Abstract
Cancer is the major leading reason of death around the world. However, the early identification and prediction of a cancer type is very critical for patient’s health. Recently, microarray gene expression data was utilized for efficient and early diagnosis of cancer. Previous work shows that microarray data has two major issues which are high dimensionality and small sample size. Several researchers have analyzed and evaluated the cancer classification problem using different statistical and machine learning-based approaches but there are still some issues with these approaches that make cancer classification a nontrivial task. Such as, the inability of certain machine learning algorithms to use unstructured data has limited their utility in the cancer classification process. Convolutional neural networks are proven to very suitable to analyze variety of unstructured data. This ability allowed the deep learning algorithms to play a vibrant part in early detection of cancer through data classification. In this research, a hybrid deep learning model based on Laplacian Score-Convolutional Neural Network (LS-CNN) is employed for the classification of given cancer’s data. The performance of the proposed system was evaluated on 10 different benchmark datasets using various performance measurement metrics such as accuracy and confusion matrix. The experimental results conclude that proposed LS-CNN model outperformed compared to traditional machine learning and recently used deep learning approaches.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Modern deep learning-based techniques has proven to very successful in dealing variety of structure and unstructured data comprising of image, audio, video, text and disease related data. In cancer disease, cells in some tissues undergo uncontrolled division in the body. Because of this condition, malignant growth occurs in the body and cancer effected cells destroy neighbor’s healthy tissues and organs. According to National Cancer Institute (NCI) report currently there are more than 200 different cancer types [1]. Recently, cancer is the major reason behind most of the deaths around the world. Generally, about 1 death from 6 total deaths is because of a cancer [2]. Thus, by 2030, the number of new cancer estimated cases per year may increase up to 25 million [3]. However, timely diagnosis of a cancer may save countless lives and billions of dollars.
The early identification and prediction of a cancer type is very critical for patient’s health and in cancer research [4]. When a cancer is diagnosed at an early stage, treatment is very effective. Previously, classification of a cancer relies on the morphological and clinical techniques [5]. The innovations in the technology have made significant improvement in accurate observation of thousands of cancer genes through gene expression data [6]. This approach provided a large amount of data to researchers by whom they can explore a lot of knowledge but it has some issues [7]. The major issues of microarray data are high dimensionality and low sample size. Also most of the microarray cancer data is noisy and may not be very informative in the cancer detection [8]. To select most significant genes related to cancer and to classify cancer type more precisely and accurately is the main challenge of research nowadays [9, 10]. The selected genes provide understanding the disease, enhance the performance of a cancer classification process and reduce the expense of medical diagnosis [11].
Gene expression data generally comprises of the huge number of genes, several researchers analyzed and evaluated the cancer classification problem using various data mining, statistical and machine learning-based approaches [12]. Many machine learning techniques have obtained good classification accuracy and got a lot of success in the cancer classification [13]. However, there are still some issues with these approaches that make the cancer classification a nontrivial task [14,15,16]. Such as there is a difficulty in performing statistical analysis on microarray data since there is a systematic bias in the output of the microarrays due to its high complexity [17]. A drawback of traditional machine learning (ML) algorithms is that they require pre-engineered organization of raw input data into structured datasets [18]. The inability of certain ML algorithms to use unstructured data has limited their utility in the cancer classification task [19]. Popular algorithms suitable to analyze unstructured data are based on deep learning method of designing neural networks [20]. Deep learning is a branch of machine learning that uses layered architecture to build sophisticated models with the capability to understand complex data [21]. These algorithms learn significant features in the training process, without pre-engineering unstructured data [22]. This ability allowed DL algorithms to outperform against traditional ML algorithms in several fields like computer vision, image classification and speech recognition, etc., [23,24,25]. Deep learning is very helpful in the diagnosis of cancer at an early stage. Published research of NVIDIA indicated that deep learning decrease error rate by 85% for diagnosis of breast cancer [26].
Since deep learning is of incredible potential to support medical and paramedical professionals by decreasing the human error rate, helping in diagnosis of cancer and in analysis of complex data. In this work, a hybrid deep learning-based LS-CNN model is designed for the classification of cancer data. First preprocessing was performed on raw input data then we selected most relevant genes from gene expression data using laplacian score and after that we classified selected genes using convolutional neural network. The objective of this research is the following.
-
To analyze the effects of the feature selection methods on the final accuracy of the convolutional neural network in order to achieve better classification results.
-
To evaluate the performance of hybrid deep learning model LS-CNN for cancer classification using microarray data.
The rest of paper is presented as follows. Section 2 provides summarized review of the relevant literature. Section 3 presents comprehensive description of the proposed methodology. In Sect. 4, experimental results, analysis and findings of the proposed study are presented. Section 5 provides conclusion, recommendations and future directions in this work.
2 Literature review
There exists a lot of problems associated during the cancer data classification. In [27], a deep learning-based CNN model is proposed as a solution in which 1D gene expression data is being transformed into 2D images. Proposed model selected optimal features from overall features of the genes and helped a lot in the classification task. During training and testing of 33 different types of cancer, proposed model achieved 95.65% accuracy. The achieved results were good and need to be enhanced.
In [28], various CNN models (e.g., 1D-CNN, 2D-Vanilla-CNN and 2D-Hybrid-CNN) were introduced for the accurate prediction of cancer genes based on their respective cancer types. Proposed models were trained and tested on multiple samples of gene expression data having 33 different cancer types. Further extension in proposed 1D-CNN model has been made for prediction of breast cancer subtypes. These models achieved prediction accuracy ranges from (93.9 to 95.0%).
In [29], hybrid model of backpropagation neural network (BPN) and fast genetic algorithm (GA) was used to detect interesting features and to characterize cancer subtypes form the gene expression data. The hybrid fast GA-BPN method detected important features and provided high-classification accuracy on different microarray datasets.
Microarray data have low size of samples and huge number of features so, selecting informative features is crucial before classification task. In [30] deep learning-based convolutional neural network (CNN) was used for feature selection and classification of microarray data. Proposed CNN provided flexibility while determining and classification of informative features. Results showed that in comparison with other similar techniques (e.g., mSVM-RFE-IRF and varSel-IRF) proposed CNN provided better accuracy.
In bioinformatics, cancer-related gene selection is important because it improves classification accuracy and reduces complexity. In [31], randomization test (RT) for selection of cancer-related genes from microarray data and sequence of partial least squares discriminant analysis (PLSDA) models is utilized for the evaluation of selected significant genes. Proposed model investigated four datasets, selected significant genes and validated results using PCA and MLR modeling.
Microarray data has millions of genes while the available number of samples are frequently fewer (< 100) so, gene selection is necessary for classification purpose. In [32] a distance-based feature selection technique (Bhattacharyya distance) and SVM for classification of selected marker genes were used. In comparison with other two techniques, e.g., SVM-RFE and SWKC/SVM proposed B-SVM seems to be very operational, selected relevant marker genes and reduced computational cost. Results show that proposed B-SVM outclassed other two techniques in terms of average misclassification and average recovery rate.
In medical field classification of cancer is a hot topic and several machine learning methods were investigated in the literature on different cancer datasets for efficient prediction of cancer. In [33], a comparison has been made among different learning algorithms for better classification of cancer using eleven different cancer datasets. Proposed study obtained maximum accuracy 90.6% using machine learning-based algorithm (Logistic Regression) and 94.43% accuracy using deep learning-based algorithm. The obtained results showed that deep learning-based algorithm are promising for microarray data and it helps in early detection and in treatment of different types of cancer.
The existing machine learning methods are good for microarray data classification but still hybrid and improved machine learning techniques are required for efficient microarray data classification. In [34], information gain (IG) and genetic algorithm (GA) was used for selection of features in the cancer classification. Proposed IG-SGA assessed using seven different gene expression datasets and compared results with other similar techniques. Results showed that IG-SGA performed better than other techniques.
Recently deep learning has gained a lot of popularity in the classification tasks of many areas but still, its use for cancer classification task is very rare. In [35], two deep learning centered techniques, Sample Expansion-Based SAE (SESAE) and Sample Expansion-Based 1DCNN (SE1DCNN) were used for the classification of microarray data. The authors tested SESAE and SE1DCNN on three cancer datasets and results showed that proposed models are effective for cancer classification task.
From the literature, it is concluded that there is need of a highly accurate technique that classify microarray data into classes. This work focuses to achieve the desired objectives.
3 Proposed methodology
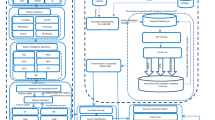
The existence of a large number of microarray datasets reduces analysis quality of a cancer disease [36]. To improve the quality, proper analysis of such datasets is essential. This section discusses thorough explanation of the experimental data, followed by the processing steps and finally the structure of proposed LS-CNN hybrid deep learning model. Figure 1 shows the various phases of the proposed system.
3.1 Experimental datasets
We have selected ten different types of benchmark datasets for the experiments. Most of these datasets were used in the relevant literature so, we preferred to choose these datasets except three datasets (e.g., Arcene, MLL, and Brain tumor) which were not vastly used in the literature. Also these datasets contain multiple records and are quite different from each other. Mostly microarray datasets are binary datasets in nature but three multi-class datasets are also included in our experiments for better understanding of results.
Table 1 outlines the particulars of the experimental datasets.
3.1.1 Data preprocessing
Preprocessing is an essential part to properly analyze the input data. Generally, we have solved three problems of the input data during the preprocessing phase. Various tasks in preprocessing include, removal of categorical row and column, removal of missing values from samples and to convert it into numeric feature matrix. We have removed categorical rows (e.g., gene IDs) and categorical columns (e.g., class labels) from the raw input data. After that we generated labels from raw data and stored in separate variables. Missing values were filled with the mean value of the respective column. Finally, we converted 1D input data into a 4D Array and stored it in numeric matrix format so that it can be passed directly to CNN model. After preprocessing of data, we split it into train and test set data with 80:20 ratio.
3.2 Feature selection
As we discussed earlier that major issues of microarray data are high dimensionality and low sample size. All genes in the microarray data are not informative and may contains noisy data as well [37]. Thus, selection of significant genes from raw microarray data greatly improves the performance of the cancer classification process and reduce the expense of medical diagnosis [38]. Different feature selection methods are used in the literature and new hybrid techniques are still emerging constantly. We have experimented different feature selection methods, e.g., PCA and Info Gain, etc., with our proposed CNN model but Laplacian Score proved itself better for feature selection with our proposed CNN model so, we preferred this method. Also our problem of feature selection for proposed model is unsupervised and in scenario of unsupervised learning when class labels are absent. So, we chose this technique because it performs better in unsupervised scenarios. Detail of Laplacian score technique is described in the following subsection.
3.2.1 Laplacian score (LS)
Fundamental basics of Laplacian Score are preserving locality projection and Laplacian eigen-maps [39]. LS evaluates features using locality preserving power. To represent locality preserving power, laplacian score is computed for each feature. LS observes that if two data points are near to each other than LS assigns them to that same topic. In LS a nearest neighbor graph is built to model the local geometric structure. LS selects only those features that belong to this graph structure. A general overview of working of LS is shown in Fig. 2.
3.2.2 LS algorithm
Let Laplacian Score of the rth feature is Lr and fri is the ith sample of that feature, where (i = 1,.…,m) [33]. Complete LS algorithm is defined in Algorithm 1:

3.3 Convolutional neural network (CNN)
A CNN is a feedforward neural network which was initially established in late 90s but at that time for problems like pattern recognition it was considered inefficient [16, 40]. However, with the development in fast and parallel processing, now this technique is better than other traditional machine learning techniques in understanding and classification of raw input data [41]. Also CNN’s is currently the dominant deep learning model for computer vision, image classification, and cancer classification tasks [42]. CNN models are effective in circumstances where the information comprises of numerous arrays, as the architecture of CNN is fit for managing information with high and different dimensionalities, for example, microarray data and 2D images, etc. [43]. Because of such reasons and popularity of CNN as compare to other machine learning-based methods, we preferred to use it in this work.
3.3.1 Proposed CNN architecture
Based on such the characteristics of CNN, it is employed for the cancer classification task. Before applying CNN on the selected set of features, we arranged cancer data as matrix vectors. Then matrix of data is processed and classified using CNN. The CNN model configured in this work consists of 27 layers. This model has three main components: input layer (layer 1), hidden/encoder layer (From layer 2 to layer 24) and output layer (From layer 25 to layer 27). The complete architecture of CNN model is presented in Fig. 3.
Figure 3 shows that the first layer is the input layer (image input layer) where the size of input data is specified. Which, in this case, is 1-by-N-by-1; where N is number of top ranked selected features. In (1 × N × 1), first 1 correspond to the height, N correspond to the width, and the last one correspond to the channel size for image input layer. In the hidden layer portion, six convolutional layers have been used where first convolutional layer contains eight different filters, while the second, third and up to sixth convolutional layers contain 16, 32, 64, 128, and 256 filters respectively. The filter size of first three convolutional layers is 2 × 2 and the filter size of other three convolutional layers is 3 × 3. The stride and padding size in all six convolutional layers is one (1). With six convolutional layers six batch normalization layers and six ReLU layers have been used. The batch normalization layers perform normalization of gradients and activations present in a network for making training of network an easier problem of optimization. To increase the training speed of network we used layers of batch normalization in between ReLU and convolutional layers. Six nonlinear functions are used after the layers of batch normalization, e.g., rectified linear unit (ReLU). Five Max pooling layers have been used followed by ReLU Layers. As Max Pooling Layers selects significant features, remove irrelevant features and hence minimize the size of total data. Each max pooling layer used in proposed model has a pool size of 1 × 1 and stride size 1 with zero padding. After hidden layer portion in output layer we used a fully connected layer, softmax layer, and a classification output layer.
All previously learned features are then combined by fully connected layer to recognize significant patterns from data for the classification. Hence, the parameter of output in fully connected layer represents the number of classes in data, e.g., output size of 2 represent that there are 2 classes in data. The output of fully connected layer is normalized by the softmax activation function. Softmax layer output contain positive values that are added to one and are used as probability values for the layer of classification. Classification layer is the final layer in the network and it uses probability values of softmax function to assign input to a specific class.
3.3.2 Proposed CNN training options
We specified training option after outlining the structure of network. The mentioned network is trained with Stochastic Gradient Descent with Momentum (SGDM) having initial learning-rate of 0.01 with max 100 number of epochs. The accuracy is monitored during the training of network by setting validation frequency to 30 for validation data. During validation, data does not update the weights of the network. At the end progress plot of training is turned on and command window output is turned off.
4 Experimental results and discussion
4.1 Environmental setup
All experiments have been performed on Intel Core i5 CPU has a processor speed of 5 GHz with 8 GB of RAM. MATLAB tool is used for the analysis and experiments. Ten different benchmark microarray gene expression datasets are observed to check the performance of the proposed system. Results are analyzed and compared on the basis of different evaluation parameters.
4.2 Evaluation parameters
After performing experiments, we accessed the performance of proposed model using classification accuracy and Confusion matrix. Accuracy measures how well the classifier predicted the classes [44]. To eliminate redundancy from network training and testing we calculated average accuracy value from accuracy values of three experiments. For proposed LS-CNN model accuracy as an evaluation parameter is defined in Eq. 3.
Confusion matrix is popular evaluation parameter used to measure the performance of a classification model [45]. It provides true values of accuracy against a classifier. It consist of four different types of values, e.g., True Positive, True Negative, False Positive and False negative values, through which one can also calculate precision, Recall and F-measure, etc. For optimal results we also used two more evaluation parameters Elapsed Time and number of iterations. The number of iterations are rounds taken by the network for training and testing of data, while elapsed time is time taken by the network for training and testing of data.
4.3 Experimental results
4.3.1 LS-CNN results
This section illustrates the results of proposed system on various datasets. We have performed three experiments on top-ranked selected features using our proposed model to obtain good results and to remove redundancy. Based on accuracies achieved after all three experiments, we calculated an average accuracy. The details of all three experiments, number of selected features and classification accuracies are given in Table 2. Selected features in Table 2 are significant and optimal features selected during feature selection process with the help of Laplacian Score. Based on these selected features, proposed LS-CNN model achieved optimal results and desired accuracies. Datasets like AML, CNS, Colon and Prostate achieved 100% accuracy in all three experiments so average accuracy value of all these datasets are also 100%. While MLL dataset achieved 89% accuracy in first, 96% accuracy in second, 100% accuracy in third experiment and its average accuracy value is 95%. SRBCT dataset achieved 88.5% accuracy in first, 93.8% accuracy in second, 93.8% accuracy in third experiment and its average accuracy value is 92%. Arcene dataset achieved 97.5% accuracy in first and second experiment, 100% accuracy in third experiment and its average accuracy value is 98.33%. DLBCL dataset achieved 94% accuracy in first and 100% accuracy in second and third experiment and its average accuracy value is 98%. Leukemia dataset achieved 97% accuracy in first and 100% accuracy in second and third experiment and its average accuracy value is 99%. Brain Tumor dataset achieved 94% accuracy in first, 97% accuracy in second, 100% accuracy in third experiment and its average accuracy value is 97%.
Overall average accuracy values of all datasets ranges from (90 to 100) which are quite high and optimal for a deep learning model. For binary class datasets (e.g., AML, CNS, Colon and Prostate), proposed LS-CNN achieved 100% accuracy except Arcene, DLBCL and Leukemia datasets. For multi-class datasets (e.g., MLL, SRBCT and Brain Tumor), proposed LS-CNN achieved accuracy values ranges from (90 to 100). Overall proposed LS-CNN provided optimal results because LS feature selection technique selected best features and proposed CNN accurately classified selected features and achieved high classification accuracies. In general, achieved results support our proposed synopsis since average accuracy values of our proposed LS-CNN model are very high which indicates toward better and exceptional performance of our proposed model.
The detail of optimal results for each dataset in form of confusion matrix, elapsed time and no. of iterations are shown in Table 3. These are best result achieved by our proposed model using selected set of features from each data set. From results it is clear that percentage accuracy value for each dataset is 100%, except SRBCT dataset for which percentage accuracy value is 93.8%. Also percentage true negative and true positive values for each dataset are different and percentage false positive and false negative values for each dataset is 0% but for SRBCT dataset percentage false negative value is 6.2%. The elapsed time for most of the datasets ranges from 20 to 45 s but for Arcene dataset took more time (91 s) and for Brain tumor dataset took minimum time (18 s) in comparison with other datasets. Similarly number of iterations w.r.t total no of iterations has also been discussed in detail. Out of total 100 number of iterations most of the datasets took 100 number of iterations, four datasets took 80 number of iteration while, brain tumor dataset took least number of iterations (70) among other datasets during training and testing.
For all datasets accuracy graph plots were created. The detail values of these graphs have been discussed in Table 3. Here graph of only two datasets (AML and SRBCT) are shown since, all other graphs were having same accuracy value of 100%. Figure 4 shows accuracy graph plot for AML data.
Figure 5 shows accuracy graph plot for SRBCT data.
4.3.2 Comparative results
Table 4 provides the comparative analysis of the LS-CNN model results with the existing techniques. For better comparison, existing results are obtained from recent and relevant literature, i.e., from 2011 to 2018. All existing studies have used accuracy as an evaluation parameter, also other evaluation parameters (e.g., Precision, Recall, F-measure, execution time, Error rate and average misclassification rate, etc.) are used in few studies. Few datasets (e.g., Arcene, MLL and Brain tumor) used in the proposed study could not be found in the relevant literature so we have not included in the comparison of results. To obtain competitive results, we made comparison on the basis of common datasets and classification accuracy values. Proposed model has been compared with nine different machine learning and deep learning based techniques, namely: Hybrid Genetic Algorithm and Backpropagation Network (HGA-BPN) [14], Convolutional Neural Network (CNN) [15], Random Test and Partial Least Square Discriminant Analysis (RT-PLSDA) [16], Bhattacharyya Distance and Support Vector Machine (B-SVM) [17], Principal Component Accumulation (PCAcc) [18], Information Gain and Standard Genetic Algorithm (IG-SGA) [19], Sample Expansion-based 1D Convolutional Neural Network (SE1DCNN) [20], Random Forest with Statistical Tests (RFST) [21] and Random Forest (RF) [22]. In comparison with all other techniques performance of proposed LS-CNN model is better. For SRBCT dataset result of CNN [15] method is better (e.g., 100% accuracy) than LS-CNN model (e.g., 92% Accuracy) but for other datasets (e.g., AML, Colon and Prostate Cancer) proposed model outperformed. For prostate cancer dataset result of IG-SGA [19] method is equal with proposed model result (e.g., 100% accuracy) but for other datasets (e.g., CNS, Colon, DLBCL, and Leukemia) proposed model outperformed the same method. For all other datasets (e.g., AML, CNS, Colon, DLBCL and Leukemia) accuracy values of proposed model are higher than all other techniques. Compare to hybrid and traditional machine learning techniques (e.g., HGA-BPN, RT-PLSDA, B-SVM, PCAcc, IG-SGA, RFST and RF) results of proposed model are quite better and accuracy value ranges from (90 to 100). Compared to recently use deep learning techniques (e.g., CNN and SE1DCNN) results of proposed model are also better except result of CNN [15] against SRBST dataset. In general, proposed LS-CNN model outperformed traditional and hybrid machine learning approaches and recently used deep learning approaches. Results showed that proposed model can be used for selection and classification of cancer related genes in accurate and efficient manner from sparse datasets. Results also support that proposed model can be used for efficient diagnosis and prognosis of cancer subtypes. Proposed model can also help in medicine discovery and in improving treatment strategies of cancer disease.
5 Conclusion
In this work, a hybrid deep learning model LS-CNN is proposed for the classification of cancer’s data. The major objective was to analyze the effects of the feature selection methods on the final accuracy of convolutional neural network. We have used ten different benchmark microarray datasets. Classification accuracy and confusion matrix are used as evaluation parameters for the comparison and better understanding of experimental results. The experimental result showed that all datasets average accuracy ranges from (90 to 100) which is quite high and optimal for a deep learning model. For binary class datasets, proposed LS-CNN achieved 100% accuracy except Arcene dataset for which average accuracy value is 98.33. For multi-class datasets, proposed LS-CNN achieved accuracy ranges from (90 to 100). We have also compared results of proposed LS-CNN model with the existing results. Our proposed LS-CNN model outperformed traditional machine learning and recently used deep learning approaches. In future, proposed LS-CNN model can be applied on multi-class image datasets in order to achieve better accuracy results.
References
NIH (2019) National Cancer Institute (NCI), cancer statistics. Available from: https://www.cancer.gov/. Accessed 23 April 2019
World Health Organization, Cancer (2018) Available from: https://www.who.int/news-room/fact-sheets/detail/cancer. Accessed 23 April 2019
Babu M, Sarkar K (2016) A comparative study of gene selection methods for cancer classification using microarray data. In: 2016 second international conference on research in computational intelligence and communication networks (ICRCICN). IEEE
Arslan MT, Kalinli A (2016) A comparative study of statistical and artificial intelligence based classification algorithms on central nervous system cancer microarray gene expression data. Int J Intell Syst Appl Eng. https://doi.org/10.18201/ijisae.267094
Bolón-Canedo V, Sánchez-Marono N, Alonso-Betanzos A, Benítez JM, Herrera F (2014) A review of microarray datasets and applied feature selection methods. Inf Sci 282:111–135
Hu H, Niu Z, Bai Y, Tan X (2015) Cancer classification based on gene expression using neural networks. Genet Mol Res 14:17605–17611
Bhola A, Tiwari AK (2015) Machine learning based approaches for cancer classification using gene expression data. Mach Learn Appl Int J 2(3/4):01–12
Singh RK, Sivabalakrishnan M (2015) Feature selection of gene expression data for cancer classification: a review. Proc Comput Sci 50:52–57
Gölcük G (2017) Cancer classification using gene expression data with deep learning. Paper presented at Department of Electronics, Informatics and Bioengineering Polytechnic University of Milan, Italy, 20 Dec 2017. http://hdl.handle.net/10589/138427
Khan MZ, Harous S, Hassan SU, Khan MUG, Iqbal R, Mumtaz S (2019) Deep unified model for face recognition based on convolution neural network and edge computing. IEEE Access 7:72622–72633
Guillen P, Ebalunode J (2016) Cancer classification based on microarray gene expression data using deep learning. In: 2016 international conference on computational science and computational intelligence (CSCI). IEEE
Bhat RR, Viswanath V, Li X (2017) DeepCancer: detecting cancer via deep generative learning through gene expressions. In: 2017 IEEE 15th international conference on dependable, autonomic and secure computing, 15th international conference on pervasive intelligence and computing, 3rd international conference on big data intelligence and computing and cyber science and technology congress (DASC/PiCom/DataCom/CyberSciTech). IEEE
Danaee P, Ghaeini R, Hendrix DA (2017) A deep learning approach for cancer detection and relevant gene identification. In: Pacific symposium on biocomputing 2017. World Scientific
Wenyan Z, Xuewen L, Jingjing W (2017) Feature selection for cancer classification using microarray gene expression data. Biostat Biom Open Access J 1(2):555557
Dang S, Wen M, Mumtaz S, Li J, Li C (2020) Enabling multi-carrier relay selection by sensing fusion and cascaded ANN for intelligent vehicular communications. IEEE Sens J. https://doi.org/10.1109/JSEN.2020.2986322
Matsubara T, Ochiai T, Hayashida M, Akutsu T, Nacher JC (2019) Convolutional neural network approach to lung cancer classification integrating protein interaction network and gene expression profiles. J Bioinform Comput Biol 17(03):1940007
Hamena S, Meshoul S (2018) Multi-class classification of gene expression data using deep learning for cancer prediction. Int J Mach Learn Comput 8(5):454–459
Luque-Baena R, Urda D, Subirats J, Franco L, Jerez J (2013) Analysis of cancer microarray data using constructive neural networks and genetic algorithms. In: Proceedings of the IWBBIO, international work-conference on bioinformatics and biomedical engineering
Natarajan A, Ravi T (2014) A survey on gene feature selection using microarray data for cancer classification. Int J Comput Sci Commun (IJCSC) 5(1):126–129
Kong Y, Yu T (2018) A deep neural network model using random forest to extract feature representation for gene expression data classification. Sci Rep 8(1):16477
Kumar M, Rath NK, Swain A, Rath SK (2015) Feature selection and classification of microarray data using MapReduce based ANOVA and K-nearest neighbor. Proc Comput Sci 54:301–310
Iqbal MS, Ahmad I, Bin L, Khan S, Rodrigues JJ (2020) Deep learning recognition of diseased and normal cell representation. Trans Emerg Telecommun Technol. https://doi.org/10.1002/ett.4017
Lyu B, Haque A (2018) Deep learning based tumor type classification using gene expression data. In: Proceedings of the 2018 ACM international conference on bioinformatics, computational biology, and health informatics. ACM
Khalifa NEM, Taha MHN, Ali DE, Slowik A, Hassanien AE (2020) Artificial intelligence technique for gene expression by tumor RNA-seq data: a novel optimized deep learning approach. IEEE Access 8:22874–22883
Khan S, Muhammad K, Mumtaz S, Baik SW, de Albuquerque VHC (2019) Energy-efficient deep CNN for smoke detection in foggy IoT environment. IEEE Internet Things J 6(6):9237–9245
Reena G (2011) A survey of human cancer classification using micro array data. Int J Comput Technol Appl 2(5):1523–1533. http://www.ijcta.com/vol2issue5-page3.php
Joseph M, Devaraj M, Leung CK (2019) DeepGx: deep learning using gene expression for cancer classification. In: 2019 IEEE/ACM international conference on advances in social networks analysis and mining (ASONAM). IEEE
Mostavi M, Chiu Y-C, Huang Y, Chen Y (2020) Convolutional neural network models for cancer type prediction based on gene expression. BMC Med Genom 13:1–13
Vimaladevi M, Kalaavathi B (2014) A microarray gene expression data classification using hybrid back propagation neural network. Genetika 46(3):1013–1026
Zeebaree DQ, Haron H, Abdulazeez AM (2018) Gene selection and classification of microarray data using convolutional neural network. In: 2018 international conference on advanced science and engineering (ICOASE). IEEE
Mao Z, Cai W, Shao X (2013) Selecting significant genes by randomization test for cancer classification using gene expression data. J Biomed Inform 46(4):594–601
Zhong W (2014) Feature selection for cancer classification using microarray gene expression data. University of Calgary, Calgary
Tabares-Soto R, Orozco-Arias S, Romero-Cano V, Bucheli VS, Rodríguez-Sotelo JL, Jiménez-Varón CF (2020) A comparative study of machine learning and deep learning algorithms to classify cancer types based on microarray gene expression data. PeerJ Comput Sci 6:e270
Salem H, Attiya G, El-Fishawy N (2017) Classification of human cancer diseases by gene expression profiles. Appl Soft Comput 50:124–134
Liu J, Wang X, Cheng Y, Zhang L (2017) Tumor gene expression data classification via sample expansion-based deep learning. Oncotarget 8(65):109646
Lee K, Man Z, Wang D, Cao Z (2013) Classification of bioinformatics dataset using finite impulse response extreme learning machine for cancer diagnosis. Neural Comput Appl 22(3–4):457–468
Wu Q, Boueiz A, Bozkurt A, Masoomi A, Wang A, DeMeo DL, Weiss ST, Qiu W (2018) Deep learning for predicting disease status using genomic data. PeerJ Preprints
Liu Y, Zhang N, He Y, Lun L (2015) Prediction of core cancer genes using a hybrid of feature selection and machine learning methods. Genet Mol Res 14(3):8871–8882
He X, Cai D, Niyogi P (2006) Laplacian score for feature selection. In: Advances in neural information processing systems
Mandal S, Banerjee I (2015) Cancer classification using neural network. Int J Emerg Eng Res Technol 3(7):172–178
Liu B, Wei Y, Zhang Y, Yang Q (2017) Deep neural networks for high dimension, low sample size data. In: IJCAI
Kim B-H, Yu K, Lee PC (2020) Cancer classification of single-cell gene expression data by neural network. Bioinformatics 36(5):1360–1366
Smolander J (2016) Deep learning classification methods for complex disorders
Fakoor R, Ladhak F, Nazi A, Huber M (2013) Using deep learning to enhance cancer diagnosis and classification. In: Proceedings of the international conference on machine learning. ACM, New York, USA
Zhou W, Dickerson JA (2014) A novel class dependent feature selection method for cancer biomarker discovery. Comput Biol Med 47:66–75
Liu J, Cai W, Shao X (2011) Cancer classification based on microarray gene expression data using a principal component accumulation method. Sci China Chem 54(5):802–811
Nagpal A, Singh V (2018) Identification of significant features using random forest for high dimensional microarray data. J Eng Sci Technol 13(8):2446–2463
Ram M, Najafi A, Shakeri MT (2017) Classification and biomarker genes selection for cancer gene expression data using random forest. Iran J Pathol 12(4):339
Acknowledgements
This work has been partially supported by FCT/MCTES through national funds and when applicable co-funded EU funds under the Project UIDB/50008/2020; and by Brazilian National Council for Scientific and Technological Development (CNPq) via Grant No. 309335/2017-5 and HEC Pakistan under NRPU funded Project No. 6338.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shah, S.H., Iqbal, M.J., Ahmad, I. et al. Optimized gene selection and classification of cancer from microarray gene expression data using deep learning. Neural Comput & Applic (2020). https://doi.org/10.1007/s00521-020-05367-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00521-020-05367-8