Abstract
Distillers’ dried grains with solubles (DDGS) is a by-product of dry-mill corn ethanol production comprising a high nutritional value due to residual fiber, protein, and lipid contents. The fiber content of DDGS is high enough to be considered a valuable source for the production of hydrolytic enzymes, such as cellulase and xylanases, which can be used for hydrolysis of lignocellulosic feedstock during ethanol production. The DDGS-based medium prepared after acid hydrolysis provides adequate sugars for enzyme production, while additional macronutrients, such as salts and nitrogen sources, can enhance the enzyme production. Therefore, this study was undertaken to evaluate the effect of salts (KH2PO4, CaCl2·2H2O, MgSO4·7H2O, FeSO4·7H2O, CoCl2·6H2O, and MnSO4·H2O), peptone, and yeast extract on enzyme secretion by four different Aspergillus niger strains and to optimize the nitrogen source for maximum enzyme production. Yeast extract improved the cellulase production (0.38 IU/ml) for A. niger (NRRL 1956) as compared to peptone (0.29 IU/ml). However, maximum cellulase productions of 0.42 IU/ml and 0.45 IU/ml were obtained by A. niger (NRRL 330) and A. niger (NRRL 567), respectively, in presence of ammonium sulfate. The optimized nitrogen amounts resulted in a significant increase in the cellulase production from 0.174 to 0.63 IU/ml on day 9 of the fermentation with A. niger (NRRL 330). The composite model improved both cellulase and xylanase production. In conclusion, the optimization of all three nitrogen sources improved both cellulase and xylanase production in the DDGS-based media.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Among various renewable energy solutions for a clean environment and energy security, biofuel is the most popular and efficient option [1]. The production of bioethanol among biofuels grows annually by 6% due to high demand and policy [2]. The bioethanol has increased from 17 billion liters to 108 billion liters in the last two decades [2, 3]. It is considered feasible in the transportation industry and has been blended at different proportions such as 10–15% in the USA. The main feedstock for bioethanol production is starch-based grains or crops, such as corn, wheat, and barley. Currently, the USA and Brazil are the two major producers of bioethanol from corn and sugarcane [1].
Bioethanol produced from corn, wheat, or sugarcane is known as the first-generation bioethanol (1G). A more common process of producing bioethanol from grains is the dry-mill process, in which grains are ground and mixed with the saccharifying enzymes and water to turn into a mesh in a consecutive step called liquefaction. After conversion of simple sugars into ethanol through fermentation, almost one-third of the grain is left as residue, which is known as distillers’ dried grains with solubles (DDGS). The final steps in this process are centrifugation of distiller grain, concentration of thin stillage, blending of syrup with distiller grain, and drying. A large portion of DDGS is composed of undigested fibers [28.2–40.3% neutral detergent (NDF) and 10.3 to 18.1% acid detergent fibers (ADF)]. The difference between NDF and ADF is an estimation of hemicellulose while ADF represents the sum of cellulose and lignin in the material [4]. DDGS is also high in protein (23.4–28.7%) and lipid (2.9–12.8%) contents [5]. The authors estimated these ranges by performing standard AOAC procedures [6]. This high nutritional profile makes DDGS an excellent source for microbial fermentation [7,8,9].
Production of biofuels from food crops is viewed controversial and therefore the demand for bioethanol from lignocellulosic biomass (known as second-generation fuel, 2G), such as crops, grasses, and agricultural waste materials, is high [10,11,12,13]. While the interest in 2G bioethanol is increasing, the industrial production of 2G ethanol is not realized yet at a large scale due to the need for hydrolytic enzymes, such as cellulases and xylanases, which are produced by expensive cellulosic materials. Since DDGS has a high nutritional profile [14], the high fiber content can be used for the production of cellulase and xylanases [15, 16]. This can combine and improve the profitability of both G1 and G2 bioethanol in near future.
The main producers of cellulases and xylanases are filamentous fungi, such as Aspergillus niger and Trichoderma reesei [17]. There are many strains of these fungi that are reported for hydrolytic enzyme productions, such as A. niger (NRRL 2001) [16], A. niger (NRRL 567) [18], T. reesei (ATCC 56765) [19], and T. reesei (NRRL 6156) [15]. On the other hand, some bacterial strains, like Clostridium thermocellum and Bacillus subtilis, are also known as cellulase and xylanase producers [20]. In our previous study, a comparison between selected fungal and bacterial strains was made and it was found that the evaluated fungal strains produced higher levels of cellulases and xylanases than the bacterial strains [21]. The DDGS was also proven as efficient carbon source for hydrolytic enzyme production in our previous work, but still improvements can be achieved with the help of different strategies, such as optimization of culture parameters and nutrient amendments [13, 19, 20]. The nutrient fraction that can affect the enzyme production are salts, such as KH2PO4, CaCl2·2H2O, MgSO4·7H2O, FeSO4·7H2O, CoCl2·6H2O, and MnSO4·H2O and nitrogen sources [23, 24]. The nitrogen source can be either organic, such as peptone or yeast extract or inorganic like ammonium sulfate [25]. Both types of nitrogen sources are of high efficacy in different metabolic processes. However, it is still important to know whether organic or inorganic sources are better for cellulase and xylanase production.
There are different strategies to determine the effect of specific medium components or culture parameters, such as carbon source, pH, temperature, and agitation, on microbial product formation. Statistical designs such as response surface design (RSM) are commonly used for optimization studies [26, 27].
Among medium ingredients, salts and nitrogen have an essential role in metabolic activity of cells, and thus in hydrolytic enzyme production [22, 28]. Therefore, enzyme production can be improved by supplementing or adjusting salts and nitrogenous substances. However, neither the main effect nor the concentrations of such ingredients have never been evaluated for the DDGS-based medium in the literature so far. Therefore, the objective of this study is to evaluate the effect of salts and nitrogen sources on cellulase and xylanase production in the DDGS-based media. In particular, optimization of peptone, yeast extract, and ammonium sulfate concentrations was studied by the response surface design method after examining their effect with or without salts. The goal of this study was to enhance the cellulase and xylanase productions by salt and nitrogen amendments of the DDGS-based fermentation media (peptone, yeast extract, and ammonium sulfate) for three A. niger strains and one T. reesei strain.
Materials and methods
Microbial strains
The following fungal strains were selected for this study based on our previous study [21]: Aspergillus niger (NRRL 330), A. niger (NRRL 567), A. niger (NRRL 1956), and Trichoderma reesei RUT-C30 (ATCC 56765). The NRRL strains were obtained from Agricultural Research Service (ARS) Culture Collection (Peoria, IL), while the ATCC strains were obtained from American Type Culture Collection (ATCC, Manassas, VA). After obtaining the strains, they were kept frozen at -80 °C in 20% glycerol solution and were revived using potato dextrose agar (PDA, BD Diagnostic Systems, Cockeysville, MD) as needed. The activated strains were sub-cultured biweekly using PDA slants by incubating at 30 °C for 5 days.
Acid hydrolysis of DDGS
The corn DDGS was obtained from Pennsylvania Grain Processing, LLC® (Clearfield, PA). The chemical composition of DDGS including NDF (28.40%) and ADF (11.20%) is available in our earlier work [29]. The pre-treatment of DDGS was carried out according to a study conducted in our lab [29]. Briefly, acid hydrolysis was undertaken by autoclaving the DDGS slurry [20% (w/v) solid load] prepared with 5% (w/w) sulfuric acid (density: 1.0317 g/ml) in DI water at 121 °C and 15 psi for 30 min in a laboratory autoclave (Model Beta Star, RV Industries, Honey Brook, PA) [29]. The DDGS slurries were then cooled to room temperature and filtered by cheesecloth (VWR, Radnor, PA) using a vacuum filter apparatus (Nalgene, Rochester, NY). The pH of the filtrate was adjusted to pH 5 by adding approximately 10 ml of 10 M NaOH solution.
Media preparation and nutrient supplementation
The DDGS filtrate was centrifuged at 1510×g for 10 min to separate the solid residues from the liquid phase and 100 ml of this filtrate was then transferred to 250-ml Erlenmeyer flasks. Finally, the salts shown in Table 1 and nitrogen sources were added to each flask according to the experimental design as explained below.
Experimental design
Evaluation of medium elements
The salts and nitrogenous substances shown in Table 1 were used to test their potential to improve enzyme production. The quantities and limits of mineral and nitrogen sources were inspired from literature [30]. Peptone (Bacto™ Life Technologies, Carlsbad, CA), yeast extract (VWR Life Sciences, Bacteriological Grade, Radnor, PA), and ammonium sulfate (VWR, Radnor, PA) concentrations were examined separately for their effect on enzyme production. The salts were added in the first phase of the study and their effect on enzyme production was evaluated prior to the optimization stage.
Optimization of nitrogen sources
The Box–Behnken response surface design for optimization of nitrogen source was constructed for peptone (0.5–5 g/l), yeast extract (2–20 g/l), and ammonium sulfate (0.5–2 g/l) using Minitab Statistic Software Package (Version 19, Minitab Inc., State College, PA) for the selected strains. After adding nitrogen sources according to the experimental design, the flasks were autoclaved for 20 min to sterilize the media. The optimized results obtained from the RSM were then repeated with three replicates for validation. The following polynomial equation was fitted to data when all factors and interactions are to be significant:
where Y is the xylanase or cellulase activity (IU/ml), b’s are the coefficients, and X1, X2, X3, are peptone, yeast extract, and ammonium sulfate in g/l, respectively.
Inoculum preparation and fermentation
The fungal strains from the PDA slants were rinsed with a pre-sterilized 1 ml 0.1% (w/w) Tween 80 (VWR) solution and 0.1 ml of the resulting spore suspension was transferred to each of PDA plates, which were then incubated for 5 days at 30 °C for sporulation [23]. The grown spores were harvested with glass Hockey stick using about 10 ml pre-sterilized 0.1% (w/w) Tween 80 solution for each plate to give spore suspension with OD600 of 0.9 for inoculation [30, 31]. The inoculated flasks were incubated at 30 °C and 180 rpm in a shaker incubator (MaxQ 5000, Thermo Scientific, Laguna Hills, CA) for enzyme production. Aliquots of samples were taken on day 9 aseptically.
Enzyme activity analysis
Samples (ca. 3 ml) were taken on day 9 and kept at 4 °C until analysis. Before analysis, the culture slurries were filtered using 0.45 µm PTFE syringe filters (VWR, Radnor, PA) to remove DDGS residues and fungal biomass from the liquid enzyme extract. The samples were analyzed for both cellulase and xylanases using the following methods.
Cellulase analysis
Cellulase activity of the culture extracts was assayed according to the dinitrosalicyclic acid (DNS) method (Miller 1959). Approximately 50 mg or 1 × 6 cm of Whatman No. 1 filter paper strips (GE Healthcare, Chicago, IL) was used as the substrate in citrate buffer (pH 4.8). The 1 ml of appropriately diluted enzyme samples, the filter paper strips, and citrate buffer were mixed and incubated at 50 °C for 1 h, along with the reagent, substrate, and enzyme blanks, and the glucose standards (0.1–0.6 mg/ml). After incubation, 3 mL of dinitrosalicyclic acid (DNS; VWR) was added and all the tubes were boiled for 15 min to stop the reaction. After cooling the tubes to room temperature, the absorbance of the samples was measured at 575 nm with a spectrophotometer (Evolution 21, Thermo Scientific, Oakwood, OH). One-unit activity or international unit (IU) of cellulase was defined as the amount of enzyme that liberates one micromole of glucose per minute under assay conditions.
Xylanase analysis
The substrate for xylanase assay was Birchwood xylan dissolved in acetate buffer (pH 5.0) at 0.5% (w/w) (Crescent Chemicals, Islandia, NY). The diluted sample with substrate and buffer along with enzyme and substrate blank tubes were incubated at 50 °C for 30 min. Immediately after incubation, DNS (VWR) solution was added to tubes and boiled for 15 min to stop the reaction. The absorbance of the samples was measured at 575 nm with a spectrophotometer and the recorded values were translated into enzyme activity using the standard curve prepared at 0.1–0.6 mg/ml of xylose (VWR). One unit of the enzyme or international unit (IU) is defined as the amount of enzyme needed to release one micromole of xylose per minute under assay conditions [32].
Statistical analysis
The treatments with varying salts and individual nitrogen sources were compared by pair-wise comparisons using Minitab Statistical Software (Version 19, Minitab Inc, State College, PA) at p < 0.05. The work of RSM optimization was also designed, analyzed, and validated using Minitab Statistical Software.
Results
Effect of salts and nitrogen sources
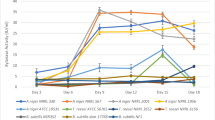
Figure 1 summarizes the effect of salt and nitrogen supplements on cellulase production. The two-way analysis of variance showed that each strain responded differently to each medium component (p < 0.05). For A. niger (NRRL 1956), the cellulase production increased from 0.14 to 0.38 IU/mL within 9 days supplementing the medium with only yeast extract. The salt addition caused the cellulase activity to decrease from 0.38 to 0.31 ml, which was found statistically insignificant (p > 0.05). A similar effect was also observed when only ammonium sulfate as an inorganic nitrogen source was used in the hydrolyzed DDGS medium for A. niger (NRRL 1956), for which cellulase significantly increased from 0.14 to 0.31 on day 9 (p < 0.05). The addition of peptone also increased cellulase production from 0.14 IU/ml to 0.29 (Fig. 1). While all three nitrogen sources had a positive effect on cellulase production by A. niger (NRRL 1956), the addition of salt did not increase cellulase production with any of the nitrogen sources for A. niger 1956. Collectively, all nitrogen sources resulted in the increase of cellulase production when added individually for this strain.
Similar trends were observed for A. niger (NRRL 330). However, the increase was not evident as much as A. niger (NRRL 1956). Only the addition of ammonium sulfate showed a significant increase in cellulase production (0.44 IU/ml) for A. niger (NRRL 330). The addition of yeast extract, ammonium salt, or peptone increased cellulase production but the change is insignificant. The addition of salt did not yield any further increase for all three nitrogen sources. All nitrogen sources were not effective on A. niger (NRRL 567), resulting in similar enzyme productions. The addition of salt had a significant negative effect on this strain as well. A. niger (NRRL 567) produced maximum cellulase without the addition of any nitrogen sources or salts as compared to all other evaluated strains. Finally, T. reesei (ATCC 56765) showed no remarkable improvement after the addition of any nitrogen sources. An interesting trend was observed with T. reesei (ATCC 56,765) showing cellulase increase from 0.19 to 0.35 IU/ml when peptone and salts were added.
Figure 2 shows the effect of different nitrogen sources along with salts on xylanase production by three A. niger strains and T. reesei. In comparison to cellulase, DDGS-based media with no salts or nitrogen sources show better enzyme production for some of the strains. In addition, the trends in the change of enzyme production after the addition of nitrogen sources and salts were very different from cellulase production. For example, A. niger (NRRL 1956) showed no significant increase by the addition of any type of nitrogen source, and the same was correct for the supplement of salts into the media. The effect of peptone on xylanase production by A. niger (NRRL 330) was evident resulting in an increase from 27.5 to 42 IU/ml (Fig. 2). The xylanase with ammonium sulfate was much better, causing an increase from 27.5 to 49 IU/ml. This increase shows the positive effect of peptone and ammonium sulfate on xylanase production. However, yeast extract had significantly negative effect on xylanase production (Fig. 2).
Similar to A. niger (NRRL 1956), A. niger (NRRL 567) was not positively affected by the addition of nitrogen sources for xylanase production. The decrease was significant for both yeast extract and peptone, which confirms no benefits of adding organic nitrogen sources to the media. Xylanase production by T. reesei (ATCC 56,765) was very low (1.4 IU/ml) compared to cellulase production. Thus, the addition of nitrogen sources and salts shows suppressing effects on xylanase production by this strain.
Optimization of nitrogen sources for cellulase production
After analyzing the effect of minerals and nitrogen sources, the three selected nitrogen sources (peptone, yeast extract, and ammonium sulfate) were optimized for cellulase production by two selected A. niger strains. The results for A. niger (NRRL 330) are given in Table 2. Maximum cellulase activity was obtained for 5 g/l peptone, 11 g/l yeast extract, and 2 g/l ammonium sulfate. The maximum cellulase activity was, thus correlated with the maximum concentration of each of the nitrogen sources. The R2 for this model was 0.957 and the lack of fit value was 0.357. The response surface plots of this model are given in Fig. 3. As it can be seen in the surface plots, both ammonium sulfate and peptone have a linear positive effect on cellulase production, while yeast extract enhances cellulase at high amounts, justifying the significant square of yeast extract term in the model.
The model also predicts maximal cellulase at maximum concentrations for each of the nitrogen sources. To evaluate the effect of a further increase in their concentration, additional runs were carried out with 30% less and 30% higher than the recommended level and the results are presented in Fig. 4. While the results of 30% lesser and higher than the optimized values were not statistically different from each other, the results of the optimized values were higher than both, which approve the validity of the obtained optimal values.
The results of response surface optimization for cellulase production are given in Table 3 for A. niger (NRRL 567). The maximal cellulase was produced at maximum peptone concentration (5 g/l) and minimum yeast extract (2 g/l) and moderate ammonium sulfate (1.25 g/l) concentrations. However, the R2 for this model was lower than that of A. niger (NRRL 330). The experimental values of optimal conditions were also lower (0.62 IU/ml) than the predicted values.
Optimization of nitrogen sources for xylanase production
Compared to cellulase, xylanase results of A. niger strains were better in terms of overall enzyme production. The results of response surface optimization for xylanase production by A. niger (NRRL 330) are given in Table 2. The maximum production of xylanase was observed to be 54.64 IU/ml with 0.5 g/l peptone, 11 g/l yeast extract, and 2 g/l ammonium sulfate. This RSM model had R2 value of 0.804. The lack of fit value was 0.109. Ammonium sulfate and its square were the most significant terms in the model and the model predicted maximal xylanase of 55.8 IU/ml. The increase in the ammonium sulfate increased xylanase production as indicated in Fig. 5. While the optimum value of yeast extract was within the concentration range that was selected for this study, the minimum amount of peptone was correlated to the highest enzyme production.
The results for xylanase optimization by A. niger (NRRL 567) are shown in Table 3. Maximum xylanase activity of 54.88 IU/ml was measured, which was not significantly different from that of A. niger (NRRL 330). In addition, the maximum xylanase production was obtained with 2.75 g/l peptone, 11 g/l yeast extract, and 1.25 g/l ammonium sulfate, which are mid-values of the tested amounts.
Optimization of nitrogen sources for enzyme cocktails
A. niger strains (NRRL 330 and NRRL 567) showed promising results for both cellulase and xylanase production separately. However, both enzymes need to be simultaneously produced to reduce the cost of enzyme during industrial hydrolysis of lignocellulosic biomass. Therefore, co-production of cellulases and xylanases was searched using the obtained individual response surface models. For A. niger (NRRL 330), the optimized values of peptone, yeast extract, and ammonium sulfate for maximal co-production were 5, 16.5, and 1.9 g/l, respectively (Fig. 6). The composite D-value for the combined model was 0.97. The model also suggested that the composite desirability for cellulase model is higher than that of xylanase production. The composite model predicts 0.56 and 48.8 IU/ml of cellulase and xylanase, respectively. The experimental counterparts were 0.54 ± 0.02 for cellulase and 48.71 ± 2.05 for xylanase (Table 4), which are quite close to the predicted values. Similarly, the composite optimized values for A. niger (NRRL 567) are shown in Fig. 7. For this strain, the optimized values were 5 g/l peptone, 12.4 g/l yeast extract, and 1.5 g/l ammonium sulfate. The experimental values of the predicted optimal conditions are also given in Table 4 and also considered close to the predicted values with small values of coefficient variation (0.345 and 0.055 for cellulase and xylanase, respectively).
Discussion
The salt and nitrogen amendment shows different results for different strains and leads to valuable results that were used for the optimization of medium components in the next phase of the study. Ammonium sulfate is an inorganic nitrogen source, which is inexpensive and preferable to organic sources. DDGS has 28–29.5% dry basis protein, which can serve as nitrogen source for enzyme production [14]. However, the amino acid profile of the protein can vary with grain type and batch operations [6, 33, 34]. Therefore, addition of nitrogenous substances is feasible for cellulase production.
Many other studies in the past have reported the effect of nitrogen sources on enzyme production. For example, Rodriguez-Gomez and Hobley [24] reported positive effects of peptone and ammonium sulfate on cellulase production by T. reesei (ATCC 56765). However, they used Avicel or microcrystalline cellulose, which is an ideal, but very expensive feedstock for enzyme production [24]. Therefore, it can be inferred that T. reesei (ATCC 56765) need specific polysaccharides for the enzyme production that were not present in acid hydrolysate of DDGS. However, the high efficacy of A. niger strains to produce cellulase and hemicellulose in the acid hydrolysate of DDGS shows that such enzymes can be produced with A. niger strains instead of T. reesei RUT-C30 using inexpensive carbon sources such as DDGS. It was also concluded that ammonium sulfate was a better source than peptone, which is similar to the results of our study. The effect of medium elements on the cellulase production has been studied with special focus on the fiber type, nitrogen source and mineral composition [30]. The nitrogen sources are needed for synthesis of essential amino acids during growth and enzyme production while the minerals provide co-factors and other micronutrients. The fiber on the other hand induces the particular enzyme production.
In comparison to cellulase, xylanase production with respect to different medium components is not studied widely in the literature. There are few studies that utilize the concept of hemicellulose present in the agricultural biomass such as sugar beet pulp [35, 36]. The DDGS has a high fiber content and therefore has been proven to be an excellent source for xylanase production. This study shows that while peptone and ammonium sulfate can be good amendment options for xylanase production, this is true only for A. niger (NRRL 330) while other fungal strains can act differently in the presence of these nitrogen sources.
In our earlier study, maximum cellulase of 0.4 IU/ml was obtained, which had been increased from 0.174 IU/ml in DDGS-based media without any supplement [21]. However, the enzyme production was not stable and decreased after three days. Therefore, it was concluded that enzymes produced on day 6 and day 9 were correlated with the components present in the media and can be optimized further to increase enzyme production. The potential of A. niger (NRRL 330) for different hydrolytic enzyme production was reported in various research articles [37,38,39]. A. niger (NRRL 330) was reported as one of the best strains for glucoamylase production [39]. The medium supplementation improved the enzyme production by 126% for this strain. This shows the potential of this strain for metabolic engineering and improvement of hydrolytic enzyme production. However, the medium optimization can vary based on the enzyme under consideration. For example, in the case of glucoamylase, malt extract, CaCl2·2H2O and FeSO4·7H2O were reported to have a significant effect on glucoamylase production [39]. For cellulase production, our study shows that salt supplements do not have a positive effect.
Enzyme production by A. niger (NRRL 330) through response surface optimization of nitrogen sources gave very promising results for cellulase and xylanase (Table 2). The individual cellulase model predicted maximum concentrations of each of the nitrogen sources as ideal for maximum enzyme activity. Similar results were also reported by Acharya et al. [40], who studied optimization of peptone and ammonium sulfate. In their study, maximum enzyme activity was obtained at 0.125% of peptone and 0.14% of ammonium sulfate. While these values are less than what was predicted by the RSM model in our study, the overall cellulase activities (0.12–0.15 IU/ml) were also lower than our study. In this study, cellulase production was increased from 0.174 to 0.63 IU/ml by optimizing the three nitrogen sources (Fig. 4). The lack of fit values also suggested that these models of nitrogen source optimization fit well and the lack of fit value is insignificant compared to the pure error. The validation experiments also show that further increase in the concentrations of these three nitrogen sources does not increase the enzyme production further. In fact, only 0.56 IU/ml was obtained by increasing the nitrogen source concentration by 30%. This cellulase activity is less than the suggested by the RSM model. Therefore, it can be concluded that RSM modeling is an effective way to optimize the concentrations of nitrogen sources in the medium, as RSM optimization resulted in the ideal concentrations of nitrogen sources for maximum cellulase production.
In case of xylanase production, the model predicted similar values for yeast extract (16.9 g/l) and ammonium sulfate (2 g/l). However, the concentration of peptone (0.5 g/l) was very different from that of cellulase as peptone negatively affected xylanase (Fig. 5). Therefore, it was disregarded while making a composite RSM model where both cellulase and xylanase were increased simultaneously. The composite model predicted concentrations of nitrogen sources that were not very different from the cellulase model (Fig. 6). The concentration of yeast extract was 16.5 g/l instead of 20 g/l and the concentration of ammonium sulfate was 1.9 g/l instead of 2 g/l, while the concentration of peptone was the same for both models.
In case of A. niger (NRRL 567), cellulase production was higher than A. niger (NRRL 330). However, the variation in the predicted and experimental values is slightly distinct (Table 4). In the case of xylanase, the experimental value obtained was 54.67 ± 2.39 IU/ml while the model predicted 50.55 IU/ml. This strain has been studied widely in terms of different enzymes, specifically hydrolytic enzymes. For example, Ghori et al. [18] used corn stover as the substrate with A. niger (NRRL 567) to make a set of three different cellulases mainly exoglucanase, endoglucanase, and β-glucosidase analyzing ammonium sulfate and urea as the nitrogen sources and obtained different values of enzymes according to their research design. Overall, urea was reported to be more efficient than ammonium sulfate [18]. Some other studies also showed the promise of this strain for other microbial products such as citric acid [41]. This strain was also tested with the validated optimized conditions of A. niger (NRRL 330) model, and it produced 0.59 ± 0.04 IU/ml cellulase and 57.6 ± 0.72 IU/ml xylanase. Therefore, it can be concluded that the optimized nitrogen concentrations obtained for A. niger (NRRL 330) are also good for enzyme production by A. niger (NRRL 567).
While both organic and inorganic nitrogen sources are utilized in the synthesis of essential amino acids required for growth and enzyme production, inorganic nitrogen sources are considered better because they are inexpensive. Similarly, cellulase production is favorable if the medium has cellulose fibers in it and the degradation of such fibers is required to produce simple sugars for energy needs [42]. Cellulase enzymes, however, are not needed once enough simple sugars are available in the medium. To induce cellulase production after this stage, it is necessary to extract cellulase from the fermentation media.
The incubation time for all experiments in this study was kept at 9 days. The results of this study show that if the right nutrients are present, the fungi can produce cellulase in the early days of fermentation. Once simple sugars such as glucose are released, the fungal strains tend to increase mycelial growth instead of hydrolytic enzyme productions [43]. The lowered tendency for enzyme can be correlated with many reasons but the most important could be the transition from enzyme production to mycelial growth after obtaining enough amounts of cellulase for biomass degradation in the media. The product inhibition in case of cellulase could be another reason for the decrease in the enzyme activity in the media [44].
As indicated earlier, A. niger (NRRL 330) is known as one of the best strains for hydrolytic enzyme production [37,38,39]. The xylanases are also hydrolytic enzymes that are necessary for industrial production of second-generation biofuels. This study confirms a promising increase in the xylanases production by optimizing the nitrogen sources in the DDGS-based media. The same is true for A. niger (NRRL 567). Both strains have the potential to be used in the industrial production of xylanases using DDGS-based media supplemented with additional nitrogen.
The production of xylanases in the early days of fermentation by A. niger (NRRL 567) was also reported by Dhillon et al. [45] However, the fermentation mode and substrate were different from our study. The solid-state fermentation was used while the main substrate was apple pomace [45]. Regardless of these two differences, the highest enzyme productions were reported within 48–72 h. Therefore, this strain has the potential to produce the xylanase enzymes in the early days of the fermentation, which can be economically beneficial.
The analysis of the developed models shows that cellulase production can be predicted more efficiently than xylanase production. The extent of improvements in cellulase production is also higher than that of xylanases. The effect of nitrogen sources on xylanase production is not studied as widely as cellulases. However, the results show that xylanase production can be increased by adding different types of nitrogen sources into the fiber-based medium. The hydrolysis of DDGS provides simple sugars to meet the energy needs of fungal strains; but the presence of fiber components in the medium correlates with the high production of xylanase enzymes [21].
Conclusion
The effect of salts and nitrogen sources on the production of cellulases and xylanases is evaluated in this study. Salts did not have a positive effect (p > 0.05) on cellulase and xylanase production for the evaluated A. niger strains. The optimization of nitrogen sources without salts improved the cellulase production from 0.174 to 0.56 IU/ml for A. niger (NRRL 330). Xylanase production was increased significantly by this strain. The results show the efficiency of using nitrogen sources to decrease the fermentation period and increase enzyme production in the early days of fermentation.
References
RFA (2019) 2019 ethanol industry outlook. https://ethanolrfa.org/wp-content/uploads/2019/02/RFA2019Outlook.pdf. Accessed 26 Dec 2019
RFA (2019) Annual world fuel ethanol production. https://ethanolrfa.org/statistics/annual-ethanol-production/. Accessed 26 Dec 2019
Liu K (2011) Chemical composition of distillers grains, a review. J Agric Food Chem 59:1508–1526
Iram A, Cekmecelioglu D, Demirci A (2020) Distillers’ dried grains with solubles (DDGS) and its potential as the fermentation feedstock. Appl Microbiol Biotechnol (in review)
Świątkiewicz S, Koreleski J (2008) The use of distillers dried grains with solubles (DDGS) in poultry nutrition. Worlds Poult Sci J 64:257–266
Cromwell GL, Herkelman KL, Stahly TS (1993) Physical, chemical, and nutritional characteristics of distillers dried grains with solubles for chicks and pigs. J Anim Sci 71:679–686
Nghiem NP, Montanti J, Kim TH (2016) Pretreatment of dried distiller grains with solubles by soaking in aqueous ammonia and subsequent enzymatic/dilute acid hydrolysis to produce fermentable sugars. Appl Biochem Biotechnol 179:237–250
Houweling-Tan GBN, Sperber BLHM, van der Wal H et al (2016) Barley distillers dried grains with solubles (DDGS) as feedstock for production of acetone, butanol and ethanol. Wageningen University, Wageningen
Brock S, Kuenz A, Prüße U (2019) Impact of hydrolysis methods on the utilization of agricultural residues as nutrient source for d-lactic acid production by Sporolactobacillus inulinus. Fermentation 5:12
Kang Q, Appels L, Tan T, Dewil R (2014) Bioethanol from lignocellulosic biomass: current findings determine research priorities. Sci World J 2014:13
Zhao X-Q, Zi L-H, Bai F-W et al (2011) Bioethanol from lignocellulosic biomass. biotechnology in China III: biofuels and bioenergy. Springer, Berlin, pp 25–51
Balat M (2011) Production of bioethanol from lignocellulosic materials via the biochemical pathway: a review. Energy Convers Manag 52:858–875
Graham-Rowe D (2011) Beyond food versus fuel. Nature 474:S6
Chrenková M, Čerešňáková Z, Formelová Z et al (2012) Chemical and nutritional characteristics of different types of DDGS for ruminants. J Anim Feed Sci 663:127
Cekmecelioglu D, Demirci A (2020) Production of cellulase and xylanase enzymes using distillers dried grains with solubles (DDGS) by Trichoderma reesei at shake-flask scale and the validation in the benchtop scale bioreactor. Waste Biomass Valoriz 1–10 (in press)
Ximenes EA, Dien BS, Ladisch MR et al (2007) Enzyme production by industrially relevant fungi cultured on coproduct from corn dry grind ethanol plants. In: Mielenz JR, Klasson KT, Adney WS, McMillan JD (eds) Applied biochemistry and biotechnology. ABAB symposium. Humana Press, Totowa, pp 171–183. https://doi.org/10.1007/978-1-60327-181-3_16
Prajapati AS, Panchal KJ, Pawar VA et al (2018) Review on cellulase and xylanase engineering for biofuel production. Ind Biotechnol 14:38–44
Ghori MI, Ahmed S, Malana MA, Jamil A (2011) Corn stover-enhanced cellulase production by Aspergillus niger NRRL 567. Afr J Biotechnol 10:5878–5886
Peterson R, Nevalainen H (2012) Trichoderma reesei RUT-C30–thirty years of strain improvement. Microbiology 158:58–68
Maki M, Leung KT, Qin W (2009) The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int J Biol Sci 5:500–516a
Iram A, Cekmecelioglu D, Demirci A (2020) Screening of bacterial and fungal strains for cellulase and xylanase production using distillers’ dried grains with solubles (DDGS) as the main feedstock. Biomass Convers Biorefin. https://doi.org/10.1007/s13399-019-00588-x
Ahamed A, Vermette P (2008) Enhanced enzyme production from mixed cultures of Trichoderma reesei RUT-C30 and Aspergillus niger LMA grown as fed batch in a stirred tank bioreactor. Biochem Eng J 42:41–46. https://doi.org/10.1016/j.bej.2008.05.007
Ahamed A, Vermette P (2008) Culture-based strategies to enhance cellulase enzyme production from Trichoderma reesei RUT-C30 in bioreactor culture conditions. Biochem Eng J 40:399–407
Rodriguez-Gomez D, Hobley TJ (2013) Is an organic nitrogen source needed for cellulase production by Trichoderma reesei Rut-C30? World J Microbiol Biotechnol 29:2157–2165
Clarke KG (2013) Bioprocess engineering: an introductory engineering and life science approach. Elsevier, Amsterdam
Izmirlioglu G, Demirci A (2015) Enhanced bio-ethanol production from industrial potato waste by statistical medium optimization. Int J Mol Sci 16:24490–24505
Hao X-C, Yu X-B, Yan Z-L (2006) Optimization of the medium for the production of cellulase by the mutant Trichoderma reesei WX-112 using response surface methodology. Food Technol Biotechnol 44:89–94
Ellilä S, Fonseca L, Uchima C et al (2017) Development of a low-cost cellulase production process using Trichoderma reesei for Brazilian biorefineries. Biotechnol Biofuels 10:30
Iram A, Cekmecelioglu D, Demirci A (2019) Optimization of dilute sulfuric acid, aqueous ammonia, and steam explosion as the pretreatments steps for distillers’ dried grains with solubles as a potential fermentation feedstock. Bioresour Technol 282:475–481
Ahamed A, Vermette P (2009) Effect of culture medium composition on Trichoderma reesei’s morphology and cellulase production. Bioresour Technol 100:5979–5987
Germec M, Turhan I (2019) Evaluation of carbon sources for the production of inulinase by Aspergillus niger A42 and its characterization. Bioprocess Biosyst Eng 42:1993–2005
Ghose TK, Bisaria VS (1987) Measurement of hemicellulase activities: part I xylanases. Pure Appl Chem 59:1739–1751
Spiehs MJ, Whitney MH, Shurson GC (2002) Nutrient database for distiller’s dried grains with solubles produced from new ethanol plants in Minnesota and South Dakota. J Anim Sci 80:2639–2645
Nuez Ortín WG, Yu P (2009) Nutrient variation and availability of wheat DDGS, corn DDGS and blend DDGS from bioethanol plants. J Sci Food Agric 89:1754–1761
Cheilas T, Stoupis T, Christakopoulos P et al (2000) Hemicellulolytic activity of Fusarium oxysporum grown on sugar beet pulp. Production of extracellular arabinanase. Process Biochem 35:557–561
Godden B, Legon T, HelvensteinPenninckx PM (1989) Regulation of the production of hemicellulolytic and cellulolytic enzymes by a Streptomyces sp. growing on lignocellulose. Microbiology 135:285–292
Ashokkumar B, Kayalvizhi N, Gunasekaran P (2001) Optimization of media for β-fructofuranosidase production by Aspergillus niger in submerged and solid state fermentation. Process Biochem 37:331–338
Barton LL, Georgi CE, Lineback DR (1972) Effect of maltose on glucoamylase formation by Aspergillus niger. J Bacteriol 111:771–777
Izmirlioglu G, Demirci A (2016) Strain selection and medium optimization for glucoamylase production from industrial potato waste by Aspergillus niger. J Sci Food Agric 96:2788–2795
Acharya PB, Acharya DK, Modi HA (2008) Optimization for cellulase production by Aspergillus niger using saw dust as substrate. Afr J Biotechnol 7:4147–4152
Barrington S, Kim J-W (2008) Response surface optimization of medium components for citric acid production by Aspergillus niger NRRL 567 grown in peat moss. Bioresour Technol 99:368–377
Teugjas H, Väljamäe P (2013) Selecting β-glucosidases to support cellulases in cellulose saccharification. Biotechnol Biofuels 6:105
Hansson L, Dostálek M (1988) Effect of culture conditions on mycelial growth and production of γ-linolenic acid by the fungus Mortierella ramanniana. Appl Microbiol Biotechnol 28:240–246
Teugjas H, Väljamäe P (2013) Product inhibition of cellulases studied with 14 C-labeled cellulose substrates. Biotechnol Biofuels 6:104
Dhillon GS, Kaur S, Brar SK, Verma M (2012) Potential of apple pomace as a solid substrate for fungal cellulase and hemicellulase bioproduction through solid-state fermentation. Ind Crops Prod 38:6–13
Funding
This study was funded in-part by FULBRIGHT Student Program and USDA National Institute of Food and Agriculture Federal Appropriations under Project PEN04671 and accession number 1017582.
Author information
Authors and Affiliations
Contributions
AD and DC conceived the idea. AI conducted the research, analyzed the data and drafted the manuscript. AD and DC critically evaluated and made significant improvements. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Attia Iram declares that she has no conflict of interest. Deniz Cekmecelioglu declares that he has no conflict of interest. Ali Demirci declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Iram, A., Cekmecelioglu, D. & Demirci, A. Salt and nitrogen amendment and optimization for cellulase and xylanase production using dilute acid hydrolysate of distillers’ dried grains with solubles (DDGS) as the feedstock. Bioprocess Biosyst Eng 45, 527–540 (2022). https://doi.org/10.1007/s00449-021-02676-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-021-02676-7