Ribes D, Abdulkadir A, Bach-Cuadra M, Krueger G (2015) An evaluation of volume-based morphometry for prediction of mild cognitive impairment and Alzheimers disease. NeuroImage Clin 7:7–17
Fischl B, Salat DH, Kouwe AJ, Makris N, Ségonne F, Quinn BT, Dale AM (2004) Sequence-independent segmentation of magnetic resonance images. NeuroImage 23:S69
Lancaster J, Kochunov P, Nickerson D, Fox P (2000) Stand-alone Java-based version of the Talairach daemon database system. NeuroImage 11(5):S923
Minoshima S, Frey KA, Foster NL, Kuhl DE (1995) Preserved pontine glucose metabolism in Alzheimer disease. J Comput Assist Tomogr 19(4):541–547
Erlandsson K, Irène Buvat P, Pretorius H, Thomas BA, Hutton BF (2012) A review of partial volume correction techniques for emission tomography and their applications in neurology, cardiology and oncology. Phys Med Biol 57(21):R119–R159. https://doi.org/10.1088/0031-9155/57/21/R119
Thomas BA, Cuplov V, Bousse A, Mendes A, Thielemans K, Hutton BF, Erlandsson K (2016) PETPVC: a toolbox for performing partial volume correction techniques in positron emission tomography. Phys Med Biol 61(22):7975–7993
Fischl B, Sereno MI, Tootell RB, Dale AM (1999) High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp 8(4):272–284
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M et al (2011) Scikit-learn: machine learning in python. J Mach Learn Res 12:2825–2830
Geurts P, Ernst D, Wehenkel L (2006) Extremely randomized trees. Mach Learn 63(1):3–42
Lebedev A, Westman E, Westen GV, Kramberger M, Lundervold A, Aarsland D, Simmons A (2014) Random forest ensembles for detection and prediction of Alzheimers disease with a good between-cohort robustness. NeuroImage Clin 6:115–125
Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehéricy S, Habert M, Colliot O (2011) Automatic classification of patients with Alzheimers disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage 56(2):766–781
Seref B, Bostanci E (2019) Performance comparison of naïve bayes and complement naïve bayes algorithms. In: 2019 6th international conference on electrical and electronics engineering (ICEEE)
Dinu A.J (2019) Early detection of alzheimers disease using predictive K-NN instance based approach and T-Test method. Int J Adv Trends Comput Sci Eng 8:29–37
Bersimis FG, Varlamis I (2019) Use of health-related indices and classification methods in medical data. Classif Tech Med Image Anal Comput Aided Diagnos 2019:31–66
Gupta Y, Lee KH, Choi KY, Lee JJ, Kim BC, Kwon GR (2019) Early diagnosis of Alzheimer’s disease using combined features from Voxel-based morphometry and cortical, subcortical, and hippocampus regions of MRI T1 brain images. Plos One 14(10):e0222446
Beheshti I, Demirel H (2016) Feature-ranking-based Alzheimer’s disease classification from structural MRI. Magn Reson Imaging 34(3):252–263
Jha D, Alam S, Pyun J, Lee KH, Kwon G (2018) Alzheimers disease detection using extreme learning machine, complex dual tree wavelet principal coefficients and linear discriminant analysis. J Med Imaging Health Inf 8(5):881–890
Gupta Y, Lee KH, Choi KY, Lee JJ, Kim BC, Kwon GR (2019) Early diagnosis of Alzheimer’s Disease using combined features from voxel-based morphometry and cortical, subcortical, and hippocampus regions of MRI T1 brain images. Plos One 14(10):e0222446
Mehmood A, Yang S, Feng Z, Wang M, Ahmad AS, Khan R, Yaqub M (2021) A transfer learning approach for early diagnosis of alzheimer’s disease on MRI images. Neuroscience 460:43–52
Furst AJ, Agarwal N, Mormino EC (2011) Amyloid vs FDG-PET in the differential diagnosis of AD and FTLD. Neurology 77(23):2034–2042
Martino-IST IT, Navarra ES (2018) Machine learning based analysis of FDG-PET image data for the diagnosis of neurodegenerative diseases. Appl Intell Syst Proc Int APPIS Conf 310:280
Gupta Y, Lama RK, Kwon G (2019) Prediction and classification of Alzheimer’s disease based on combined features from apolipoprotein-E genotype, cerebrospinal fluid, MR, and FDG-PET imaging biomarkers. Front Comput Neurosci. https://doi.org/10.3389/fncom.2019.00072/full
Lesman-Segev OH, La Joie R, Iaccarino L, Lobach I, Rosen HJ, Seo SW, Janabi M et al (2021) Diagnostic accuracy of amyloid versus 18f-fluorodeoxyglucose positron emission tomography in autopsy-confirmed dementia. Ann Neurol 89(2):389–401
Li Y, Rinne JO, Mosconi L, Pirraglia E, Rusinek H, Desanti S, Leon MJ (2008) Regional analysis of FDG and PIB-PET images in normal aging, mild cognitive impairment, and Alzheimer’s disease. Eur J Nucl Med Mol Imaging 35(12):2169–2181
Giacomucci G, Mazzeo S, Bagnoli S, Casini M, Padiglioni S, Polito C, Bessi V (2021) Matching clinical diagnosis and amyloid biomarkers in Alzheimer’s disease and frontotemporal dementia. J Personal Med 11(1):47
Salvatore C, Cerasa A, Castiglioni I (2018) MRI characterizes the progressive course of ad and predicts conversion to Alzheimer’s dementia 24 months before probable diagnosis. Front Aging Neurosci. https://doi.org/10.3389/fnagi.2018.00135
Liu K, Chen K, Yao L, Guo X (2017) Prediction of mild cognitive impairment conversion using a combination of independent component analysis and the cox model. Front Hum Neurosci. https://doi.org/10.3389/fnhum.2017.00033
Zhu X, Suk H, Wang L, Lee S, Shen D (2017) A novel relational regularization feature selection method for joint regression and classification in AD diagnosis. Med Image Anal 38:205–214
Lu D, Popuri K, Ding GW, Balachandar R, Beg MF (2018) Multimodal and multiscale deep neural networks for the early diagnosis of Alzheimer’s Disease using structural MR and FDG-PET images. Sci Rep. https://doi.org/10.1038/s41598-018-22871-z
Li W, Shen Y, Tian D, Bu X, Zeng F, Liu Y, Wang Y (2019) Brain Amyloid-β deposition and blood biomarkers in patients with clinically diagnosed Alzheimer’s disease. J Alzheimers Dis 69(1):169–178
Vandenberghe R, Adamczuk K, Dupont P, Laere KV, Chételat G (2013) Amyloid PET in clinical practice: its place in the multidimensional space of Alzheimers disease. NeuroImage Clin 2:497–511
Lowe VJ, Kemp BJ, Jack CR, Senjem M, Weigand S, Shiung M, Petersen RC (2009) Comparison of 18F-FDG and PiB PET in Cognitive Impairment. J Nucl Med 50(6):878–886
Zhang D, Wang Y, Zhou L, Yuan H, Shen D, Initiative ADN (2011) Multimodal classification of Alzheimer’s Disease and mild cognitive impairment. Neuroimage 55(3):856–867
Rallabandi VS, Tulpule K, Gattu M (2020) Automatic classification of cognitively normal, mild cognitive impairment and Alzheimers disease using structural MRI analysis. Inf Med Unlocked 18:100305
Kitajima K, Abe K, Takeda M, Yoshikawa H, Ohigashi M, Osugi K, Yamakado K (2021) Clinical impact of 11C-Pittsburgh compound-B positron emission tomography in addition to magnetic resonance imaging and single-photon emission computed tomography on diagnosis of mild cognitive impairment to Alzheimers disease. Medicine 100(3):e23969
Martínez G, Vernooij RW, Padilla PF, Zamora J, Flicker L, Cosp XB (2017) 18F PET with flutemetamol for the early diagnosis of Alzheimers disease dementia and other dementias in people with mild cognitive impairment (MCI). Cochrane Database Syst Rev. https://doi.org/10.1002/14651858.CD012884/full
Ding Y, Sohn JH, Kawczynski MG, Trivedi H, Harnish R, Jenkins NW, Franc BL (2019) A deep learning model to predict a diagnosis of alzheimer disease by using 18F-FDG PET of the brain. Radiology 290(2):456–464
Zhang S, Han D, Tan X, Feng J, Guo Y, Ding Y (2012) Diagnostic accuracy of 18F-FDG and 11C-PIB-PET for prediction of short-term conversion to Alzheimer’s disease in subjects with mild cognitive impairment. Int J Clin Pract 66(2):185–198
Ma Y, Zhang S, Li J, Zheng D, Guo Y, Feng J, Ren W (2014) Predictive accuracy of amyloid imaging for progression from mild cognitive impairment to alzheimer disease with different lengths of follow-up. Medicine 93(27):e150
Peng J, Zhu X, Wang Y, An L, Shen D (2019) Structured sparsity regularized multiple kernel learning for Alzheimer’s disease diagnosis. Pattern Recogn 88:370–382
Suk H-I, Lee S-W, Shen D (2015) Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Struct Funct 220(2):841–859
Zheng N, Qiu H, Wang Z, Liu W, Zhang H, Li Y (2018) A new switching-delayed-PSO-based optimized SVM algorithm for diagnosis of Alzheimer’s Disease. Neurocomputing 320:195–202
Kim J, Lee B (2018) Identification of Alzheimers disease and mild cognitive impairment using multimodal sparse hierarchical extreme learning machine. Hum Brain Mapp 39(9):3728–3741
Chételat G, Arbizu J, Barthel H, Garibotto V, Law I, Morbelli S, Drzezga A (2020) Amyloid-PET and 18F-FDG-PET in the diagnostic investigation of Alzheimers disease and other dementias. Lancet Neurol 19(11):951–962
Suppiah S, Ching SM, Mfammed AJ, Mrad SV (2018) The role of PET/CT amyloid imaging compared with Tc99m-HMPAO SPECT imaging for diagnosing Alzheimer’s. Med J Malaysia 73(3):147
Zhang D, Shen D (2012) Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PLoS ONE 7(3):e33182
Suk H, Lee S, Shen D (2014) Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. Neuroimage 101:569–582
Wang P, Chen K, Yao L, Hu B, Wu X, Zhang J, Guo X (2016) Multimodal classification of mild cognitive impairment based on partial least squares. J Alzheimers Dis 54(1):359–371
Devanand D, Mikhno A, Pelton GH, Cuasay K, Pradhaban G, Kumar JD, Parsey RV (2010) Pittsburgh compound B (11C-PIB) and fluorodeoxyglucose (18 F-FDG) PET in patients with alzheimer disease, mild cognitive impairment, and healthy controls. J Geriatr Psychiatry Neurol 23(3):185–198
Yang Z, Liu Z (2020) The risk prediction of Alzheimer’s Disease based on the deep learning model of brain 18F-FDG positron emission tomography. Saudi J Biol Sci 27(2):659–665
Ortiz A, Munilla J, Górriz JM, Ramírez J (2016) Ensembles of deep learning architectures for the early diagnosis of the Alzheimer’s disease. Int J Neural Syst 26(07):1650025
Vandenberghe R, Nelissen N, Salmon E, Ivanoiu A, Hasselbalch S, Andersen A, Dupont P (2013) Binary classification of 18F-flutemetamol PET using machine learning: comparison with visual reads and structural MRI. Neuroimage 64:517–525
Sivapriya TR, Kamal AR, Thangaiah PR (2015) Ensemble merit merge feature selection for enhanced multinomial classification in Alzheimer’s Dementia. Comput Math Methods Med 2015:1–11
Cheng Bo, Liu M, Zhang D, Munsell BC, Shen D (2015) Domain transfer learning for MCI conversion prediction. IEEE Trans Biomed Eng 62(7):1805–1817
Mosconi L, McHugh PF (2011) FDG-and amyloid-PET in Alzheimer’s Disease: Is the whole greater than the sum of the parts. Q J Nucl Med Mol Imaging 55(3):250
Rabinovici GD, Rosen HJ, Alkalay A, Kornak J, Furst AJ, Agarwal N, Jagust WJ (2011) Amyloid vs FDG-PET in the differential diagnosis of AD and FTLD. Neurology 77(23):2034–2042
Tong T, Gray K, Gao Q, Chen L, Rueckert D (2017) Multi-modal classification of Alzheimer‘s disease using nonlinear graph fusion. Pattern Recognit 63:171–181. https://doi.org/10.1016/j.patcog.2016.10.009
Young J, Modat M, Cardoso MJ, Mendelson A, Cash D, Ourselin S (2013) Accurate multimodal probabilistic prediction of conversion to Alzheimer’s Disease in patients with mild cognitive impairment. NeuroImage Clin 2:735–745. https://doi.org/10.1016/j.nicl.2013.05.004
Mora Moradi E, Pepe A, Gaser C, Huttunen H, Tohka J (2015) Machine learning framework for early MRI-based Alzheimers conversion prediction in MCI subjects. Neuroimage 104:398–412
Teng L, Li Y, Zhao Y, Hu T, Zhang Z, Yao Z, Hu B (2020) Predicting MCI progression with FDG-PET and cognitive scores: a longitudinal study. BMC Neurol. https://doi.org/10.1186/s12883-020-01728-x
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflict of interest related to this review. This article does not contain any studies with human or animal subjects performed by the any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kumari, R., Nigam, A. & Pushkar, S. An efficient combination of quadruple biomarkers in binary classification using ensemble machine learning technique for early onset of Alzheimer disease. Neural Comput & Applic 34, 11865–11884 (2022). https://doi.org/10.1007/s00521-022-07076-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-022-07076-w