Summary
Background
Colorectal cancer (CRC) is among the most widespread malignancies in the world. MicroRNA (miRNA) has been identified as an important modulator of the biological processes of the cells. This group of noncoding RNAs also has a pivotal function in the growth and development of human cancers, including CRC. Among these miRNAs, miR-196, miR-132, miR-146a, and miR-134 have fundamental impacts on the regulation of cancers. The current study aimed to investigate the involvement of these miRNAs in CRC patients.
Methods
In this study, 50 pairs of tumor and tumor margin samples of CRC patients were investigated to assess the expression levels of miR-196, miR-132, miR-146a, and miR-134 in this cancer. For this purpose, firstly, quantitative real-time PCR (qRT-PCR) was applied. Also, KRAS mutation and clinicopathological characteristics of the CRC patients were analyzed in the study groups.
Results
The findings demonstrated the overexpression of miR-196 (P-value = 0.0045) and miR-146a (P-value = 0.0033) in tumor tissues compared to controls. Conversely, the expression levels of miR-132 (P-value = 0.00032) and miR-134 (P-value < 0.0001) were downregulated in tumor tissues. Also, miR-146a was the only miRNA with significant expression change in the case of the KRAS gene mutation. Interestingly, the expression ratio of these miRNAs was significantly associated with some of the clinicopathological features of the patients, such as lymph node and distant metastases.
Conclusion
Our data demonstrated that these miRNAs appear to be promising novel biomarkers for early diagnosis of CRC and may pave the way for the future establishment of novel therapeutic options for CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is the third most widespread cancer and its rate is increasing especially in developing countries [1, 2]. The CRC is a heterogeneous cancer, which happens after the increase of tumor cells in the colon, rectum, and appendix [3]. The two main factors that affect the occurrence of this cancer are environmental and genetic [4]. Prevalence and mortality rates reported for CRC are about 1,400,000 and 700,000 cases per year, respectively [1]. Overall, about 40–50% of patients die because of distant metastasis, thus irrespective of the diagnosis and standard therapy of this type of cancer, patients’ survival rate depends on tumor stage at the diagnosing time [5, 6]. MicroRNAs (miRNAs) are a group of non-coding RNA that contains 18–25 nucleotides that influence post-translational gene expression [7, 8]. It is approximated that the miRNAs control about 30% of the genome encoding various proteins [9].
Although some therapeutic options such as surgery and chemotherapy are available for CRC patients, the prophecy of the disease is inauspicious, and in numerous cases who have undergone surgery, the cancer recurrence and metastasis happen. Late diagnosis of this malignancy is one of the great challenges, and clinically important issue. Therefore, molecular studies that result in the recognition of biological biomarkers should have a clinical emphasis. To date, expansive research has shown that miRNAs are unique and helpful biomarkers for early determination and therapy of CRC [10]. The effective functioning of miRNAs in cancer is confirmed through the interaction of miRNAs with targeted genes. This has effects on various cancer properties including apoptosis, differentiation, and cell proliferation [11]; however, for tumor classification in various groups with distinct features, such as etiology and cancer cell types, miRNAs are even more accurate than messenger RNA (mRNA) [10]. In cancer samples, many miRNAs are abnormally expressed which is indicated by miRNA structure and their functions. The stages of cancer are in association with miRNAs expression patterns [10].
Some variants of miRNAs have recently been introduced as potential biomarkers. Our target in this study is to come up with more precise identification of expression patterns of 4 selected miRNAs (miR-196, miR-132, miR-146a, and miR-134). MiR-196 is reportedly overexpressed in various human malignancies, including CRC [12]. It has been suggested that miR-196a and miR-196b participate in tumor progression and tumorigenesis of CRC [13]. Studies revealed that miR-132 is downregulated in CRC patients with metastases and that the expression of miR-132 particularly inhibits cell migration in CRC cell lines [14]. MiR-132 also has a key function in both prostate and pancreatic cancers [15, 16]. Moreover, miR-132 represents a promising biomarker for the targeted therapy of CRC [17]. MiR-146a has been particularly studied due to its function in tumor progression. MiR-146a has been indicated to be overexpressed in tumor samples and cancer cell lines [18]. Dysregulated miR-146a, however, is implicated in tumorigenesis. Reports demonstrated the downregulation of miR-146a in various malignancies [19,20,21]. MiR-134 was determined to be capable to suppress the migration of cancer cells. It has been investigated that miR-134 induces apoptosis in CRC cells as well [22]. Subsequently, our main goal was to clarify the selected miRNAs expression profiles in tumor tissue and marginal tissue (i.e. control samples). Selective targeting of these miRNAs in combination with standard treatments of CRC may be useful for achieving better results in CRC therapy [23]. In the current study, we also studied the expression changes concerning the mutations detected in KRAS. Moreover, we aimed to do a wide comparison of clinical relevance with these expression patterns in all these miRNAs.
Material and methods
Study population
The study included 50 patients with primary CRC diagnosed at Imam Reza Hospital in Tabriz, Iran during 2017–2018. To achieve a genetically pure population, all the patients were selected from those native to East Azerbaijan, northwest Iran. Patients who received chemotherapy and radiotherapy were excluded during sample gathering. The clinicopathological parameters of the subjects are presented in Table 1. The samples from the primary tumor and the matched normal marginal tissues were gathered from each case. All collected tissues were histologically confirmed to be an adenocarcinoma of the colon. These samples were immediately frozen in liquid nitrogen and stored at −80 °C until RNA extraction and subsequent implementations.
DNA extraction
DNA from tissues was isolated using the Tripure isolation kit (Roche, Germany) considering the manufacturer’s manual. The DNA quality and purity were evaluated using a NanoDrop spectrophotometer (Thermo Fisher Scientific).
KRAS mutation detection
High resolution melting (HRM) analysis was done by Step One plus real-time PCR system (Applied Biosystems, Foster City, CA, USA). Mutation detection for exon2 of the KRAS gene in tumor samples was carried out using gene-specific primers and the SYBR green master mix (Takara Bio, Shiga, Japan). All susceptible samples were sent to sequencing to confirm the mutation. The primer sequences are available on request.
RNA isolation and transcriptase reactions
As DNA, total RNA content of tumor and the matched normal tissues was isolated using Tripure isolation kit (Roche) considering the manufacturer’s manual. A NanoDrop Spectrophotometer was used to check the RNA quality and purity. Besides, in order to generate complementary DNA (cDNA) of extracted RNAs, the TAKARA cDNA synthesis kit (Takara Bio, Shiga, Japan) was used according to the instruction guide.
Quantitative real-time PCR
Quantitative analysis was done by the real-time PCR system [24]. Relative quantification of the expression level of genes from tumor and normal marginal tissue samples was carried out using gene-specific primers and the SYBR green master mix. Expression of the U6 gene was also measured in each sample as the housekeeping gene. Primers were designed using oligo 7 software. The average score of duplicated Ct values was measured for each sample and the comparative Ct (2ΔΔCt) method was used to determine the relative expression level of target genes [25]. Real-time PCR primers are represented in Table 2.
Description analysis of miR-196, miR-132, miR-146a, and miR-134 as tumor markers
The expression analysis of miR-196, miR-132, miR-146a, and miR-134 was done by a randomization test applying the Relative Expression Software Tool (REST) [26]. The receiver operating characteristic (ROC) curve was plotted to assess the specificity and sensitivity of predicting CRC from normal tissues by these miRNA expression ratios. For this purpose, the sensitivity in the function of the specificity at the different cut-off grades was studied using Sigma Plot 12.5 software. This plot is a graph of the true positive rate (TPR) and false positive rate (FPR) for different descriptors threshold [26]. Hence, the vertical axis in the ROC curve represents TPR and the horizontal axis indicates FPR. The bigger vertical axis values for a hypothetical horizontal axis value show a better description of a descriptor.
Statistical analysis
GraphPad Prism 6 (Graph Pad Software Inc., San Diego, CA, USA) was used for statistical analysis. The data were shown as mean ± standard deviation (SD) based on three independent experiments. An independent sample t-test was carried out to determine the difference in gene expression level between primary tumor tissues and corresponding normal tissues. For evaluation of the association between the clinical feature of the patients with relative expression of genes, a cross tab (Eta) analysis was performed. P < 0.05 was considered as a statistically significant level.
Results
In this study, the HRM technique was used to identify KRAS gene mutations and 16 subjects with mutations in the KRAS gene were detected. The expression of our target miRNAs was assessed both in tumor and marginal tissues and their expression concerning KRAS mutation was analyzed. Moreover, the expression patterns were compared in association with clinical features such as age, sex, tumor cell differentiation, and tumor stages.
MiR-196 expression in CRC
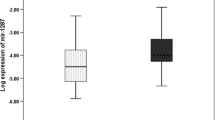
We determined the expression of miR-196a in 50 CRC cancer tissues compared with adjacent tumor-free normal tissues. The U6 gene expression was used as an internal control for normalizing these data. Our data revealed the overexpression of miR-196 in tumor tissues (P-value = 0.0045). Also, the relationship analysis of miR-196 expression with clinicopathological traits of patients showed a significant correlation between the expression of miR-196a with lymph node (P‑value = 0.042) and distant metastases (P‑value < 0.0001). Due to the KRAS mutation, there is no significant change in KRAS positive samples in comparison to KRAS negative in our study (Fig. 1).
MiR-132 expression in CRC
According to our data, the expression ratio of miR-132 is downregulated in tumor samples compared to tumor-free marginal normal tissues (P‑value = 0.00032). In relationship analysis, we found that there is a significant association between the expression of miR-132 and tumor stages (P‑value = 0.0067). Similar to miR-196a expression, the expression level of miR-132 was not related to the KRAS mutation too (Fig. 2).
MiR-146a expression in CRC
Our data revealed the up-regulation of miR-146 in tumor tissues in comparison to matched tumor-free normal tissues (P‑value = 0.0033). In relationship analysis, we discovered a significant correlation between miR-146a expression level and distant metastases (P‑value = 0.0016). In our study, miR-RNA146a was the only microRNA with significant expression change in the case of the KRAS gene mutation. This miRNA expression was significantly upregulated in KRAS positive samples in compression to KRAS negative samples (P‑value = 0.0102) (Fig. 3).
MiR-134 expression in CRC
We ascertained the expression of miR-134 in CRC participant’s tissues in comparison with marginal tissues of the subjects and the results demonstrated significant downregulated expression of miR-134 in tumor samples (P‑value < 0.0001). Also, relationship analysis showed the correlation among the expression of this gene and tumor stages (P‑value = 0.0026), lymph node (P‑value = 0.021), and distant metastasis (P‑value = 0.035) of samples. On the other hand, in the case of KRAS mutation, this miRNA was not significantly changed among the two studied groups (Fig. 4). All the relevant clinical data regarding the entitled miRNAs expression profiles are presented in Table 3 respectively.
The capability of miRNAs for the diagnosis of CRC
The ROC curve was applied to estimate the sensitivity and specificity of miR-196, miR-132, miR-146a, and miR-134 as candidates and promising biomarkers for CRC. The ROC curve data are shown in Fig. 5. The vertical axis in the ROC curves of Fig. 5 represents the sensitivity, which is the ability of a test to correctly identify those with the disease (TPR [true positive rate]) and the horizontal axis indicates specificity, which is the ability of the test to correctly identify those without the disease (FPR [false positive rate]). The bigger vertical axis values for a hypothetical horizontal axis value show a better description of a descriptor. Area under the curve values for both miR-196 and miR-132 were 0.7551 (P-value = 0.02164), miR-146a was 0.1164 (P-value = 0.1585), and miR-134 was 0.7396 (P-value = 0.03786). The detailed results of the statistical analysis of ROC curves for diagnostic evaluation are presented in Table 4.
Receiver operating characteristic (ROC) curve for miR-196a (a), miR-132 (b), miR-146 (c) and miR-134 (d) which differentiates CRC cases from normal controls. Area under the curve values for both miR-196 and miR-132 were 0.7551 (P-value = 0.02164), miR-146a was 0.6627 (P-value = 0.1585), and miR-134 was 0.7396 (P-value = 0.03786)
Discussion
The activation of the immune system, regulation of cholesterol homeostasis, ontogenesis, and other numerous functions are some of the biological processes that are controlled by miRNAs [27,28,29]. The patterns of expression of miRNAs are distinct in tumor tissues and body fluids like saliva, serum, urine, and plasma [27]. The variations among the cancer cells types and their surrounding stromal tissue could be results of diversity in miRNAs expression profiles in various cancers [10]. These expression patterns can shed light on the application of miRNAs as informative agents for diagnosis and prognosis. For this reason, we concentrated on the expression patterns of 4 hotspot miRNAs (miR-196, miR-132, miR-146a, and miR-134) in the tissue of CRC patients regarding their KRAS mutational background. It has been reported that 12 miRNAs (including miR-196a and miR-134) are overexpressed and 8 miRNAs (including miR-146a) are downregulated in the stool of CRC patients [30]. Also, Iannone et al. could differentiate CRC incidences from healthy controls by analyzing these 20 miRNAs [31]. They also managed to clarify different TNM stages with high specificity and sensitivity [31].
MiR-196 upregulation has been designated in several malignancies including CRC [32]. Additionally, it has been demonstrated that miR-196 is upregulated in colon cancer tissues [13]. We also designated the upregulation of miR-196 in tumor samples in comparison with adjacent tumor-free normal tissues. In research by Ge et al., it has been indicated that miR-196 is significantly highly expressed in CRC compared to corresponding normal colorectal tissue. The results were also aligned with the prognosis in CRC subjects. These observations revealed that mir-196 may be correlated with a more aggressive clinical outcome in patients with CRC [33]. There are also articles demonstrating that miR-196a may have pro-oncogenic effects in CRC [13, 32]. Despite all the studies, miR-196’s mechanisms of action in different cancers are unclear. It largely depends on the molecules that miR-196 targets. If miR-196 shows a dominant effect on the blockade of oncogenic molecules, this means that miR-196 is playing a tumor suppressor role but if miR-196 targets the tumor suppressors, it mainly will perform oncogenic effects [34].
One study claimed the role of miR-196 in CRC because of its overexpression. Besides, the expression of miR-196 was meaningfully linked with the stage as well as lymph node and distant metastasis. The same study indicated that the expression patterns were not associated with depth of invasion, size, location, differentiation, gender, and age [35]. We observed that there is a statistically significant correlation between the expression of miR-196a with lymph node metastasis and distant metastasis. One study provided evidence that miR-132 acts as a tumor suppressor through enhancing apoptosis as well as inhibiting cancer cell proliferation and migration [36]. In pancreatic cancer progression, however, the expression of miR-132 suppressed the proliferation and colony formation of cancer cells [37]. Taken together, these findings illustrated that miR-132 behaves as a tumor suppressor [16]. Our results revealed that miR-132 is downregulated in tumor tissues compared to tumor-free marginal tissues. Increased expression of miR-132 reduces the incidence of colitis-related tumors [38]. MiR-132 can modulate cell proliferation, differentiation, apoptosis, metabolism, and growth [39, 40]. Confirming our results, Zheng et al. also reported that miR-132 was downregulated in CRC patients with distant metastases. This downregulation was correlated with adverse prognosis in CRC patients and more aggressive tumor phenotypes [14]. Moreover, we found that there is a significant association between the expression of miR-132 and tumor stages.
MiR-146a expression can reduce the proliferation of many types of cancer cells [41, 42] except melanoma, cervix, and thyroid carcinoma in which miR-146a is upregulated [19,20,21]. Our data revealed the upregulation of miR-146 in tumor tissues in comparison to matched tumor-free tissues. Several studies suggested that miR-146a is potentially involved in CRC. Pizzini et al. indicated that miR-146a is downregulated in the metastasis tumor tissue in comparison to primary CRC tissue. On the contrary, as mentioned before Ahmed et al. reported that decreased miR-146 expression in the stool of CRC subjects in comparison with normal subjects [30, 43]. Taken together it has been indicated that miR-146a may be included in CRC development.
Previous studies have shown that rs2910164 polymorphism in miR-146a may be related to CRC risk [30, 43, 44]. Wang et al. observed that miR-146a was reduced in cancer tissue and that it was interestingly related to the TNM stage, which suggested the potential connection between miR-146a and the disease pathogenesis [45]. We discovered a significant association between miR-146a expression and distant metastasis. Besides miR-RNA146a was the only miRNA among the selected four miRNAs that had a significant expression change regarding KRAS gene mutation. The expression was significantly upregulated in KRAS positive samples in comparison to KRAS negative samples. The expression of miR-134 is downregulated in glioma tissues [46] as well as breast cancer tissues [47]. A study also revealed that miR-134 can be considered as a marker for oral cell carcinoma diagnosis [48]. As mentioned above miR-134 is upregulated in the stool of CRC patients [30, 31]. Our results have shown the significant downregulated expression of miR-134 in tumor tissues in comparison with marginal tissues of the subjects. To the best of our knowledge, miR-134’s association with CRC and its clinical relevance remains unclear. We observed a meaningful correlation among the expression level of this gene and tumor stages as well as lymph node metastasis. According to our study, these miRNAs are among the most important miRNAs in CRC development and metastasis, but, of course, other miRNAs must be considered in future experimental and clinical studies.
Despite the elevating number of successful therapeutic options for CRC therapy, the clinical achievements concerning inhibition of recurrence and metastasis are still poor. This results in low long-term survival rates. Over the past two decades, a growing list of biomarkers associated with the prognosis and progression of CRC have been attributed; however, the majority of these biomarkers have been reduced in clinical validation studies. Thus, there is an urgent demand for novel biomarkers to diagnose early metastasis as well as to predict recurrence in CRC subjects. Although the expression patterns of several miRNAs in CRC are partly studied, the relationship between these expression profiles and the clinical features of CRC remains to be determined; however, in the early research stage, miRNAs present great potential for detection and therapy. Therefore, further investigations regarding their expression profiles in CRC tissue will shed a light on their role as biomarkers for early diagnosis and as agents for monitoring the disease.
Conclusion
The diagnosis and therapeutic outcomes for cancer could be improved by the availability of miRNAs. Moreover, some of the biological and clinical characteristics of the tumors are associated with the expression of miRNAs. Subsequently, these miRNAs appear to be novel biomarkers for the diagnosis of CRC. Our data serves as a basis for further research before these miRNAs can be used as biomarkers for CRC. Detection of miRNA target molecules and demonstrating their expression patterns concerning specific mutations will help discover the mechanisms of cancer. Last but not least, taking a vital step for early detection and proper monitoring of CRC, using miRNAs as diagnostic and prognostic markers is very beneficial.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. Ca A Cancer J Clin. 2015;65:87–108.
Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–53.
Shirafkan N, Mansoori B, Mohammadi A, Shomali N, Ghasbi M, Baradaran B. MicroRNAs as novel biomarkers for colorectal cancer: new outlooks. Biomed Pharmacother. 2018;97:1319–30.
Esmailzadeh S, Mansoori B, Mohammadi A, Shanehbandi D, Baradaran B. siRNA-mediated silencing of HMGA2 induces apoptosis and cell cycle arrest in human colorectal carcinoma. J Gastrointest Cancer. 2017;48:156–63.
Corté H, Manceau G, Blons H, Laurent-Puig P. MicroRNA and colorectal cancer. Dig Liver Dis. 2012;44:195–200.
Gonzalez-Pons M, Cruz-Correa M. Colorectal cancer biomarkers: Where are we now? Biomed Res Int. 2015; https://doi.org/10.1155/2015/149014.
Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta. 2010;1803:1231–43.
Ghasabi M, Mansoori B, Mohammadi A, Duijf PH, Shomali N, Shirafkan N, et al. MicroRNAs in cancer drug resistance: basic evidence and clinical applications. J Cell Physiol. 2019;234:2152–68.
Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–78.
Mohammadi A, Mansoori B, Baradaran B. The role of microRNAs in colorectal cancer. Biomed Pharmacother. 2016;84:705–13.
Karami H, Baradaran B, Esfahani A, Estiar MA, Naghavi-Behzad M, Sakhinia M, et al. siRNA-mediated silencing of survivin inhibits proliferation and enhances etoposide chemosensitivity in acute myeloid leukemia cells. Asian Pac J Cancer Prev. 2013;14:7719–24.
Suzuki H, Takatsuka S, Akashi H, Yamamoto E, Nojima M, Maruyama R, et al. Genome-wide profiling of chromatin signatures reveals epigenetic regulation of microRNA genes in colorectal cancer. Cancer Res. 2011;71:5646–58.
Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen XM, Gao HJ. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis. 2010;11:50–4.
Bin Zheng Y, Luo HP, Shi Q, Hao ZN, Ding Y, Wang QS, et al. miR-132 inhibits colorectal cancer invasion and metastasis via directly targeting ZEB2. World J Gastroenterol. 2014;20:6515–22.
Formosa A, Lena AM, Markert EK, Cortelli S, Miano R, Mauriello A, et al. DNA methylation silences miR-132 in prostate cancer. Oncogene. 2013;32:127–34.
Zhang S, Hao J, Xie F, Hu X, Liu C, Tong J, et al. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32:1183–9.
Qin J, Ke J, Xu J, Wang F, Zhou Y, Jiang Y, et al. Downregulation of microRNA -132 by DNA hypermethylation is associated with cell invasion in colorectal cancer. Onco Targets Ther. 2015;8:3639–48.
Chae YS, Kim JG, Lee SJ, Kang BW, Lee YJ, Park JYJS, et al. A miR-146a polymorphism (rs2910164) predicts risk of and survival from colorectal cancer. Anticancer Res. 2013;33:3233–40.
Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, Muller A, et al. Signatures of MicroRNAs and selected MicroRNA target genes in human melanoma. Cancer Res. 2010;70:4163–73.
Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. Plos One. 2008;3(7):e2557.
Sun M, Fang S, Li W, Li C, Wang L, Wang F, et al. Associations of miR-146a and miR-146b expression and clinical characteristics in papillary thyroid carcinoma. Cancer Biomark. 2015;15:33–40.
Ye Q, Su L, Chen D, Zheng W, Liu Y. Astragaloside IV induced MIR-134 expression reduces EMT and increases chemotherapeutic sensitivity by suppressing CREB1 signaling in colorectal cancer cell line SW-480. Cell Physiol Biochem. 2017;43:1617–26.
Aghaee F, Pirayesh Islamian J, Baradaran B. Enhanced radiosensitivity and chemosensitivity of breast cancer cells by 2‑deoxy-D-glucose in combination therapy. J Breast Cancer. 2012;15:141.
Hajiasgharzadeh K, Tavangar SM, Javan M, Dehpour AR, Mani AR. Does hepatic vagus nerve modulate the progression of biliary fibrosis in rats? Auton Neurosci. 2014;185:67–75.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–8.
Daryabari SS, Safaralizadeh R, Hosseinpourfeizi M, Moaddab Y, Shokouhi B. Overexpression of SSH1 in gastric adenocarcinoma and its correlation with clinicopathological features. J Gastrointest Oncol. 2018;9:728–33.
Madhavan D, Cuk K, Burwinkel B, Yang R. Cancer diagnosis and prognosis decoded by blood-based circulating micro RNA signatures. Front Genet. 2013;4:116.
Thome AD, Harms AS, Volpicelli-Daley LA, Standaert DG. MicroRNA-155 regulates alpha-synuclein-induced inflammatory responses in models of Parkinson disease. J Neurosci. 2016;36:2383–90.
Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328(5985):1570–3.
Ahmed FE, Ahmed NC, Vos PW, Bonnerup C, Atkins JN, Casey M, et al. Diagnostic MicroRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genomics Proteomics. 2013;10:93–113.
Iannone A, Losurdo G, Pricci M, Girardi B, Massaro A, Principi M, et al. Stool investigations for colorectal cancer screening: from occult blood test to DNA analysis. J Gastrointest Cancer. 2016;47:143–51.
Schimanski CC, Frerichs K, Rahman F, Berger M, Lang H, Galle PR, et al. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;15:2089–96.
Ge J, Chen Z, Li R, Lu T, Xiao G. Upregulation of microRNA-196a and microRNA-196b cooperatively correlate with aggressive progression and unfavorable prognosis in patients with colorectal cancer. Cancer Cell Int. 2014;14:128.
Chen CJ, Zhang Y, Zhang L, Weakley SM, Yao Q. MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med. 2011;15:14–23.
Shen S, Pan J, Lu X, Chi P. Role of miR-196 and its target gene HoxB8 in the development and proliferation of human colorectal cancer and the impact of neoadjuvant chemotherapy with FOLFOX4 on their expression. Oncol Lett. 2016;12:4041–7.
Geng F, Wu JL, Lu GF, Liang ZP, Duan ZL, Gu X. MicroRNA-132 targets PEA-15 and suppresses the progression of astrocytoma in vitro. J Neurooncol. 2016;129:211–20.
Zhang B, Lu L, Zhang X, Ye W, Wu J, Xi Q, et al. Hsa-miR-132 regulates apoptosis in non-small cell lung cancer independent of acetylcholinesterase. J Mol Neurosci. 2014;53:335–44.
Alzahrani AM, Hanieh H, Ibrahim H‑IM, Mohafez O, Shehata T, Bani Ismail M, et al. Enhancing miR-132 expression by aryl hydrocarbon receptor attenuates tumorigenesis associated with chronic colitis. Int Immunopharmacol. 2017;52:342–51.
Xu G, Zhu H, Zhang M, Xu J. Histone deacetylase 3 is associated with gastric cancer cell growth via the miR-454-mediated targeting of CHD5. Int J Mol Med. 2018;41:155–63.
Wu J, He D, Yue B, Zhang C, Fang X, Chen H. miR-101‑1 expression pattern in Qinchuan cattle and its role in the regulation of cell differentiation. Gene. 2017;636:64–9.
Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, et al. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8:e60317.
Yao Q, Cao Z, Tu C, Zhao Y, Liu H, Zhang S. MicroRNA-146a acts as a metastasis suppressor in gastric cancer by targeting WASF2. Cancer Lett. 2013;335:219–24.
Pizzini S, Bisognin A, Mandruzzato S, Biasiolo M, Facciolli A, Perilli L, et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genomics. 2013;14:589.
Mao Y, Li Y, Jing F, Cai S, Zhang Z, Li Q, et al. Association of a genetic variant in microRNA-146a with risk of colorectal cancer: a population-based case-control study. Tumor Biol. 2014;35:6961–7.
Wang C, Guan S, Liu F, Chen X, Han L, Wang D, et al. Prognostic and diagnostic potential of miR-146a in oesophageal squamous cell carcinoma. Br J Cancer. 2016;114:290–7.
Niu CS, Yang Y, Cheng CD. MiR-134 regulates the proliferation and invasion of glioblastoma cells by reducing Nanog expression. Int J Oncol. 2013;42:1533–40.
Leivonen SK, Sahlberg KK, Mäkelä R, Due EU, Kallioniemi O, Børresen-Dale AL, et al. High-throughput screens identify microRNAs essential for HER2 positive breast cancer cell growth. Mol Oncol. 2014;8:93–104.
Liu CJ, Shen WG, Peng SY, Cheng HW, Kao SY, Lin SC, et al. MiR-134 induces oncogenicity and metastasis in head and neck carcinoma through targeting WWOX gene. Int J Cancer. 2014;134:811–21.
Acknowledgements
The authors would like to thank the Immunology Research Center, Tabriz University of Medical Sciences for providing facilities to carry out this work.
Author information
Authors and Affiliations
Contributions
MM, DS, MA, SH, and HMA performed the experiments. BB provided biological materials and reagents. MM and KH wrote the initial draft of the article. MM, DS, MA, and BB performed data analysis. BB and MP reviewed and edited the article. BB supervised the study.
Corresponding author
Ethics declarations
Conflict of interest
M. Maralani, D. Shanehbandi, M. Asadi, S. Hashemzadeh, K. Hajiasgharzadeh, H. Mashhadi Abdolahi, B. Baradaran and M. Peeters declare that they have no competing interests.
Ethical standards
All procedures performed in studies involving human participants or on human tissue were in accordance with the ethical standards of the institutional and/or national research committee (Ethics committee of Tabriz University of Medical Sciences, Tabriz, Iran) and with the 1975 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Rights and permissions
About this article
Cite this article
Maralani, M., Shanehbandi, D., Asadi, M. et al. Expression profiles of miR-196, miR-132, miR-146a, and miR-134 in human colorectal cancer tissues in accordance with their clinical significance. Wien Klin Wochenschr 133, 1162–1170 (2021). https://doi.org/10.1007/s00508-021-01933-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00508-021-01933-9