Abstract
Key message
Polyploidy (diploid to octoploid) was evidenced from seven Psidium species, besides the outcomes of the whole-genome duplication about the nuclear DNA content, DNA sequence, and distribution.
Abstract
The previous studies have reported the occurrence of polyploid species in Psidium, all deriving from the basic chromosome number x = 11, which is conserved in Myrtaceae. Here, we aimed to assess the ploidy levels of seven Psidium species and to investigate the genomic outcomes of this karyotype change. Data on chromosome number, ploidy level, nuclear DNA content, and DNA sequence (SSR markers) were sought, quantified, and compared to geographical distribution of the studied Psidium species. A euploid series based on x = 11 was evidenced, with diploid, tetraploid, hexaploid, and octoploid species. These species also differed regarding at least one of the other analyzed traits, especially the hexaploids and the octoploid in relation to the others. Diploid species show restricted geographical distribution in the Atlantic Forest, differently from the polyploid species, which occur in several biomes in Brazil. Ploidy level of the Psidium species is related with the nuclear genome size and both seems to be related with species’ geographical distribution. Besides polyploidy, the genetic changes associated with numerical chromosome shift shown in this study, which increases the knowledge about the diversification and distribution of Psidium species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyploidy, euploidy—a numerical chromosome rearrangement characterized by whole-genome duplication (Stebbins 1950; Edger and Pires 2009; Marchant et al. 2016; Spoelhof et al. 2017)—is arguably the most important karyotype change that increases the diversification and drive speciation in plants (Edger and Pires 2009; Madlung 2013; Alix et al. 2017; Slijepcevic 2018). Polyploidy directly leads to extensive genomic (of the chromosome number to the DNA sequence), epigenetic, and transcriptomic changes (Dhooghe et al. 2011; Marchant et al. 2016; Segraves 2017; Spoelhof et al. 2017). Due to these changes, polyploid taxa can exhibit new phenotypes or even attributes (morphologic, phenologic, physiologic, and reproductive) in relation to their counterparts (Levin 2002; Dhooghe et al. 2011; Segraves 2017; Spoelhof et al. 2017; Shu et al. 2018), within only one or few generations (Otto and Whitton 2000; Beest et al. 2012).
These novelties potentially influence the ecology (Otto and Whitton 2000; Soltis and Soltis 2000; Segraves 2017), as the increased ecological tolerance, allowing the polyploids overlap the niche of their ancestors (Marchant et al. 2016), as well as to colonize new habitats (Stebbins 1985; Soltis and Soltis 2000; Segraves 2017; Spoelhof et al. 2017). This hypothesis is supported, among others, by several studies on polyploid cytotypes of the genera Fragaria (Hancock and Bringhurst 1981), Eupatorium (Watanabe 1986), Plantago (Van Dijk and Bakx-Schotman 1997), and Aster (Münzbergová 2007). In these studies, diploids are found to have restricted spatial distributions, contrary to the widely dispersed polyploids. Cytogeographical studies have shown that diploids and polyploids often occupy different regions of the landscape along ecological gradients, such as moisture, whereby polyploids are generally capable of occurring in drier habitats when compared to diploids (Kay 1969; Watanabe 1986; Maherali et al. 2009; Treier et al. 2009).
The polyploidy was estimated to account for the speciation of 2–4% of today’s flowering plant species, with woody plants representing a lower fraction (Otto and Whitton 2000). After a century of study (Barker et al. 2016), the polyploidy has been identified in several taxa, mainly crops (Alix et al. 2017), being currently considered that the whole-genome duplication probably occurred in the ancestor of all angiosperm plants (Alix et al. 2017; Spoelhof et al. 2017). Nevertheless, the knowledge about the role of this genomic change in the diversification, speciation, and ecology in tropical lineages is still scarce (Husband et al. 2013; Spoelhof et al. 2017), especially for trees.
One of the polyploidy outcomes is the nuclear DNA content change (Kron et al. 2007; Slijepcevic 2018), which can promote modifications in the size and/or number of vegetative and/or reproductive structures of a new plant when compared to its ancestors (Stebbins 1950). These changes can affect fitness, and include alterations in growth rates, seed production, the so-called hybrid vigor or heterosis, coupled with effective dispersal, and higher germination rates (Baker 1965; Bretagnolle and Lumaret 1995; Otto and Whitton 2000; Soltis and Soltis 2000; Comai 2005; Sattler et al. 2016).
In the Angiosperm family Myrtaceae, polyploidy has mainly been evidenced by chromosome counting in fleshy-fruited species of the clade Myrtoideae (Andrade and Forni-Martins 1998; Costa and Forni-Martins 2006a, b, 2007). This includes the Neotropical Psidium, a monophyletic group (Lucas et al. 2007; Rivero et al. 2012; Murillo et al. 2012) with rapid diversification rates (Vasconcelos et al. 2017). The genus comprises at least 100 species, distributed from Mexico and the Caribbean to Argentina and Uruguay (WCSP 2017). Sixty percent of the Psidium species occur in Brazil, being found in different biomes, such as evergreen tropical rain forests (Amazon and Atlantic Forest), savannas (Cerrado), and semi-arid forests (Caatinga) (BFG 2015). The large geographical distribution of many Psidium species is suggested to result from their superior competitive ability (Soares-Silva and Proença 2006). Staggemeier et al. (2016) have demonstrated that species of Myrtaceae exhibit a wide variety of fruit morphology and phenological strategies that support a variety of frugivorous sizes while retaining overall ecosystem functionality. Polyploidy can also explain this success. Psidium species have chromosome numbers of 2n = 22, 33, 44, 55, 66, 77, and 88, deriving from the basic chromosome number x = 11 (Atchison 1947; Andrade and Forni-Martins 1998; Bolkhovskikh et al. 1969; Goldblatt 1981; Goldblatt and Johnson 1996; Moore 1977; Costa et al. 2008; Marques et al. 2016). Despite the existence of many polyploidy events in Psidium, the origin of polyploidy and its effects on the species’ diversification and geographical distribution have not been investigated so far. Besides, the possible relationships among ploidy and geographical ranges in Psidium may offer a better understanding about how speciation affects the dispersal and establishment abilities of tropical species.
The previous studies have suggested the relationships between polyploidy and different measures of ecological ‘success’ (Stebbins 1947, 1950; Ehrendorfer 1980; Lewis 1980; Thompson and Lumaret 1992; Soltis and Soltis 2000; Alix et al. 2017; Segraves 2017). However, these hypotheses have rarely been tested (Segraves 2017) in the tropics, and the factors that contribute to the success of polyploids have seldom been identified. The consequences of whole-genome duplication on species’ geographical distribution remain marginally explored. Therefore, the main goal of this study was to expand the knowledge about the Psidium genome, including the chromosome number, nuclear DNA content, DNA sequence, and the geographic distribution of six species indigenous to Brazil.
Materials and methods
Sampling
This study included six Psidium species indigenous to Brazil, as well as the naturalized P. guajava L. The selection was based on differences in the geographical distribution of the species using the BFG (2015) database. Five of the indigenous species occur in two or more Brazilian biomes, and two are restricted to the Brazilian Atlantic Forest: P. guineense Sw. is widely distributed across the different Brazilian biomes (except the Pampas); P. myrtoides O. Berg and P. cattleyanum Sabine occur in the Cerrado, Caatinga and Atlantic Forest; P. longipetiolatum D. Legrand is found in the Cerrado and Atlantic Forest; and P. oblongatum O. Berg and P. cauliflorum Landrum & Sobral are restricted to the Atlantic Forest. Young and healthy leaves, from five individuals for each species, were collected in field expeditions and stored in silica gel for molecular analysis. As the number of fruits varied between the Psidium individuals, all fruits were collected for flow cytometry and cytogenetic analyses, being: 29 for P. guajava, nine for P. oblongatum, six for P. cauliflorum, 55 for P. guineense, 50 for P. cattleyanum, 38 for P. myrtoides, and 18 for P. longipetiolatum. One voucher per population was collected, dried (Peixoto and Maia 2013), and deposited at the RB herbarium of the Botanical Garden of Rio de Janeiro: P. guajava (Tuler, A 445), P. oblongatum (Carrijo, T 2105), P. cauliflorum (Tuler, A 511), P. guineense (Tuler, A 487), P. cattleyanum (Tuler, A 427), P. myrtoides (Tuler, A 451), and P. longipetiolatum (Tuler, A 450).
In vitro establishment, nuclear 2C value measurement, and chromosome number determination
Seeds of the seven Psidium species and Solanum lycopersicum Mill ‘Stupické’ (reference standard, 2C = 2.00 pg; Praça-Fontes et al. 2011) were disinfested under laminar flow hood (Oliveira et al. 2013) and inoculated into flasks containing MS medium (Murashige and Skoog 1962) supplemented with 3.0% (w/v) sucrose and 0.7% (w/v) type A agar, pH 5.7. The flasks were maintained at 25 °C under a 16/8 h light/dark regimen, with 36 µmol m−2 s−1 light radiation provided by two fluorescent lamps (20 W, Osram®). As performed by Marques et al. (2016), from the in vitro plantlets, leaves were collected for 2C value measurement, and roots were collected for 2n chromosome number determination. The use of in vitro plantlets was important owing to the unavailability of fruits during all months of the year in which the study was executed.
Nuclear 2C value measurement by flow cytometry is relevant to screen the polyploid taxa and record ploidy changes, as well as the increase and decrease in genome size that occurs after this event (Bennetzen and Kellogg 1997; Petrov 2002; Soltis et al. 2003). Therefore, leaf fragments of S. lycopersicum ‘Stupické’ plantlets (reference standard) grown in vitro and of each Psidium species (samples) were chopped together, and the nuclei were extracted and isolated (Otto 1990; Coser et al. 2012; Marques et al. 2016). The resulting suspensions were stained with buffer containing propidium iodide (Praça-Fontes et al. 2011; Coser et al. 2012) and analyzed in a Partec PAS® flow cytometer (Partec® GmbH, Munster, Germany) (Coser et al. 2012; Marques et al. 2016). The FlowMax® software (Partec®) was used to analyze the histograms. The mean nuclear genome size (2C) was measured by dividing the mean channel of the fluorescence peak corresponding to the standard’s G0/G1 nuclei by that of each sample. At least 20 in vitro plantlets were used for each species.
Just as for flow cytometry, in vitro plantlets were fundamental to accomplish the cytogenetic evaluation. From these plantlets, roots were removed and immediately treated with 4 µM amiprophos-methyl (APM, Nihon Bayer Agrochem K. K.®) for 5 h at 30 °C. The roots were washed with distilled water (dH2O) for 20 min, fixed in fresh methanol:acetic acid (Merck®) solution (3:1), stored at -20 °C for at least 24 h, washed again with dH2O for 20 min, and macerated with enzymatic solution for 2 h at 34 °C (Coser et al. 2012; Marques et al. 2016). Root meristem dissociation and air-drying (Carvalho et al. 2007) procedures were used to prepare the slides, which were analyzed using a Nikon 80i microscope (Nikon, Japan). Metaphase images were captured with a Media Cybernetics® Evolution™ charge-coupled device (CCD) video camera (Nikon, Japan) coupled to this microscope.
Molecular analysis
The transferability rates of 141 SSR (simple sequence repeat) markers were obtained from a previous study (Tuler et al. 2015). Of these, 32 SSR were selected which amplified for the seven species analyzed in this study (Table 1). Details of DNA extraction and SSR amplification are available in Tuler et al. (2015). The number of alleles per locus and observed heterozygosis per primer was estimated. Data obtained for alleles of each individual were subjected to dissimilarity index analysis using the weighted index.
Results
In vitro establishment, nuclear 2C value measurement, and chromosome number determination
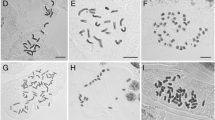
All seeds of Psidium and S. lycopersicum germinated after 30 days, providing morphologically normal plantlets, which were maintained under controlled environmental in vitro conditions. From the in vitro plantlets, nuclear genome size analysis evidenced high intrageneric variation of mean 2C values among the seven Psidium species. Psidium cauliflorum showed the lowest nuclear genome size, 2C = 0.93 ± 0.002 pg (1C = 0.465 pg), followed by P. guajava, 2C = 0.95 ± 0.021 pg (1C = 0.475 pg) and P. oblongatum, 2C = 0.98 ± 0.004 pg (1C = 0.490 pg). In comparison, the other species presented significantly greater mean values: P. guineense exhibited 2C = 1.86 ± 0.003 pg (1C = 0.930 pg), a nuclear genome size approximately twofold higher than in P. cauliflorum, P. guajava, and P. oblongatum. P. myrtoides showed 2C = 3.07 ± 0.0045 pg (1C = 1.535 pg), or 3.30 times higher; P. cattleyanum had 2C = 3.57 ± 0.00 pg (1C = 1.785 pg), or 3.84 times higher; and P. longipetiolatum displayed 2C = 5.12 ± 0.002 pg (1C = 2.560 pg), or 5.51 times higher than in the three above-mentioned species. Thus, mean 2C value data suggest that karyotype alterations occurred during evolution, promoting a strong variation in nuclear genome size (Fig. 1; Table 2).
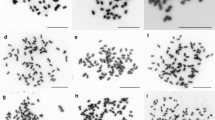
Schematic histogram and karyotype of the seven Psidium species. Flow cytometry was executed separately for each species using the internal standard S. lycopersicum (2C = 2.00 pg; Praça-Fontes et al. 2011). G0/G1 nuclei peaks of each Psidium species are represented in the same histogram, as follows: P. cauliflorum (2C = 0.93 pg), P. guajava (2C = 0.95 pg) and P. oblongatum (2C = 0.98 pg) in channel 100; P. guineense (2C = 1.86 pg) in channel 200; P. myrtoides (2C = 3.07 pg) in channel 323; P. cattleyanum (2C = 3.57 pg) in channel 376; and P. longipetiolatum (2C = 5.12 pg) in channel 539. Following the lines from each G0/G1 peak, the karyotype of each species is shown: aP. cauliflorum, bP. guajava and cP. oblongatum with 2n = 2x = 22 chromosomes; dP. guineense with 2n = 4x = 44 chromosomes; eP. myrtoides and fP. cattleyanum with 2n = 6x = 66 chromosomes; and gP. longipetiolatum with 2n = 8x = 88 chromosomes. Note the euploid series in these Psidium species based on x = 11, the similar nuclear genome size of the diploid species, and the clear nuclear DNA content difference between hexaploid species. Bars = 5 µm
Based on this hypothesis, a cytogenetic approach was performed from the same in vitro plantlets to assess the chromosome number of each species, as well as polyploidy in P. guineense, P. myrtoides, P. cattleyanum, and P. longipetiolatum. Psidium cauliflorum, P. guajava and P. oblongatum presented 2n = 22 chromosomes; P. guineense 2n = 44; P. myrtoides and P. cattleyanum 2n = 66; and P. longipetiolatum 2n = 88. Therefore, cytogenetics revealed a euploid series comprising diploid (P. cauliflorum, P. guajava, and P. oblongatum), tetraploid (P. guineense), hexaploid (P. myrtoides and P. cattleyanum), and octoploid species (P. longipetiolatum) (Fig. 1; Table 2).
Considering the non-replicated monoploid genome x = 11 for the Psidium species sampled here, the 1Cx DNA value (DNA content of basic chromosome number x; Greilhuber et al. 2005) was: 0.465 pg for P. cauliflorum and P. guineense; 0.475 pg for P. guajava; 0.490 pg for P. oblongatum; 0.512 pg for P. myrtoides; 0.595 pg for P. cattleyanum; and 0.640 pg for P. longipetiolatum.
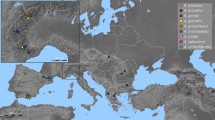
The diploid species, P. cauliflorum and P. oblongatum, are endemic to the Atlantic Forest, restricted to a few locations, mainly in rainforest regions. The octoploid P. longipetiolatum is also restricted to the Atlantic Forest, occurring in Ombrophilous Forest, Semideciduous forest in the states of southeastern (Espírito Santo, Minas Gerais, Rio de Janeiro, and São Paulo states) and Mixed Ombrophilous Forest in south (Paraná, Rio Grande do Sul, and Santa Catarina states). The tetraploid (P. guineense) and hexaploid (P. myrtoides and P. cattleyanum) are widely distributed in Brazil, occurring under different environmental conditions in the Atlantic Forest, Caatinga, Cerrado, and Amazon Rainforest (Fig. 2).
Distribution of the seven Psidium species in Brazilian biomes based on data from the literature, herbaria, and the present study. Distribution of the diploid species (2n = 2x = 22 chromosomes): P. cauliflorum (blue circle), P. guajava (black circle), and P. oblongatum (yellow circle). Note that P. cauliflorum and P. oblongatum (yellow circle) only occur in the Atlantic Forest. Tetraploid species (2n = 4x = 44 chromosomes): distribution of P. guineense (black square) was not registered in Pantanal and the Pampas. Hexaploid species (2n = 6x = 66 chromosomes): P. cattleyanum (green triangle) and P. myrtoides (red triangle) occur in the Atlantic Forest, Cerrado, Caatinga, and the Pampas. Octoploid species (2n = 8x = 88 chromosomes): P. longipetiolatum (blue hexagon) is found distributed in the Atlantic Forest from Minas Gerais to Rio Grande do Sul. The coordinates used to define the species’ geographical distribution were obtained with the application Google Earth, using the locations indicated on the labels of the herbarium specimens. The map of geographical distribution was made using the program DIVA GIS 5.4
Molecular analysis
Thirty-two SSR markers were chosen amongst the 132 SSR markers developed for P. guajava, according to Tuler et al. (2015), based on transferability in the six Psidium species (Table 2). Diploid species showed the lowest heterozygosity rates, with P. cauliflorum (2x = 22) having all 32 loci in homozygosis (heterozygosity rate of 0.00%), corresponding to a mean allele number per locus equivalent to 1.00. In contrast, the octoploid P. longipetiolatum exhibited the highest heterozygosity rate (50.00%), as well as the highest mean number of alleles per locus among all species (1.81) (Table 2).
The 32 SSR loci differed among the species. A total of 149 alleles were amplified, with a mean of 4.6 alleles per locus. The SSR loci mPgCIR 26, mPgCIR 94, mPgCIR 99, and mPgCIR 148 produced the largest number of alleles (8 or 9), whereas mPgCIR 158, mPgCIR 188, and mPgCIR 233 generated the smallest (1 or 2). In general, SSR from transcribed regions showed more allelic forms (5.5 alleles per locus) than those from non-transcribed regions (4.8 alleles per locus) (Table 1).
Discussion
Nuclear 2C value, chromosome number, and molecular data evidenced the euploid, and dynamic and progressive genomic modifications in the seven Psidium species, expanding the data about tropical tree species (Husband et al. 2013; Spoelhof et al. 2017). The four polyploid species of Psidium in this study are an example of natural euploidy derived from whole-genome duplication. Therefore, these species were originated from intraspecific whole-genome duplication (autopolyploidy—endoreplication or endomitosis of the zygote or fusion of non-reduced reproductive cells), or from the interspecific crossing (allopolyploidy—hybridization) involving or not the whole-genome duplication (Stebbins 1950; Sattler et al. 2016; Shu et al. 2018). This is a dramatic quest that remains open, which must be look for each species. Due to the relatively recent (~ 9.9–20.8 Ma, Oligocene–Miocene) radiation of the tribe Myrteae (Thornhill et al. 2015; Berger et al. 2016), these Psidium species represent recent polyploids.
The progressive increase in nuclear DNA content, from the diploid species P. cauliflorum, P. guajava, and P. oblongatum to the polyploid species P. guineense, P. cattleyanum, P. myrtoides, and P. longipetiolatum, indicates strong karyotype differences related to numerical changes (euploidy). Therefore, there is a relation between the chromosome number and the nuclear genome size of the seven species. Interspecific and intraspecific variations in nuclear 2C value in Psidium have been reported for P. guajava ‘White’—2C = 0.507 pg and P. guajava ‘Red’—2C = 0.551 pg (Costa et al. 2008), P. acutangulum—2C = 1.167 pg (Costa et al. 2008), P. australe—2C = 2.97 pg (Souza et al. 2015), P. guineense—2C = 1.85 pg (Marques et al. 2016) and 2C = 2.02 pg (Souza et al. 2015), and P. cattleyanum—2C = 1.053 pg (Costa et al. 2008) and 2C = 1.99–5.47 pg (Souza et al. 2015).
Chromosome counting corroborated the obtained 2C values and shed light on the karyotype divergences (Fig. 1). Diploidy was confirmed for P. guajava (Costa et al. 2008; Souza et al. 2015; Marques et al. 2016), as well as tetraploidy for P. guineense (Souza et al. 2015; Marques et al. 2016) and hexaploidy for P. cattleyanum (Souza et al. 2015). Nuclear genome size and chromosome number were characterized for the first time in P. cauliflorum and P. oblongatum, which exhibited the same nuclear 2C value and chromosome number as P. guajava. The family Myrtaceae is basically diploid (2n = 2x = 22), as illustrated by Australasian species of the genera Eucalyptus and Melaleuca (Atchison 1947; Brighton and Ferguson 1976; Rye 1979). The tribe Myrteae also displays a predominance of 2n = 2x = 22, except for Eugenia, Myrcia, and Psidium, in which polyploid species are also found (Costa and Forni-Martins 2006a, b, 2007; Silveira et al. 2017).
Considering polyploid species, P. myrtoides shows the same 2n = 6x = 66 chromosomes as P. cattleyanum, but its 2C value is 0.50 pg lower than in the latter. These results suggest the occurrence of structural chromosome changes during the karyotype evolution in Psidium. Besides euploidy, the karyotype evolution also involves aneuploidy, which did not observed in the Psidium species of this study, and structural chromosome rearrangements (Sattler et al. 2016; Slijepcevic 2018). Alternatively, P. cattleyanum and P. myrtoides may have originated from distinct progenitors (auto- or allopolyploids). The number of 2n = 8x = 88 chromosomes reported here for P. longipetiolatum has been previously reported for P. cattleyanum (Atchison 1947), reinforcing that polyploidy occurs in this genus. Contrary to the previous studies (Atchison 1947; Costa and Forni-Martins 2006a, b; Costa et al. 2008; Souza et al. 2015), no variation in 2C value or chromosome number was found between the distinct individuals of P. guajava and P. cattleyanum.
The interspecific differences in the nuclear DNA content among the Psidium species (Fig. 1) represent outcomes of the polyploid origin due that the basic chromosome number x = 11, which is conserved in Myrtaceae. A polyploid series from x = 11 was confirmed for Psidium, as well as the mean nuclear 2C value was showed for each ploidy level (Fig. 1). The polyploidy has mainly been reported for the tribe Myrteae (Silveira et al. 2017; Costa and Forni-Martins 2006a, b, 2007), which includes fleshy-fruited species in South America. According to the phylogenetic relationships proposed for Myrtaceae (Vasconcelos et al. 2017), the tribe Eucalypteae is a basal clade in the subfamily Myrtoideae. The diploid species of the tribes Eucalypteae present 2C = 1.13 pg (Eucalyptus globulus, Azmi et al. 1997). Differently, for the Psidium sampled here, the mean value for nuclear genome size (2C = 1.85 pg for P. guineense to 2C = to 2C = 5.75 pg for P. longipetiolatum) increased through polyploidy events.
The SSR markers showed that polyploidy in Psidium also resulted in higher heterozygosity rate and mean number of alleles. This is a direct effect of this karyotype change. The high polymorphism of the primers mPgCIR 26, 94, 99, and 148 was a result of the higher number of alleles present in polyploid species (P. cattleyanum: 3 alleles for mPgCIR 94; P. myrtoides: 4 alleles for mPgCIR 26; P. guineense: 4 alleles for mPgCIR 148; and P. longipetiolatum: 4 alleles for mPgCIR 99 and 4 alleles for mPgCIR 94). The occurrence of more than two alleles per SSR locus has also been reported in accessions of P. guajava (mPgCIR 253, Aranguren et al. 2010), P. guineense, P. cattleyanum, and P. friedrichsthalianum (mPgCIR 255, Costa and Santos 2013).
Most of the polymorphic primers are derived from functional regions (Table 1; e.g., mPgCIR 94, 99, and 148). Functional regions are associated with control and variation of adaptive characteristics and/or important traits for occupation of new habitats, thus affecting the species’ distribution (Grattapaglia et al. 2012). Based on nuclear 2C value, chromosome number, and molecular data, we suggest that the large ecological and geographical amplitudes that the four polyploid Psidium species occupy can be linked to their polyploid condition.
The polyploid Psidium species (P. guineense, P. cattleyanum, P. myrtoides, and P. longipetiolatum) have large geographical distribution compared to the endemic and diploid species (P. cauliflorum and P. oblongatum). For instance, P. guineense occurs in all Brazilian biomes (Atlantic Forest, Caatinga, Cerrado, and Amazon Rainforest), and this broad geographical distribution comprises a wide range of environmental conditions. Psidium guajava is the only diploid species presenting large geographical distribution in Brazil. This can be explained by its cultivation for economic purposes. Considering that polyploids show broader ecological tolerances and higher colonization abilities in comparison to diploids (Stebbins 1950; Grant 1981), it is possible that the phenotypical diversity presented by the four evaluated polyploid Psidium species enables their exploration of new habitats.
Conclusion
Euploidy, based on the basic chromosome set x = 11, was confirmed for four of the seven Psidium species studied here. The chromosome number explains the increase in nuclear genome size and genetic diversity, discriminating the tetraploid and octoploid species. Therefore, polyploidy contributed to the diversification in the studied Psidium species, representing an important mechanism of speciation. As a challenging field, further understanding of the evolutionary history and diversification of Psidium will probably require approaches including the understanding of the phenotypic variation associated with the species’ geographic distributions, and the development of phylogenetic studies. Such studies, however, will be better understood in the light of the cytogenetic and molecular patterns revealed in this study.
Author contribution statement
ACT, TTC, MLG, and WRC conceived, designed, and conducted the study. ACT, ALP, and TTC identified the species of Psidium. ACT, MSS, and WRC carried out the cytogenetic analyses. WRC and CRC performed the flow cytometry analysis. ACT and MFSF conceived and conducted the molecular marker analysis. All authors equally contributed to the writing, editing and revision of the manuscript, and approved the final version for submission.
References
Alix K, Gérard PR, Schwarzacher T et al (2017) Polyploidy and interspecific hybridization: partners for adaptation, speciation and evolution in plants. Ann Bot 120:183–194. https://doi.org/10.1093/aob/mcx079
Andrade FG, Forni-Martins ER (1998) Estudos cromossômicos em espécies de Myrtaceae. Genet Mol Biol 21(Suppl.):166
Aranguren Y, Valecillos C, Fermín G (2010)Variability of Venezuelan guava geographic landraces employing phenotypic markers. In: Proceedings of the second international guava symposium, Mexico, Acta horticulturae, vol 849, pp 87–95
Atchison E (1947) Chromosome numbers in the Myrtaceae. Am J Bot 34:159–164
Azmi A, Noin M, Landré P, Prouteau M, Boudet AM, Chriqui AM D (1997) High frequency plant regeneration from Eucalyptus globulus Labill. hypocotyls: ontogenesis and ploidy level of the regenerants. Plant Cell 51:9–16
Baker HG (1965) Characteristics and modes of origin of weeds. In: Baker HG, Stebbins GL (eds) The genetics of colonizing species. Academic Press, New York, pp 147–168
Barker MS, Husband BC, Pires JC (2016) Spreading winge and flying high: the evolutionary importance of polyploidy after a century of study. Am J Bot 103:1139–1145. https://doi.org/10.3732/ajb.1600272
Beest MT, Roux JJL, Richardson DM, Brysting AK, Suda J, Magdalena Kubesová M, Pysek P (2012) The more the better? The role of polyploidy in facilitating plant invasions. Ann Bot 109:19–45. https://doi.org/10.1093/aob/mcr277
Bennetzen JL, Kellogg EA (1997) Do plants have a-oneway ticket to genomic obesity? Plant Cell 9:1509–1514
Berger BA, Kriebel R, Spalink D, Sytsma KJ (2016) Divergence times, historical biogeography, and shifts in speciation rates of Myrtales. Mol Phylogenet Evol 95:116–136
BFG (2015) Growing knowledge: an overview of seed plant diversity in Brazil. Rodriguésia 66:1085–1113. https://doi.org/10.1590/2175-7860201566411
Bolkhovskikh Z, Matvejeva VG, Zakharyeva O (1969) Chromosome numbers of flowering plants. Academy of Sciences of the USSR, Saint Petersburg
Bretagnolle F, Lumaret R (1995) Bilateral polyploidization in Dactylis glomerata L. subsp. lusitanica: occurrence, morphological and genetic characteristics of first polyploids. Euphytica 84:197–207
Brighton CA, Ferguson IK (1976) Chromosome counts in the genus Melaleuca (Myrtaceae). Kew Bull 31:27–33
Carvalho CR, Clarindo WR, Almeida PM (2007) Plant cytogenetics: still looking for the perfect mitotic chromosomes. Nucleus 50:453–462
Comai L (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6:836–846. https://doi.org/10.1038/nrg1711
Coser SM, Ferreira MFS, Ferreira A, Mitre LK, Carvalho CR, Clarindo WR (2012) Assessment of genetic diversity in Psidium guajava L. using different approaches. Sci Hortic 148:223–229. https://doi.org/10.1016/j.scienta.2012.09.030
Costa IR, Forni-Martins ER (2006a) Chromosome studies in species of Eugenia, Myrciaria and Plinia (Myrtaceae) from southeastern Brazil. Aust J Bot 54:409–415. https://doi.org/10.1071/BT04199
Costa IR, Forni-Martins ER (2006b) Chromosome studies in Brazilian species of Campomanesia Ruiz & Pávon and Psidium. L. (Myrtaceae Juss.). Caryologia 59:7–13. https://doi.org/10.1080/00087114.2006.10797891
Costa IR, Forni-Martins ER (2007) Karyotype analysis in South American species of Myrtaceae. Bot J Linn Soc 155:571–580. https://doi.org/10.1111/j.1095-8339.2007.00704.x
Costa SR, Santos CAF (2013) Allelic database and divergence among Psidium accessions by using microsatellite markers. Genet Mol Res 12:6802–6812
Costa IR, Dornelas MC, Forni-Martins ER (2008) Nuclear genome size variation in fleshy-fruited neotropical Myrtaceae. Plant Syst Evol 276:209–217. https://doi.org/10.1007/s00606-008-0088-x
Dhooghe E, Van Laere K, Eeckhaut T, Leus L, Huylenbroeck JV (2011) Mitotic chromosome doubling of plant tissues in vitro. Plant Cell Tissue Organ Cult 104:359–373. https://doi.org/10.1007/s11240-010-9786-5
Edger PP, Pires JC (2009) Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res 17:699–717. https://doi.org/10.1007/s10577-009-9055-9
Ehrendorfer F (1980) Polyploidy and distribution. In: Lewis WH (ed) Polyploidy: biological relevance. Plenum, New York, pp 45–60
Goldblatt P (1981) Index to plant chromosome numbers 1975–1978. Monographs in systematic botany from the Missouri Botanical Garden, Saint Louis
Goldblatt P, Johnson DE (1996) Index to plant chromosome numbers 1992–1993. Missouri Botanical Garden, Saint Louis
Grant V (1981) Plant speciation, 2nd edn. Columbia University Press, New York
Grattapaglia D, René E, Vaillancourt RE, Shepherd M, Thumma BR, Foley W, Külheim C, Potts BM, Myburg AA (2012) Progress in Myrtaceae genetics and genomics: Eucalyptus as the pivotal genus. Tree Genet Genomes 8:463–508. https://doi.org/10.1007/s11295-012-0491-x
Greilhuber J, Doleźel J, Lysák M, Bennett MD (2005) The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Ann Bot 95:255–260. https://doi.org/10.1093/aob/mci019
Hancock JF, Bringhurst RS (1981) Evolution in California populations of diploid and octoploid Fragaria (Rosaceae): a comparison. Am J Bot 68:1–5
Husband BC, Baldwin SJ, Suda J (2013) The incidence of polyploidy in natural plant populations: major patterns and evolutionary processes. In: Leitch IJ, Greilhuber J, Doležel J, Wendel JF (eds) Plant genome diversity 2: physical structure, behaviour and evolution of plant genomes. Springer, Wien, pp 255–276
Kay QON (1969) The origin and distribution of diploid and tetraploid Tripleurospermum inodorum (L.) Schultz Bip. Watsonia 7:130–141
Kron P, Suda J, Husband BC (2007) Applications of flow cytometry to evolutionary and population biology. Annu Rev Ecol Evol Syst 38:847–876. https://doi.org/10.1146/annurev.ecolsys.38.091206.095504
Levin DA (2002) The role of chromosomal change in plant evolution. Oxford University Press, Oxford
Lewis WH (1980) Polyploidy in angiosperms: dicotyledons. In: Lewis WH (ed) Polyploidy: biological relevance. Plenum Press, New York, pp 241–268
Lucas EJ, Harris SA, Mazine FF, Belsham SR, Nic Lughadha EM, Telford A, Peter E. Gasson PE, Chase MW (2007) Suprageneric phylogenetics of Myrteae, the generically richest tribe in Myrtaceae (Myrtales). Taxon 56:1105–1128. https://doi.org/10.2307/25065906
Madlung A (2013) Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity 110:99–104. https://doi.org/10.1038/hdy.2012.79
Maherali H, Walden AE, Husband BC (2009) Genome duplication and the evolution of physiological responses to water stress. New Phytol 184:721–731. https://doi.org/10.1111/j.1469-8137.2009.02997.x
Marchant DB, Soltis DE, Soltis PS (2016) Patterns of abiotic niche shifts in allopolyploids relative to their progenitors. New Phytol 212:708–718. https://doi.org/10.1111/nph.14069
Marques AM, Tuler AC, Carvalho CR, Carrijo TT, Ferreira MFS, Clarindo WR (2016) Refinement of the karyological aspects of Psidium guineense (Swartz, 1788): a comparison with Psidium guajava (Linnaeus, 1753). Comp Cytogenet 10:117–128. https://doi.org/10.3897/CompCytogen.v10i1.6462
Moore RJ (1977) Index to plant chromosome numbers 1973–1974. Regnum Veg 96:1–257 p
Münzbergová Z (2007) No effect of ploidy level in plant response to competition in a common garden experiment. Biol J Linn Soc 92:211–219. https://doi.org/10.1111/j.1095-8312.2007.00820.x
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Murillo -AJ, Ruiz- PE, Landrum LR, Stuessy TF, Barfuss MHJ (2012) Phylogenetic relationships in Myrceugenia (Myrtaceae) based on plastid and nuclear DNA sequences. Mol Phylogenet Evol 62:764–776. https://doi.org/10.1016/j.ympev.2011.11.021
Oliveira SC, Nunes ACP, Carvalho CR, Clarindo WR (2013) In vitro polyploidization from shoot tips of Jatropha curcas L.: a biodiesel plant. Plant Growth Regul 69:79–86. https://doi.org/10.1007/s10725-012-9749-4
Otto FJ (1990) DAPI staining of fixed cells for high-resolution flow 670 cytometry of nuclear DNA. In: Darzynkiewiez Z, Crissman HA, 671 Robinson JP (eds) Methods in cell biology, vol 33. Academic 672, San Diego, pp 105–110
Otto SP, Whitton J (2000) Polyploid incidence and evolution. Annu Rev Genet 34:401–437. https://doi.org/10.1146/annurev.genet.34.1.401
Peixoto AL, Maia LC (2013) Manual de procedimentos para herbários. Recife. Editora Universitária UFPE
Petrov DA (2002) DNA loss and evolution of genome size in Drosophila. Genetica 115:81–91
Praça-Fontes MM, Carvalho CR, Clarindo WR (2011) C-value reassessment of plant standards: an image cytometry approach. Plant Cell Rep 30:2303–2312. https://doi.org/10.1007/s00299-011-1135-6
Rivero G, Salazar G, Pacheco D, Sánchez A, Quirós M, Sthormes G (2012) Relaciones filogeneticas entre especies de Psidium (Myrtaceae) presentes en el occidente de venezuela a partir de secuencias de adn nuclear (ITS) y plastidial (trnH-psbA). Interciencia 37:838–844
Rye BL (1979) Chromosome number variation in the Myrtaceae and its taxonomic implications. Aust J Bot 27:547–573. https://doi.org/10.1071/BT9790547
Sattler MC, Carvalho CR, Clarindo WR (2016) The polyploidy and its key role in plant breeding. Planta 243:281–296. https://doi.org/10.1007/s00425-015-2450-x
Segraves KA (2017) The effects of genome duplications in a community context. New Phytol 215:57–69. https://doi.org/10.1111/nph.14564
Shu Z, Row S, Deng W (2018) Endoreplication: the good, the bad, and the ugly. Trends Cell Biol 28:465–474. https://doi.org/10.1016/j.tcb.2018.02.006
Silveira RM, Machado RM, Forni-Martins ER, Verola CF, Costa IR (2017) Environmental variations drive polyploid evolution in neotropical Eugenia species (Myrtaceae). Genet Mol Res 15:gmr15048842
Slijepcevic P (2018) Genome dynamics over evolutionary time: “C-value enigma” in light of chromosome structure. Mutat Res Genet Toxicol Environ. https://doi.org/10.1016/j.mrgentox.2018.05.005
Soares-Silva LH, Proença CE (2006) An old species revisited and a new combination proposed in Psidium (Myrtaceae). Kew Bull 61:199–204
Soltis PS, Soltis DE (2000) The role of genetic and genomic attributes in the success of polyploids. Proc Natl Acad Sci USA 97:7051–7057. https://doi.org/10.1073/pnas.97.13.7051
Soltis DE, Solti PS, Tate JA (2003) Advances in the study of polyploidy since plant speciation. New Phytol 161:173–191. https://doi.org/10.1046/j.1469-8137.2003.00948.x
Souza AG, Resende LV, Lima IP, Martins LSS, Techio VH (2015) Chromosome number and nuclear DNA amount in Psidium spp. resistant and susceptible to Meloidogyne enterolobii and its relation with compatibility between rootstocks and commercial varieties of guava tree. Plant Syst Evol 301:231–237. https://doi.org/10.1007/s00606-014-1068-y
Spoelhof JP, Soltis PS, Soltis DE (2017) Pure polyploidy: closing the gaps in autopolyploid research. J Syst Evol 55:340–352. https://doi.org/10.1111/jse.12253
Staggemeier VG, Cazetta E, Morellato LPC (2016) Hyperdominance in fruit production in the Brazilian Atlantic rain forest: the functional role of plants in sustaining frugivores. Biotropica 49:71–82. https://doi.org/10.1111/btp.12358
Stebbins GL (1947) The origin of the complex of Bromus carinatus and its phylogeographic implications. Contr Gray Herb Harv Univ 165:42–55
Stebbins GL (1950) Variation and evolution in plants. Oxford University Press, London
Stebbins GL (1985) Polyploidy, hybridization, and the invasion of new habitats. Ann Mo Bot Gard 72:824–832
Thompson JD, Lumaret R (1992) The evolutionary dynamics of polyploidy plants: origins, establishment and persistence. Trends Ecol Evol 7:302–307. https://doi.org/10.1016/0169-5347(92)90228-4
Thornhill AH, Ho SYW, Külheim C, Crisp MD (2015) Interpreting the modern distribution of Myrtaceae using a dated molecular phylogeny. Mol Phylogenet Evol 93:29–43. https://doi.org/10.1016/j.ympev.2015.07.007
Treier UA, Broennimann O, Normand S et al (2009) Shift in cytotype frequency and niche space in the invasive plant Centaurea maculosa. Ecology 90:1366–1377
Tuler AC, Carrijo TT, Nóia LR, Ferreira A, Peixoto AL, Ferreira MFS (2015) SSR markers: a tool for species identification in Psidium (Myrtaceae). Mol Biol Rep 42:1501–1513. https://doi.org/10.1007/s11033-015-3927-1
Van Dijk P, Bakx-Schotman T (1997) Chloroplast DNA phylogeography and cytotype geography in autopolyploid Plantago media. Mol Ecol 6:345–352. https://doi.org/10.1046/j.1365-294X.1997.00199.x
Vasconcelos TNC, Proença CEB, Ahmad B et al (2017) Myrteae phylogeny, calibration, biogeography and diversification patterns: increased understanding in the most species rich tribe of Myrtaceae. Mol Phylogenet Evol 109:113–137. https://doi.org/10.1016/j.ympev.2017.01.002
Watanabe K (1986) The cytogeography of the genus Eupatorium (Compositae): a review. Plant Spec Biol 1:99–116. https://doi.org/10.1111/j.1442-1984.1986.tb00019.x
WCSP (2017) World checklist of selected plant families. http://apps.kew.org/wcsp/. Accessed 13 May 2017
Acknowledgements
We would like to thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília—DF, Brazil; Grants 443801/2014-2 and 308828/2015-1, 305821/2016-4), Fundação de Amparo à Pesquisa do Espírito Santo (FAPES/VALE, Vitória—ES, Brazil; Grant 75516586/16), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ, Rio de Janeiro—RJ, Brazil) and VALE for financial support. This study was financid in part by the Coordernação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Alia.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tuler, A.C., Carrijo, T.T., Peixoto, A.L. et al. Diversification and geographical distribution of Psidium (Myrtaceae) species with distinct ploidy levels. Trees 33, 1101–1110 (2019). https://doi.org/10.1007/s00468-019-01845-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00468-019-01845-2