Abstract
Classification is one of the most important task in hyperspectral image processing. In the last few decades, several classification techniques have been introduced. However, most of them could not efficiently extract features from hyperspectral images (HSI). A novel deep learning framework is proposed in this paper which efficiently utilises convolutional neural network (CNN) and spatial pyramid pooling (SPP) for extracting both the spectral–spatial features for classification. The proposed hybrid framework uses principal component analysis (PCA), 3D-CNN, 2D-CNN and SPP. The proposed CNN-based model is applied on three benchmark hyperspectral datasets, and subsequently the performance is compared with state-of-the-art methods in the same field. The obtained results reveal the superiority of the proposed model in effectively classifying HSI.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Hyperspectral image (HSI) contains more than hundreds of spectral bands for each pixel [1]. In HSI, for every pixel, a spectrum of wavelengths is captured, which represents the material properties, i.e. the spectral signatures. The spectral information of HSI is added as the third-dimension to the two-dimensional (2D) spatial image and generating a three-dimensional (3D) data cube [2]. With the increase in spectral information, HSI finds its application in various fields like agriculture [3], land-cover mapping [4], surveillance [5], physics, mineralogy [6], chemical imaging, environment monitoring, etc. However, HSI processing suffers from many issues, viz. noise, computational complexity, poor contrast, huge dimensionality and insufficient training samples. To overcome the dimensionality problem, preprocessing techniques such as randomised principal component analysis (R-PCA) [7] and minimum noise fraction (MNF) [8], are employed that can extract the top features of HSI. However, the number of features to be considered for classification of HSI is decided manually.
In the past two decades, HSI classification remained as one of the active research topics as surveyed by Camps-Valls et al. [9]. The main aim of classification of HSI is to assign a label to each pixel. The HSI classification has been mainly performed using handcrafted features viz. multiscale joint collaborative representation with locally adaptive dictionary (MLJCRC) [10], feature extraction by local covariance matrix representation (LCMR) [11], histograms of directional map (HoDM) [12] approach and learning-based techniques. Many machine learning techniques have been proposed till date for pixel-wise spectral classification, viz. support vector machines (SVM) [13] and random forests [14]. However, these methods are very much sensitive to the number of training samples and they only take the spectral information into consideration for classifying HSI. To improve the classification performance, many spectral–spatial classification methods, which jointly utilise both the spectral and spatial information, have been proposed till date. This category of method includes extended morphological attribute profile (EMAP) [15] to model the spatial information according to different attributes, edge-preserving filtering (EPF) to construct the spectral–spatial features of HSIs [16] and extended random walker (ERW) to optimise the results of SVM [17]. However, the limitation of this method is that it extracts the spectral–spatial features of the HSI in a shallow fashion and the classification result is also reliant on the segmentation scale.
Recently, deep learning (DL) techniques have gained immense popularity in HSI processing due to its efficient feature extraction and classification ability that could effectively outperform the traditional techniques [18]. The widely used deep neural network (DNN) [19] architecture includes deep convolutional neural networks (CNNs) [20], stacked autoencoder networks (SAEs) [21], deep Boltzmann machines (DBMs) [22], deep belief networks (DBNs) [23] implemented as in capsule network [24], deep laboratory [25], deep pyramidal residual networks (DPRN) [26] and deep deconvolution using skip architecture [27]. CNN has come up to the forefront due to its better performance over handcrafted techniques and other DL [28] techniques. CNN has found its application in remote sensing research domains like image classification [29], semantic segmentation [30], etc. CNN is characterised by its shared weights, local connection and shift invariance that help in reducing the computational cost. CNN is the building block of the dual-path network (DPN) [31] which utilises the properties of both the residual network (RESNET) [32], i.e. the interconnection between the layers and the dense convolutional network (DenseNet) [33] for HSI classification. Deep belief network [34] is proposed to effectively extract 3D spectral–spatial features of HSI, which combines Gabor filters [35] with convolutional filters to mitigate the problem of overfitting. Spectral–spatial residual network (SSRN) [36] uses identity mapping for connecting convolutional layers. In all of these techniques, either 2D or 3D convolution is considered while designing the model which made the model either very complex or may suffer from loss of information. 2D-CNN alone cannot extract features from the spatial dimension. Similarly 3D-CNN is very much computationally complex and it cannot accurately classify classes having similar texture.
The HybridSN [37] overcomes such shortcomings as it combines 3D-CNN and 2D-CNN to extract spectral and spatial features respectively. This hybrid model utilises both the spatio-spectral features, thereby producing good classification result. However, the usage of flatten layer made the model inefficient both in terms of computation time and classification accuracy. The spatial pyramid pooling (SPP) [38] extracts spatial features in different scales, in contrast to the traditional pooling which can only extract features of the same scale. Hence, the CNN model with SPP is more robust to object distortions [39]. Therefore, in this paper a novel architecture called spectral–spatial network (SSNET) is proposed by utilising SPP in hybrid CNN. In SSNET, SPP is placed between the hybrid convolutional layer and the fully connected dense layer for extracting the spectral–spatial features effectively.
2 Proposed SSNET model
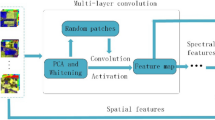
1D CNN and 2D CNN extract spectral and local spatial features of each pixel, respectively [40]. Unlike 1D and 2D CNN, the proposed model is based on 3D local convolutional filters which learn both spatio-spectral contents of the same channel simultaneously and hence is more efficient in extracting information from HSI. The overall architecture of the proposed SSNET model is depicted in Fig. 1. It includes four major components, namely PCA, 3D-CNN, 2D-CNN and SPP. These components are described in the following subsections.
2.1 PCA
The input of the model is an HSI data cube of dimension M × N × D, where M, N and D represent the width, height and number of bands, respectively. The spectral redundancy of the HSI data cube is reduced by applying principal component analysis (PCA) technique. PCA only reduces the spectral bands so as to condense the whole image such that only very important information for recognising any object is present in the resultant image cube. The reduced HSI cube can be represented as X ∈ RM×N×B, where B is the number of selected principal components. When PCA is applied on widely used HSI dataset, it is experimentally observed that the input dimension can be reduced up to 15 times while preserving 99.9% of initial information and the first 10 to 30 principal components contain the maximum amount of information [16]. A 3D patch of dimension K × K × B, centred at the spatial location (i, j) and covering the K × K spatial extent, is generated from X. The total number of such 3D patches is given by \(\frac{M}{K} \times \frac{N}{K}\). The target label is represented as one hot encoded vector y = (y1, y2,…yC) ∈ R1×1×C, where C being the land-cover classes. As neighbouring pixels of hyperspectral image is considered as the input to the model, the 3-D local convolutional filters can learn spectral–spatial features in the same channel very easily.
2.2 3D CNN
Subsequently, the spectral and spatial features are integrated together to construct a joint spatio-spectral classification framework using 3-D CNN. In 3D-CNN, the value of a neuron, i.e. activation value \(v_{ij}^{xyz}\) at position (x, y, z) of the jth feature map in the ith layer is generated using Eq. 1.
where g is the activation function, v is the output variable in the feature map, m indexes the feature map in the (i−1)th layer connected to the current (jth) feature map, and Pi and Qi are the height and the width of the spatial convolution kernel. Ri is the size of the kernel along the spectral dimension, \(w_{ijm}^{pqr}\) is the value of position (p, q, r), i.e. the weight parameter connected to the mth feature map, and bij is the bias parameter of the jth feature map in the ith layer. The high dimensionality of the input HSI data may lead to an overfitting situation so to handle such issue a regularisation strategy is implemented, i.e. nonlinear function ReLU (rectified linear unit) is introduced. The ReLU function (\(\sigma\)) is given in Eq. (2).
2.3 2D CNN
After 3D convolution, the learnt feature 3 (Fig. 1) vector is sent to the 2D-CNN. In 2D-CNN, the input feature vector is convolved with the 2D 3 × 3 kernel. The convolution is computed by the sum of the dot product between input vector and the kernel. The kernel strode over the input feature vector to cover full spatial dimension and is then passed through nonlinear activation function ReLU. In 2D-CNN, the value of a neuron, i.e. the activation value \(v_{ij}^{xy}\), at spatial position (x, y) of the jth feature map in the ith layer is expressed in Eq. 3.
where m, g, v, Pi, Qi and bij are similar to Eq. 1, and \(w_{ijm}^{pq}\) is the weight of position (p, q) connected to the mth feature map. After 2D convolution, the feature vector captured the spatial information contained in the K × K neighbourhood region of the input feature vector from 3D-CNN.
2.4 SPP
Subsequently, the learned features are fed to the pooling layers. Then spatial pyramid pooling (SPP) is introduced to the feature map 4 (Fig. 1) of two-dimensional local convolutional filters, so that the proposed model can learn these spectral–spatial features easily and generate a fixed feature vector. Three different sizes of pooling windows (l, m, n) are chosen for SPP and the features so obtained are concatenated to form a 1D vector which is fed to the input of the fully connected layer regardless of the size of the feature maps. To prevent overfitting, dropout is introduced into the fully connected network. Hence, the total number of parameters in the proposed model has reduced considerably, thereby reducing the training time. Finally, the learned features are fed to probabilistic logistic regression function softmax for classification. The bias and the weight parameters are trained using supervised approach, i.e. by using gradient descent mechanism.
In the proposed architecture (Fig. 1), there are three 3D-CNN layers consisting of kernels of size 8 × 3× 3 × 7 (where 8 is the number of 3D kernels of dimension 3 × 3 × 7, \(K_{1}^{1}\) = 3, \(K_{2}^{1}\) = 3, \(K_{3}^{1}\) = 7), 16 × 3×3 × 5 (where 16 is the number of 3D kernels of dimension 3 × 3×5, \(K_{1}^{2}\) = 3, \(K_{2}^{2}\) = 3, \(K_{3}^{2}\) = 5) and 32 × 3×3 × 3 (where \(K_{1}^{3}\) = 3, \(K_{2}^{3}\) = 3, \(K_{3}^{3}\) = 3), followed by one 2D-CNN layer of size 64 × 3×3 (where 64 is the number of 2D kernels for \(K_{1}^{4}\) = 3,\(K_{2}^{4}\) = 3). Mainly a spatial dimension of 3 × 3, 5 × 5 and 7 × 7 convolutional filters are preferred for a high-dimensional image [41]. So, after an exhaustive analysis by comparing with multiple filter size, 3 × 3 is chosen as the height and width, whereas for the depth varying kernel depths such as (3,5 , 7), (7, 5, 3), (5, 7, 3) and (3, 7, 5) have been experimented with and (7, 5, 3) is found to be the best. In order to facilitate a very deep model with reasonably reduced numbers of parameters, multiple convolutional layers have been stacked together [42] with an increasing number (8, 16, 32, 64) of feature maps. The multiple pooling layers of different scales (viz. 1, 2 and 4 represented as l, m and n, respectively, in Fig. 1) is chosen such that it can extract features with 1 × 1, 2 × 2, 4 × 4 max pooling. The multiscale filtered feature map contains rich complementary information which helps to improve the classification performance [43]. The window size and the number of principal components (PCs), i.e. the parameters K × K and B in Fig. 1, play an important role in proposed SSNET, and hence, optimum values of these parameters are chosen based on sensitivity analysis on real HSIs as presented in Sect. 4.
3 Dataset
Indian Pines (IN), University of Pavia (UP, Pavia, Italy) and Salinas Scene (SA) datasets are used for the experimental set-up. Footnote 1
-
A.
The IN data (Fig. 3a) were obtained by the Airborne Visible/Infrared Imaging Spectrometer (AVIRIS) in North-western Indiana in June 12, 1992, by NASA with 20 m spatial resolutions and 10 nm spectral resolutions covering a spectrum range of 200–2400 nm and 220 bands. The subset used for classification is of size 145 × 145 × 200, with 16 kinds of ground cover where most of them are vegetation which are nearly similar to each other due to the shared spectral characteristic of vegetation. Moreover, several mixed pixels are found due to course spatial resolution. In total, 200 bands were left after radiometric corrections and bad band removal.
-
B.
B. The UP data (Fig. 4a) are taken from the flights of the Reflective Optics System Imaging Spectrometer (ROSIS) sensor over Pavia in Northern Italy in 2003 with spatial resolution of 1.3 m in the range of 0.43–0.86 μm for 115 bands and with nine kinds of land cover. After removing low-SNR bands, 103 bands were used in the present experiment; dimension of the present dataset is 610 × 340 × 103 pixels.
-
C.
The SA data (Fig. 5a), captured by AVIRIS over Salinas valley, CA, USA, in 1998, contain 512 × 217 pixels and 224 spectral bands covering from 400 to 2500 nm. The spatial resolution is 3.7 m. Twenty bands are discarded due to water absorption. In total, 16 classes are labelled as the ground truth, where most of them are agriculture, mainly vegetable field, vineyard and bare soil
4 Experimental results and discussion
In the present experiment, all network weights are randomly initialised and trained using back-propagation algorithm with Adam optimiser by using the categorical cross-entropy loss function. Mini-batches of size 256 are used, and the network is trained for 100 epochs with an optimal learning rate of 0.001. The window size K × K and number of PCs play an important role in the results of classification. In [7], it is illustrated that the first 10 to 30 principal components contain the maximum information of the widely used HSI dataset. Hence, in order to decide an optimum number of PCs as well as spatial window size, a sensitivity analysis is carried out with varying number of PCs and window sizes. Figure 2 depicts the rescaled values (between 0 and 1) of overall accuracy (OA) observed in this analysis for three benchmark datasets. As the spatial context changes with data, different datasets performed differently for varying window size and PC number which is also evident in Fig. 2. Figure 2 shows that 17 × 17 window size is found to be most suitable for the proposed method with high classification accuracy without overburdening the model. A reasonably low test loss is also observed while using 17 × 17 window size and first 15 PCs of each dataset which are therefore chosen and subsequently used. With the window size of 17 × 17 × 15, the convolutional kernel becomes small [41] that enables efficient processing and learning distinctive features from local regions. Layer-wise detailed information of the proposed model is illustrated in Table 1 for UP dataset.
In this experiment, state-of-the-art supervised methods, i.e. SVM [13], 2D CNN [7], 3D CNN [44], SPP [39] and HybridSN [38], are compared with the proposed model on the same HSI datasets. Labelled samples are split into training (30%) and testing (70%), and subsequently, aforementioned classifiers are trained and HSI scenes are classified. The experiment is carried out ten times, and the average classification accuracies are recorded to evaluate the performance of each method. In order to quantitatively compare the performance of classifier models, overall accuracy (OA), average accuracy (AA) and kappa coefficient are measured from the confusion matrix using Eqs. 4 to 6, respectively, and listed in Table 2.
For the IN dataset, the 3D patches of 17 × 17 × 15 input volume are considered. In Table 2 and Fig. 3, the classification result for different classifier models is demonstrated. It can be observed that the proposed model attains a greater accuracy than the other tested models. The average test loss and test accuracy of the proposed model are observed as 0.52% and 99.85%, respectively, using the testing data.
Table 2 and Fig. 4 show the classification result for the UP dataset with similar spatial window of size. The average test loss and test accuracy achieved using the SSNET are 0.08% and 99.98%, respectively.
The classification results of SA dataset given in Table 2 and Fig. 5 clearly reflect the effectiveness of the proposed model. The spatial window considered is similar to the IN and UP datasets for a fair comparison. The average test loss and test accuracy of the proposed model attained using testing data are 0.027% and 99.99%, respectively. In the present experiment, it is also observed that as the number of training and test samples is increasing the test loss is decreasing along with an increase in the test accuracy.
The experimental results reveal the superiority of the proposed model among all the compared models which are commonly used for HSI classification. The training process for the proposed method on aforementioned dataset nearly converges in almost 20 epochs as clearly shown in Fig. 6. Therefore, early stopping criteria may be considered during the training procedure, in order to reduce computational cost, without deteriorating classification performance.
The computational efficiency of the proposed SSNET in terms of normalised training and testing time is shown in Fig. 7a and b, respectively. Figure 7 shows that the relative training and testing time follow almost same pattern on all the test datasets and are proportional to the size of the dataset. Among all compared methods, 3D-CNN takes the maximum time, whereas SVM takes the minimum time. As anticipated, the proposed model shows its efficiency over the HybridSN model both in training and testing phases for all the tested datasets. Hence, from the experimental results, as given in Table 2 and Fig. 7, it can be concluded that the proposed SSNET provides more accurate classification result with a moderate computation time and is certainly an improvement over the existing HybridSN model.
5 Conclusion
Classification is an essential part in remotely sensed HSI analysis. Therefore, a novel classification architecture, SSNET is proposed that combines spectral–spatial information of HSI in the form of 3D and 2D convolutions, respectively that includes SPP for generating spatial features in different scales. As the SPP is more robust to object distortions, it is introduced in two-dimensional local convolutional filters for HSI classification. SPP layer generates a fixed feature vector output that reduces the number of trainable parameters without adversely affecting the classification performance. The experiments are carried out over three benchmark datasets and compared with recent state-of-the-art methods. Experimental results confirm the superiority of the proposed SSNET model in terms of classification accuracy and execution time among other tested methods. This encourages exploration of the proposed model on other hyperspectral datasets in future to further check its effectiveness. As future work, the pooling strategy of the SPP layer can be improved and the parameters used can be more optimised so as to make the architecture more efficient. The proposed model deals with remote sensing image classification, particularly for hyperspectral imagery. However, with a nominal modification, the proposed architecture can also be applied in multispectral image classification.
References
Chang C (2007) Hyperspectral data exploitation: theory and applications. Wiley, New York
Ghamisi P, Yokoya N, Li J et al (2017) Advances in hyperspectral image and signal processing: a comprehensive overview of the state of the art. IEEE Geosci Remote Sens Mag 5:37–78. https://doi.org/10.1109/MGRS.2017.2762087
Mishra NB, Crews KA (2014) Mapping vegetation morphology types in a dry savanna ecosystem: integrating hierarchical object-based image analysis with random forest. Int J Remote Sens 35:1175–1198. https://doi.org/10.1080/01431161.2013.876120
Cheng G, Han J, Lu X (2017) Remote sensing image scene classification: benchmark and state of the art. Proc IEEE 105:1865–1883. https://doi.org/10.1109/JPROC.2017.2675998
Chen Y, Liu L, Gong Z, Zhong P (2017) Learning CNN to pair UAV video image patches. IEEE J Sel Top Appl Earth Obs Remote Sens 10:5752–5768. https://doi.org/10.1109/JSTARS.2017.2740898
Horig B, Kuhn F, Oschutz F, Lehmann F (2001) HyMap hyperspectral remote sensing to detect hydrocarbons. Int J Remote Sens 22:1413–1422. https://doi.org/10.1080/01431160120909
Makantasis K, Karantzalos K, Doulamis A, Doulamis N (2015) Deep supervised learning for hyperspectral data classification through convolutional neural networks. In: 2015 International geoscience and remote sensing symposium. pp 4959–4962. https://doi.org/10.1109/IGARSS.2015.7326945
Lixin G, Weixin X, Jihong P (2015) Segmented minimum noise fraction transformation for efficient feature extraction of hyperspectral images. Pattern Recognit 48:3216–3226. https://doi.org/10.1016/j.patcog.2015.04.013
Camps-valls G, Tuia D, Bruzzone L, Benediktsson JA (2013) Advances in hyperspectral image classification. IEEE Signal Process Mag 31:45–54. https://doi.org/10.1109/MSP.2013.2279179
Yang J, Qian J (2018) Hyperspectral image classification via multiscale joint collaborative representation with locally adaptive dictionary. IEEE Geosci Remote Sens Lett 15:112–116. https://doi.org/10.1109/lgrs.2017.2776113
Fang L, He N, Li S et al (2018) A new spatial-spectral feature extraction method for hyperspectral images using local covariance matrix representation. IEEE Trans Geosci Remote Sens 56:3534–3546. https://doi.org/10.1109/TGRS.2018.2801387
Fu Z, Qin Q, Luo B et al (2019) A local feature descriptor based on combination of structure and texture information for multispectral image matching. IEEE Geosci Remote Sens Lett 16:100–104. https://doi.org/10.1109/LGRS.2018.2867635
Melgani F, Bruzzone L (2004) Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans Geosci Remote Sens 42:1778–1790. https://doi.org/10.1109/TGRS.2004.831865
Ham JS, Chen Y, Crawford MM, Ghosh J (2005) Investigation of the random forest framework for classification of hyperspectral data. IEEE Trans Geosci Remote Sens 43:492–501. https://doi.org/10.1109/TGRS.2004.842481
Marpu PR, Pedergnana M, Dalla Mura M et al (2013) Automatic generation of standard deviation attribute profiles for spectral-spatial classification of remote sensing data. IEEE Geosci Remote Sens Lett 10:293–297. https://doi.org/10.1109/LGRS.2012.2203784
Kang X, Li S, Benediktsson JA (2013) Spectral–spatial hyperspectral image classification with edge-preserving filtering. IEEE Trans Geosci Remote Sens 52:2666–2677. https://doi.org/10.1109/TGRS.2013.2264508
Kang X, Li S, Fang L et al (2015) Extended random walker-based classification of hyperspectral images. IEEE Trans Geosci Remote Sens 53:144–153. https://doi.org/10.1109/TGRS.2014.2319373
Li S, Song W, Fang L et al (2019) Deep learning for hyperspectral image classification: an overview. IEEE Trans Geosci Remote Sens 57:6690–6709. https://doi.org/10.1109/TGRS.2019.2907932
Shi C, Pun CM (2018) Superpixel-based 3D deep neural networks for hyperspectral image classification. Pattern Recognit 74:600–616. https://doi.org/10.1016/j.patcog.2017.09.007
Nogueira K, Penatti OAB, dos Santos JA (2017) Towards better exploiting convolutional neural networks for remote sensing scene classification. Pattern Recognit 61:539–556. https://doi.org/10.1016/j.patcog.2016.07.001
Ribeiro M, Lazzaretti AE, Lopes HS (2018) A study of deep convolutional auto-encoders for anomaly detection in videos. Pattern Recognit Lett 105:13–22. https://doi.org/10.1016/j.patrec.2017.07.016
Salakhutdinov R, Hinton G (2009) Deep Boltzmann machines. J Mach Learn Res 5:448–455
Hinton GE, Osindero S, Teh Y-W (2006) A fast learning algorithm for deep belief nets. Neural Comput 18:1527–1554. https://doi.org/10.1162/neco.2006.18.7.1527
Paoletti ME, Haut JM, Fernandez-Beltran R et al (2019) Capsule networks for hyperspectral image classification. IEEE Trans Geosci Remote Sens 57:2145–2160. https://doi.org/10.1109/TGRS.2018.2871782
Niu Z, Liu W, Zhao J, Jiang G (2019) DeepLab-based spatial feature extraction for hyperspectral image classification. IEEE Geosci Remote Sens Lett 16:251–255. https://doi.org/10.1109/LGRS.2018.2871507
Paoletti ME, Haut JM, Fernandez-Beltran R et al (2019) Deep pyramidal residual networks for spectral-spatial hyperspectral image classification. IEEE Trans Geosci Remote Sens 57:740–754. https://doi.org/10.1109/TGRS.2018.2860125
Ma X, Fu A, Wang J et al (2018) Hyperspectral image classification based on deep deconvolution network with skip architecture. IEEE Trans Geosci Remote Sens 56:4781–4791. https://doi.org/10.1109/IGARSS.2016.7729850
Li Y, Xie W, Li H (2017) Hyperspectral image reconstruction by deep convolutional neural network for classification. Pattern Recognit 63:371–383. https://doi.org/10.1016/j.patcog.2016.10.019
Chen Y, Li C, Ghamisi P et al (2017) Deep fusion of remote sensing data for accurate classification. IEEE Geosci Remote Sens Lett 14:1253–1257. https://doi.org/10.1109/LGRS.2017.2704625
He K, Gkioxari G, Dollar P, Girshick R (2017) Mask R-CNN. In: Proceedings of the IEEE international conference on computer vision. pp 2980–2988. https://doi.org/10.1109/ICCV.2017.322
Kang X, Zhuo B, Duan P (2019) Dual-path network-based hyperspectral image classification. IEEE Geosci Remote Sens Lett 16:447–451. https://doi.org/10.1109/LGRS.2018.2873476
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE computer society conference on computer vision and pattern recognition. pp 770–778. https://doi.org/10.1109/CVPR.2016.90
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: 2017 IEEE conference on computer vision and pattern recognition (CVPR). pp 2261–2269. https://doi.org/10.1109/CVPR.2017.243
Chen Y, Zhao X, Jia X (2015) Spectral-spatial classification of hyperspectral data based on deep belief network. IEEE J Sel Top Appl Earth Obs Remote Sens 8:2381–2392. https://doi.org/10.1109/JSTARS.2015.2388577
Chen Y, Zhu L, Ghamisi P et al (2017) Hyperspectral images classification with gabor filtering and convolutional neural network. IEEE Geosci Remote Sens Lett 14:2355–2359. https://doi.org/10.1109/LGRS.2017.2764915
Zhong Z, Li J, Luo Z, Chapman M (2018) Spectral-spatial residual network for hyperspectral image classification: a 3-D deep learning framework. IEEE Trans Geosci Remote Sens 56:847–858. https://doi.org/10.1109/TGRS.2017.2755542
Roy SK, Krishna G, Dubey SR, Chaudhuri BB (2019) HybridSN: exploring 3-D-2-D CNN feature hierarchy for hyperspectral image classification. IEEE Geosci Remote Sens Lett 17:277–281. https://doi.org/10.1109/lgrs.2019.2918719
Yue J, Mao S, Li M (2016) A deep learning framework for hyperspectral image classification using spatial pyramid pooling. Remote Sens Lett 7:875–884. https://doi.org/10.1080/2150704X.2016.1193793
Li N, Wang C, Zhao H et al (2018) A novel deep convolutional neural network for spectral-spatial classification of hyperspectral data. Int Arch Photogramm Remote Sens Spat Inf Sci - ISPRS Arch 42:897–900. https://doi.org/10.5194/isprs-archives-XLII-3-897-2018
Hu W, Huang Y, Wei L et al (2015) Deep convolutional neural networks for hyperspectral image classification. J Sens 2015:1–12. https://doi.org/10.1155/2015/258619
Khan S, Rahmani H, Shah SAA, Bennamoun M (2018) A Guide to convolutional neural networks for computer vision. Morgan & Claypool Publishers, San Rafael
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv Prepr arXiv14091556 1–14
Han X, Zhong Y, Cao L, Zhang L (2017) Pre-trained alexnet architecture with pyramid pooling and supervision for high spatial resolution remote sensing image scene classification. Remote Sens 9:848. https://doi.org/10.3390/rs9080848
Ben Hamida A, Benoit A, Lambert P, Ben Amar C (2018) 3-D deep learning approach for remote sensing image classification. IEEE Trans Geosci Remote Sens 56:4420–4434. https://doi.org/10.1109/TGRS.2018.2818945
Acknowledgements
The authors acknowledge the support of CGM RCs, NRSC, ISRO, and Head (applications), RRSC-East, NRSC, ISRO, for carrying out the present work. The authors also acknowledge the collaboration extended by VC, MAKAUT, towards the work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Paul, A., Bhoumik, S. & Chaki, N. SSNET: an improved deep hybrid network for hyperspectral image classification. Neural Comput & Applic 33, 1575–1585 (2021). https://doi.org/10.1007/s00521-020-05069-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-020-05069-1