Abstract
Jatropha curcas L. has been considered one of the most promising alternatives for biofuel production and, thus, a relevant economic crop. In this context, in vitro tissue culture techniques such as organogenesis and embryogenesis have been conducted for mass clonal propagation of elite J. curcas lines. However, despite advancements, in vitro induction of polyploids has not yet been related for this crop. In this sense, the present study attempted to induce polyploidy in plantlets generated from shoot tips of J. curcas ‘Gonçalo’ (2n = 2× = 22 chromosomes, 2C = 0.85 pg). For this purpose, some criteria were adopted for selection of the most adequate colchicine treatment: (a) survival rate of the explants, and (b) number of tetraploid and (c) mixoploid plantlets. Tetraploid and mixoploid plantlets were obtained from different treatments, with 0.5 mM colchicine for 96 h being the most efficient. The plantlets were recovered and clonally propagated in tissue culture medium supplemented with indole-3-acetic acid and 6-benzylaminopurine. These results show that the tissue culture procedures were adequate for induction, propagation and recovery of tetraploid and mixoploid plantlets. Moreover, DNA ploidy level screening by flow cytometry was a practical and rapid strategy for selection of diploid, mixoploid and tetraploid plantlets. The tissue culture system presented here represents a reliable methodology for in vitro polyploid induction of this and other elite lines of J. curcas.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Jatropha (family Euphorbiaceae, subfamily Crotonoideae, tribe Joannesieae) comprises over 175 native species, occurring in South to Central America (Mukherjee et al. 2011), Asia and Africa (Kumar and Reddy 2012). In particular, the species Jatropha curcas L. has recently been pointed out as a relevant economic crop (Kumar and Reddy 2012). The most important aspect of this species is its large potential for biofuel production, owing to high oil content of the seed, rapid growth and stiffness of the plant (Mukherjee et al. 2011) and the low oil production cost (Jha et al. 2007).
Due to the increasing demand for biofuel, breeding programs of J. curcas have been established in distinct countries, for instance Brazil, India, Senegal and Cape Verde (Divakara et al. 2010). In this context, in vitro tissue culture techniques have mainly been performed for mass clonal propagation of J. curcas elite lines (Kalimuthu et al. 2007). For this purpose, in vitro regeneration of J. curcas plantlets has been achieved mainly through organogenesis (Rajore and Batra 2005; Jha et al. 2007; Kalimuthu et al. 2007; Kumar and Reddy 2012) and embryogenesis procedures (Jha et al. 2007).
For some species, new elite plants have been obtained by in vitro polyploidization, which has received special attention as an important tool for plant breeding programs (Zhang et al. 2008). Polyploid plantlets, especially tetraploid ones, have been successfully recovered mainly for vegetable, ornamental and medicinal crops (Dhooghe et al. 2011). Chromosome doubling efficiency, and ultimately polyploidy induction, depends on various factors, such as: (a) species, (b) type of explant, (c) type and concentration of the anti-tubulinic agent, as well as exposure time, and (d) tissue culture medium for polyploidization and propagation (Dhooghe et al. 2011).
An efficient in vitro polyploidization system also requires effective methods for direct screening and monitoring of DNA ploidy level. Flow cytometry (FCM) is the quickest and most reliable method for this purpose, given that the DNA ploidy level can be determined for several plantlets in a short time (Clarindo et al. 2008). According to Ochatt (2008), application of FCM in plant science increased significantly with the use of innovative and interesting techniques for basic research and commercial breeding.
Considering the relevance of protocols for in vitro polyploidization of J. curcas, the present study attempted to induce polyploidy from shoot tips of this relevant biofuel species.
Materials and methods
Plant material
Seeds of J. curcas ‘Gonçalo’ were collected at a crop area located in Resende Costa, Minas Gerais, Brazil. Solanum lycopersicum L. ‘Stupické’ (primary standard for FCM, 2C = 2.00 pg; Praça-Fontes et al. 2011b) were kindly supplied by Dr. Jaroslav Doležel (Experimental Institute of Botany, Czech Republic).
2C-value and karyotype of J. curcas
The genome size of J. curcas was measured from nuclei suspensions extracted and stained according to procedure described by Carvalho et al. (2008). The suspensions were analyzed with a Partec PAS® cytometer (Partec® GmbH, Munster, Germany), equipped with a laser source (488 nm). Flow cytometer parameters (i.e. gain and channel) were determined before each measurement, based on external FCM analyses of primary standard (S. lycopersicum ‘Stupické’) and sample (J. curcas). Six independent repetitions, accounting for more than 10,000 nuclei, were carried out at each analysis.
2C-value of J. curcas was calculated by dividing the mean channel of the G0/G1 fluorescence peak from the primary standard by the mean channel of the G0/G1 peak from each sample. Genome size mean value in picograms (pg) was converted to base pairs (bp), considering that 1 pg DNA corresponds to 0.978 × 109 bp (Doležel et al. 2003).
For karyotype characterization of J. curcas explant donor, metaphasic chromosomes were obtained from root tips treated according to Carvalho et al. (2008). The slides were prepared using the cell dissociation method, followed by air-drying technique (Carvalho et al. 2008). Subsequently, the slides were stained with a 5 % Giemsa solution (Merck®) in phosphate buffer (pH 6.8) for 5 min, washed twice in distilled water (dH2O), air-dried and placed on a hot plate at 50 °C for 3 min.
Images of metaphase chromosomes were captured with a Media Cybernetics® Camera Evolution™ charge-coupled device (CCD) video camera, mounted on a Nikon 80i microscope (Nikon, Japan).
In vitro recovery of J. curcas shoot tips
For shoot tip recovery, J. curcas seeds were decontaminated by immersion in 70 % ethanol (Merck®) for 1 min, followed by 2.5 % sodium hypochlorite for 20 min and rinsing three times with autoclaved dH2O. After pericarp removal under laminar flow hood, the seeds were decontaminated again for 20 min with 2.5 % sodium hypochlorite solution containing one drop of Tween (Merck®) per 100 ml. Next, the seeds were washed with autoclaved dH2O and inoculated into medium M1 (Table 1).
In vitro multiplication and assessment of DNA ploidy level
After 30 days in medium M1, shoot tips of the seedlings germinated in vitro were excised and inoculated into bottles containing 100 ml of medium M2 (Table 1). These cultures were maintained at 25 ± 1 °C under 16/8 h light/dark regime, with 36 μmol m−2 s−1 light radiation.
As medium M2 (Table 1) was supplemented with indole-3-acetic acid (IAA) and 6-benzylaminopurine (BAP) (Rajore and Batra 2005; Mukherjee et al. 2011), the shoot tips were used to verify the occurrence of numeric chromosomal aberration, eu- and aneuploidy, associated with somaclonal variation.
For this purpose, leaf samples were collected from J. curcas plants seed-raised in greenhouse (control) and from plantlets propagated in medium M2 for 120 days (samples). Nuclei suspensions of control and samples were prepared as described by Clarindo et al. (2008) and CyStain UV Ploidy Partec® protocols. These suspensions were analyzed with a Partec-PAS® flow cytometer, equipped with an UV lamp emitting at 388 nm and a TK 420 filter. The FlowMax® software (Partec®) was used for data analyses. More than 10,000 nuclei were analyzed, and three independent replications were performed for determination of DNA ploidy level in each J. curcas plantlet.
In vitro polyploidization
Shoot tip explants were excised from the plantlets obtained in medium M1 and placed in Erlenmeyer flasks containing 10 ml of liquid polyploidization medium (Praça et al. 2009) M3 (Table 1). This medium was supplemented with different concentrations (Table 2) of filter-sterilized colchicine (Sigma®). The flasks were shaken (40 rpm, at 25 ± 1 °C) in a growth room for different time periods (Table 2). Each flask contained three shoot tip explants, corresponding to three replicates for each treatment. After the set times, the explants were rinsed five times with autoclaved dH2O and inoculated into medium M2 (Table 1). Cultures were maintained at 25 °C under a 16/8 h light/dark regime, with 36 μmol m−2 s−1 light radiation provided by two fluorescent lamps (20 W, Osram®).
The shoot tips were propagated for 4 months, being subcultured every 30 days into fresh medium M2. During each subculture, the leaves were excised and the new shoots isolated. For polyploidy screening, leaves were excised from all J. curcas plantlets (samples; Table 2) recovered in medium M2. J. curcas plants seed-raised in greenhouse were used as diploid standards (control). Nuclei suspensions of controls and samples were prepared and analyzed by FCM, as described above.
Polyploidization efficiency was statistically analyzed by the t test method, using the Assistat 7.6 beta statistical software (Silva 2012). Based on statistical comparisons, a second in vitro polyploidization procedure was performed using the most efficient colchicine concentration (Table 2).
Results
2C-value and karyotype of J. curcas
The fluorescence peaks of G0/G1 nuclei of J. curcas and S. lycopersicum showed coefficient of variation (CV) lower than 5 %. With the fluorescence peaks of G0/G1 nuclei from the standard S. lycopersicum (2C = 2.00 pg; Praça-Fontes et al. 2011b) being turned to channel 200 (data not shown), the mean 2C value for all J. curcas ‘Gonçalo’ plants was equivalent to 0.85 pg ± 0.01 (0.83 × 109 bp).
Jatropha curcas root tips treated with 4 μM amiprophos-methyl (APM, microtubule inhibitor) and macerated in 1:10 pectinase solution for 2.5 h provided metaphases adequate for morphometric analysis. The metaphases presented well-spread chromosomes with well-defined constrictions, without cytoplasmic background and chromatin damage (data not shown). Morphometric analysis showed that J. curcas ‘Gonçalo’ has 2n = 22 chromosomes, with total length ranging from 1.60 to 1.14 μm.
In vitro recovery of J. curcas shoot tips
Owing to the decontamination procedure and handling of seeds under aseptic conditions, no contamination was detected during the in vitro process. After 8 days in medium M1 (Table 1), J. curcas seeds germinated with a rate of 86.66 % and speed of germination (SG) of 4.86. Consequently, plantlets were regenerated after 30 days, providing enough shoot tips for propagation and multiplication in medium M2 and in vitro polyploidization in medium M3.
In vitro multiplication and assessment of DNA ploidy level
Shoot tips were transferred to medium M2 and cultivated for 120 days, with monthly subcultures. After four subcultures in M2, the shoot tips showed a mean multiplication rate of 4.5 shoots per explant.
Leaves from the plantlets were collected for analysis of DNA ploidy level stability by FCM. In vitro (samples) and greenhouse (control; 2C = 0.85 pg; 2× = 22 chromosomes) plantlets exhibited G0/G1 peaks in the same channel (Fig. 1a). This way, it could be verified that numeric chromosomal aberration (eu- and/or aneuploidy) has not occurred during propagation and multiplication of J. curcas plantlets in medium M2.
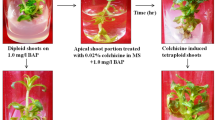
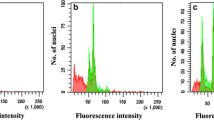
J. curcas plantlets obtained by in vitro polyploidization, and cultivated in M2 medium (Table 1), and respective FCM histograms. a Histogram showing G0/G1 peak (channel 200) of the diploid samples (2C = 2X). b Diploid plantlet representing the regenerants that showed the same DNA ploidy level of the control plants (2C = 2X). c Histogram showing G1/G0 peak (channel 200 and 400) of the mixoploid samples (2C = 2X and 2C = 4X, respectively). d Mixoploid plantlet representing the regenerants that showed DNA ploidy level 2C = 2X and 2C = 4X. e Histogram showing G1/G0 peak (channel 400) of the tetraploid samples (2C = 4X). f Tetraploid plantlet representing the regenerants that showed DNA ploidy level 2C = 4X. Bar 1 cm
In vitro polyploidization
In vitro polyploidization was conducted in two approaches. The first was performed to select the most adequate colchicine concentration. For this purpose, some criteria were adopted: (a) survival rate of the explants, and (b) number of tetraploid and (c) mixoploid plantlets.
The survival rate of explants not treated with the anti-mitotic agent colchicine, or treated with 0.5 mM for 24 h, was 100 %. With increasing concentration, the survival rate statistically decreased, reaching 0 % when the explants were treated with a colchicine concentration of 1.5 mM for 72 h (Table 2).
Shoot tips treated with colchicine showed a lower growth and multiplication rate, yielding few regenerated plantlets in comparison to explants not treated with colchicine. In consideration of the vegetative development, DNA ploidy level of the plantlets was assessed by FCM only after 4 subcultures in medium M2, totaling 120 days.
Jatropha curcas plantlets cultured in medium M3 without colchicine, as well as greenhouse plants (control; 2C = 0.85 pg; 2× = 22 chromosomes), exhibited G0/G1 peaks in the same channel (Fig. 1a). Therefore, these plantlets (Fig. 1b) showed the same DNA ploidy level. Similarly, some explants treated with colchicine generated seedlings that provided FCM histograms with the same profile in comparison to the control (Table 2; Fig. 1a). Based on this, these plantlets were also considered diploid.
Mixoploid plantlets of J. curcas (Fig. 1c, d) were obtained in colchicine concentration of 0.5 mM for 72 h (10.00 %) and 0.5 mM for 96 h (28.57 %). These plantlets exhibited G0/G1 peaks equivalent to nuclei 2C = 2X and 2C = 4X (Fig. 1c).
FCM analysis also evidenced tetraploid plantlets of J. curcas (Fig. 1e, f). In comparison to control and plantlets cultured in medium M3 without colchicine, these plantlets exhibited G0/G1 peaks equivalent to nuclei 2C = 4X (Fig. 1f). Tetraploids were observed in the treatment with 0.5 mM colchicine for 96 h (14.28 %), and 1.5 mM with exposure time of 48 h (16.66 %) (Table 2).
In regard of the three criteria (number of tetraploid and mixoploid plantlets, and survival rate), the treatment with 0.5 mM colchicine for 96 h was considered the most adequate for polyploidy induction in J. curcas shoot tips. Considering this result, a second polyploidization approach was set (Table 2), increasing the treatment time (96, 120, 144, 168 h) and maintaining the colchicine concentration (0.5 mM).
As in the first polyploidization procedure, approximately 65 % of the explants survived in the 0.5 mM/96 h treatment. Survival rate statistically decreased with the increase in exposure time (Table 2). The second polyploidization experiment confirmed that the 0.5 mM/96 h treatment is efficient for the production of tetraploids (Table 2). Analysis of DNA ploidy level by FCM evidenced that this treatment provided 53.85 % of tetraploids. This treatment also generated 46.15 % of mixoploid regenerants. Tetraploids were also obtained in the exposure time of 120 h, though with smaller percentage (42.85 %) (Table 2). The mixoploid and tetraploid plantlets have been in vitro multiplied, and some ex vitro acclimatized.
Discussion
2C-value and karyotype of J. curcas
Considering that some authors have reported the occurrence of J. curcas tetraploid plants (Soontornchainaksaeng and Jenjittikul 2003; Mukherjee et al. 2011), the first concern was to measure the 2C-value and characterize the karyotype of J. curcas ‘Gonçalo’ explant donors.
As reported by Carvalho et al. (2008), the FCM procedure provided fluorescence peaks of G0/G1 nuclei showing CVs below 5 %. This value has been considered adequate for FCM assessments in plants (Praça-Fontes et al. 2011a). In accordance with Praça-Fontes et al. (2011a), an appropriate preparation of nuclei suspensions is imperative to provide stoichiometrically stained nuclei and, consequently, low CV values, as obtained in the present work.
Solanum lycopersicum ‘Stupické’ was chosen as primary standard for 2C-value measurement. This standard has been regarded as suitable for analyses involving plants rich in phenolic compounds (Clarindo et al. 2012), such as J. curcas. Using FCM, Praça-Fontes et al. (2011a) revisited the DNA C-value of seven species often used as primary standard. In a cascade-like manner, from Arabidopsis thaliana to Allium cepa, these authors demonstrated that S. lycopersicum is an ideal primary reference standard.
The mean nuclear DNA content value of 2C = 0.85 pg found here for J. curcas was identical to the value reported by Carvalho et al. (2008). This result confirms that the nuclear genome size of J. curcas, as well as that of Euphorbia peplus L. (2C = 0.7 pg), is relatively small compared to other Euphorbiaceae species (Bennett and Leitch 2011). Corroborating with the FCM data, karyotype analysis showed that J. curcas ‘Gonçalo’ had 2n = 2× = 22 chromosomes, which is considered relatively small (Carvalho et al. 2008).
The previous FCM and cytogenetic approaches showed that J. curcas ‘Gonçalo’ has a stable genome (2× = 22 chromosomes, 2C = 0.85 pg). Since no pre-existing ploidy variation was evidenced, this line was considered adequate for in vitro polyploidy induction.
In vitro recovery of J. curcas shoot tips
The decontamination process was efficient to provide aseptic cultures of J. curcas. Seeds of this species have a high incidence of fungi (e.g. Alternaria sp., Macrophomina sp., Cladosporium sp., Fusarium spp.), even after a surface decontamination process (Vanzolini et al. 2010). Considering this, the removal of the pericarp, followed by decontamination in laminar flow, was crucial to ensure totally aseptic seeds.
As attested by the high germination rate (86.66 %), medium M1 yielded sufficient amount of seedlings for subsequent excision of shoot tips. Distinct authors (Kalimuthu et al. 2007; Kaewpoo and Te-chato 2010) have also considered M1 an efficient medium for germination of zygotic embryos and seeds of J. curcas.
Besides the high germination rate, J. curcas seeds showed a SG of 4.86 after 8 days in medium M1. This data reflects the efficiency of medium M1 to promote healthy seedlings. Gairola et al. (2011) reported a germination rate of 62.50 % and a SG of 4.85 in J. curcas seeds sown in vermiculite substrate. Thus, the in vitro conditions were adequate for seed germination of this species.
In vitro multiplication and assessment of DNA ploidy level
In accordance with Rajore and Batra (2005), medium M2 was ideal for clonal propagation and multiplication of J. curcas shoot tips. After four subcultures (120 days) in M2, the mean multiplication rate was 4.5 shoots per explant. This result indicates that the BAP/IAA combination in medium M2 increased shoot frequency, being crucial for the success of in vitro propagation and multiplication of J. curcas.
The type and concentration of growth regulators, as well as subculture frequency, can promote the occurrence of numeric chromosomal aberrations (eu- and/or aneuploidy) associated to somaclonal variation. Auxins and cytokinins, such as BAP and IAA, are the main growth regulators that act to control cell division and tissue differentiation (Fehér et al. 2003). These regulators interfere in cell cycle control and may lead to genetic variability (Bairu et al. 2011).
As the medium M2 was supplemented with auxin and cytokinin regulators, leaves from plantlets propagated in this medium were used to assess DNA ploidy level. The nuclei suspension of these plantlets provided G0/G1 peaks in the same channel as the control plants (Fig. 1a). In this sense, medium M2 (Rajore and Batra 2005) did not promote any numeric chromosomal aberration (eu- and/or aneuploidy) during propagation and multiplication of J. curcas plantlets. Therefore, the in vitro tissue culture conditions conserved the nuclear genome stability and homogeneity of propagated and multiplied J. curcas plantlets up to the fourth subculture. As related by Fiuk et al. (2010), ploidy maintenance during in vitro condition is a relevant prerequisite for clonal propagation.
Kour et al. (2009) mentioned that variability can manifest at cytological level. For this reason, FCM is commonly used for detection of DNA ploidy level variation (Clarindo et al. 2008). Besides, FCM analysis is the most practical, reliable and efficient method for this purpose (Clarindo et al. 2008; Praça et al. 2009). Therefore, DNA ploidy level screening of J. curcas plantlet samples could be done rapidly, in a working day, without the requirement of dividing cells.
In vitro polyploidization
The polyploidization experiments were conducted to find colchicine concentrations with limited toxic effect. The effect of colchicine on the survival rate depended upon concentrations of this compound and duration of treatment (Samala and Te-chato 2012).
Previous experiments performed in our laboratory showed that colchicine concentrations of 3.5, 5.0 and 6.5 mM at treatment periods of 24–72 h were highly toxic to shoot tips (data not shown). These explants died in the first week in medium M2. Thereafter, other concentrations and exposure times were tested (Table 2) so as to obtain surviving explants, as well as tetraploid and mixoploid plantlets.
Considering the three adopted criteria, treatment with 0.5 mM colchicine for 96 h was considered the most adequate for polyploidization (Table 2). Based on this result, a new polyploidization approach was performed, in which the colchicine concentration was maintained (Table 2).
The survival rate of J. curcas explants decreased with increasing colchicine concentration and exposure time (Table 2). This result was also found in other polyploidization approaches from woody plants (Gu et al. 2005; Stanys et al. 2006; Xi-Ling et al. 2011; Samala and Te-chato 2012).
Tetraploid plantlets were obtained from different treatments, with 0.5 mM colchicine for 96 h being statistically the most efficient (Table 2). From this treatment, 53.85 % of J. curcas tetraploid plantlets were recovered (Fig. 1f; Table 2). Tetraploid plantlets of the woody species Phlox subulata L. were regenerated by Zhang et al. (2008) also using 0.5 mM colchicine.
Most of the mixoploid plantlets were also obtained in the treatment with 0.5 mM colchicine for 96 h (Table 2). Though in vitro polyploidization generally yields mixoploid plantlets (Chen and Gao 2007), the mixoploid state has been considered reversible, since some plants return to the diploid condition or become tetraploid plants (Väinölä 2000). Due to the simultaneous occurrence of cells with varying ploidy, embryogenesis systems can be established to enable recovery of plantlets showing a single and stable ploidy level (Chen and Gao 2007).
In this study, a tissue culture procedure was adapted for induction, propagation and recovery of tetraploid plantlets of J. curcas. Moreover, nuclear DNA ploidy screening by FCM was a practical and rapid strategy for selection of diploid, mixoploid and tetraploid plantlets. The method presented here is also reliable for routine in vitro production of polyploids of other J. curcas elite lines.
References
Bairu MW, Aremu AO, Van Staden J (2011) Somaclonal variation in plants: causes and detection methods. Plant Growth Regul 63:147–173. doi:10.1007/s10725-010-9554-x
Bennett MD, Leitch IJ (2011) Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann Bot 107:467–590. doi:10.1093/aob/mcq258
Carvalho CR, Clarindo WR, Praça MM, Araújo FS, Carels N (2008) Genome size, base composition and karyotype of Jatropha curcas L., an important biofuel plant. Plant Sci 174:613–617. doi:10.1016/j.plantsci.2008.03.010
Chen L, Gao S (2007) In vitro tetraploid induction and generation of tetraploids from mixoploids in Astragalus membranaceus. Sci Horticult 112:339–344
Clarindo WR, Carvalho CR, Araújo FS, Abreu IS, Otoni WC (2008) Recovering polyploid papaya in vitro regenerants as screened by flow cytometry. Plant Cell Tiss Organ Cult 92:207–214. doi:10.1007/s11240-007-9325-1
Clarindo WR, Carvalho CR, Mendonça MAC (2012) Cytogenetic and flow cytometry data expand knowledge of genome evolution in three Coffea species. Plant Syst Evol 298:835–844. doi:10.1007/s00606-012-095-7
Dhooghe E, Van Laere K, Eeckhaut T, Leus L, van Huylenbroeck J (2011) Mitotic chromosome doubling of plant tissues in vitro. Plant Cell Tiss Organ Cult 104:359–373. doi:10.1007/s11240-010-9786-5
Divakara BN, Upadhyaya HD, Wani SP, Gowda CLL (2010) Biology and genetic improvement of Jatropha curcas L.: a review. Appl Energy 87:32–742. doi:10.1016/j.apenergy.2009.07.013
Doležel J, Bartoš J, Voglmayr H, Greilhuber J (2003) Nuclear DNA and genome size of trout and human. Cytometry 51:127–128
Fehér A, Pasternak TP, Dudits D (2003) Transition of somatic plant cells to an embryogenic state. Plant Cell Tiss Organ Cult 74:201–228. doi:10.1023/A:1024033216561
Fiuk A, Bednarek PT, Rybczyński JJ (2010) Flow cytometry, HPLC-RP, and metAFLP analyses to assess genetic variability in somatic embryo-derived plantlets of Gentiana pannonica scop. Plant Mol Biol Rep 28:413–420. doi:10.1007/s11105-009-0167-3
Gairola KC, Nautiyal AR, Dwivedi AK (2011) Effect of temperatures and germination media on seed germination of Jatropha curcas Linn. Adv Biores 2:66–71
Gu XF, Yang AF, Meng H, Zhang JR (2005) In vitro induction of tetraploid plants from diploid Zizyphus jujuba Mill. cv. Zhanhua. Plant Cell Rep 24:671–676. doi:10.1007/s00299-005-0017-1
Jha TB, Mukherjee P, Datta MM (2007) Somatic embryogenesis in Jatropha curcas Linn., an important biofuel plant. Plant Biotechnol Rep 1:135–140. doi:10.1007/s11816-007-0027-2
Kaewpoo M, Te-chato S (2010) Study on ploidy level of micropropagated Jatropha curcas L. via flow cytometry. J Agric Technol 6(2):391–400
Kalimuthu K, Paulsamy S, Senthilkumar R, Sathya M (2007) In vitro propagation of the biodiesel plant Jatropha curcas L. Plant Tiss Cult Biotech 17(2):137–147
Kour G, Kour B, Kaul S, Dhar MK (2009) Genetic and epigenetic instability of amplification-prone sequences of a novel B chromosome induced by tissue culture in Plantago lagopus L. Plant Cell Rep 28:1857–1867. doi:10.1007/s00299-009-0789-9
Kumar N, Reddy MP (2012) Thidiazuron (TDZ) induced plant regeneration from cotyledonary petiole explants of elite genotypes of Jatropha curcas: a candidate biodiesel plant. Ind Crop Prod 39:62–68. doi:10.1016/j.indcrop.2012.02.011
Mukherjee P, Varshney A, Johnson TS, Jha TB (2011) Jatropha curcas: a review on biotechnological status and challenges. Plant Biotechnol Rep 5:197–215. doi:10.1007/s11816-011-0175-2
Ochatt SJ (2008) Flow cytometry in plant breeding. Cytometry 73:581–598. doi:10.1002/cyto.a.20562
Praça MM, Carvalho CR, Clarindo WC (2009) A practical and reliable procedure for in vitro induction of tetraploid tomato. Sci Horticult 122:501–505. doi:10.1016/j.scienta.2009.05.032
Praça-Fontes MM, Carvalho CR, Clarindo WR, Cruz CD (2011a) Revisiting the DNA C-values of the genome size-standards used in plant flow cytometry to choose the ‘‘best primary standards’’. Plant Cell Rep 30:1183–1191. doi:10.1007/s00299-011-1026-x
Praça-Fontes MM, Carvalho CR, Clarindo WR (2011b) C-value reassessment of plant standards: an image cytometry approach. Plant Cell Rep 30:2303–2312. doi:10.1007/s00299-011-1135-6
Rajore S, Batra A (2005) Efficient plant regeneration via shoot tip explant in Jatropha curcas. J Plant Biochem Biotechnol 14:73–75
Samala S, Te-chato S (2012) Ploidy induction through secondary somatic embryo (SSE) of oil palm by colchicine treatment. J Agric Technol 8:337–352
Silva FAS (2012) Programa ASSISTAT—Aplicativo Computacional em Estatística. DEAG-CTRN-UFCG, Campina Grande-PB, Brasil
Soontornchainaksaeng P, Jenjittikul T (2003) Karyology of Jatropha (Euphorbiaceae) in Thailand. Thai For Bull (Bot) 31:105–112
Stanys V, Weckman A, Staniene G, Duchovskis P (2006) In vitro induction of polyploidy in Japanese quince (Chaenomeles japonica). Plant Cell Tiss Organ Cult 84:263–268. doi:10.1007/s11240-005-9029-3
Väinölä A (2000) Polyploidization and early screening of Rhododendron hybrids. Euphytica 112:239–244
Vanzolini S, Meorin EBK, Silva RA, Nakagawa J (2010) Qualidade sanitária e germinação de sementes de pinhão-manso. Revista Brasileira de Sementes 32:1–9
Xi-Ling W, Jin-Xing Z, Mao-De Y, Zhen-Gang L, Xiao-Yun J, Qi-You L (2011) Highly efficient plant regeneration and in vitro polyploid induction using hypocotyl explants from diploid mulberry (Morus multicaulis Poir.). In Vitro Cell Dev Biol Plant 47:434–440. doi:10.1007/s11627-010-9328-1
Zhang Z, Dai H, Xiao M, Liu X (2008) In vitro induction of tetraploids in Phlox subulata L. Euphytica 159:59–65. doi:10.1007/s10681-007-9457-8
Acknowledgments
The authors are grateful to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, DF, Brazil), Fundação de Amparo à Pesquisa do Espírito Santo (FAPES, Vitória, ES, Brazil), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG, Belo Horizonte, MG, Brazil), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brasília, DF, Brazil) for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de Oliveira, S.C., Nunes, A.C.P., Carvalho, C.R. et al. In vitro polyploidization from shoot tips of Jatropha curcas L.: a biodiesel plant. Plant Growth Regul 69, 79–86 (2013). https://doi.org/10.1007/s10725-012-9749-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-012-9749-4