Abstract
Transporter proteins are of great importance for improving the tolerance of fermentation strains to lignocellulose-derived furans and phenolic inhibitors. Different from the documented transporter proteins responsible for the tolerance of furfural and 5-hydroxymethyl-furfural (HMF), transporters responsible for that of varied phenolic aldehyde inhibitors were less investigated and elucidated. Here, an interesting phenomenon was found that no phenolic alcohols were accumulated from phenolic aldehydes degradation in Zymomonas mobilis. A transcriptional profiling of transporter genes was established in Z. mobilis ZM4 under phenolic aldehydes stress using DNA microarray. Six transporter proteins were identified as the potential candidates responsible for the tolerance of phenolic aldehydes including ABC transporter (ZMO0799 and ZMO0800), MFS transporter (ZMO1288 and ZMO1856), and RND transporter (ZMO0282 and ZMO0798). Furthermore, the analysis showed that the key transporters were significantly correlated with oxidoreductases and transcriptional regulators. This work would provide several important transporter genes serving as synthetic biology tools for improving the robustness of biorefinery strains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phenolic compounds derived from partial breakdown of lignin components, a heterogeneous aromatic polymer, inhibit fermenting strains and cellulase activities during fermentation by disrupting cell membrane and enzyme hydrophobic sites [1]. 4-Hydroxybenzaldehyde, syringaldehyde, and vanillin are the typical model phenolic compounds separately with p-hydroxyphenyl group (H), syringyl group (S), and guaiacyl group (G) according to methoxyl and functional groups [2]. The growing evidence had proved that phenolic aldehydes are the most toxic inhibitors in biofuel fermentation [3,4,5]. Tolerance and detoxification of phenolic inhibitors are the intractable barriers because of their poor water solubility and large numbers of derivatives. Expression of transporter genes is one of the practical methods to overcome the damage of toxic phenolic inhibitors [3, 6].
Efflux of toxic substances is an important way to enhance inhibitor tolerance of microbial organisms [7]. Bacterial multidrug resistance (MDR) efflux pump proteins include five active transporters, adenosine triphosphate (ATP)-binding cassette (ABC), major facilitator superfamily (MFS), multidrug and toxin extrusion (MATE) families, resistance-nodulation cell division (RND), and small multidrug resistance (SMR) [8]. Modifying microbial efflux systems is one way to reduce cytotoxicity of lignocellulose-derived inhibitors like furfural [9,10,11] and vanillin [6].
Ethanologenic bacterium Zymomonas mobilis ZM4 behaves high tolerance to the phenolic acids and is able to thrive in the presence of high concentrations of phenolic aldehydes and produce the corresponding phenolic alcohols [3, 12, 13]. Carbohydrate transporters in Z. mobilis have been applied for ethanol production and sugar transportation [14,15,16,17]. In the previous study, it confirmed that the overexpression of ZMO1288 encoding a MFS transporter protein contributed to the increase of cell growth, glucose consumption, ethanol productivity, and phenolic aldehyde conversion in Z. mobilis ZM4 [3].
In this study, it showed no phenolic aldehydes and phenolic alcohols accumulated in the intracellular of Z. mobilis. Just as good as the bioinformatic data available for the genome of Z. mobilis ZM4, the reliable and more cost effective high-throughput sequencing technology DNA microarray was used to achieve the gene transcriptional profiling of the stress response to phenolic aldehydes. The comprehensive gene transcriptional landscapes of transporter proteins responsible for the tolerant mechanism of phenolic aldehydes were established in Z. mobilis ZM4 using DNA microarray. ABC transporter (ZMO0799 and ZMO0800), MFS transporter (ZMO1288 and ZMO1856), and RND transporter (ZMO0282 and ZMO0798) were identified as the potential candidates for improving the tolerance of phenolic aldehydes in Z. mobilis ZM4. Meanwhile, the significant correlation was also constructed between the key transporter proteins with oxidoreductases and transcriptional regulators. This study would provide the important transporter genes serving as the synthetic tools for the robustness strengthening of biorefinery strains.
Materials and methods
Strain and medium
Z. mobilis ZM4 (ATCC 31821) was from ATCC (American Type Culture Collection, Manassas, VA, USA). Yeast extract was purchased from Oxoid, Hampshire, UK. Phenolic aldehydes, including 4-hydroxybenzaldehyde, syringaldehyde, vanillin, and the corresponding phenolic alcohols were from Sangon Biotech Co. Ltd., Shanghai, China. All the other analytical grade chemicals were from Sinopharm Chemical Reagents (Shanghai, China).
Inoculum preparation and cell disrupt
Z. mobilis ZM4 was cultured in Rich medium (RM) containing 20.0 g/L glucose, 10.0 g/L yeast extract, and 2.0 g/L KH2PO4 at 30 °C without shaking. The degradation assays of phenolic aldehydes were carried out with the middle inhibitory concentration of 5.0 mM 4-hydroxybenzaldehyde, syringaldehyde, and vanillin separately added in RM. To determine the transport of phenolic compounds, inocula of Z. mobilis ZM4 were grown in 100 mL RM medium in a 250 mL flask and harvested at 0 and 24 h of fermentation. Cell growth was determined at 600 nm.
The supernatants of Z. mobilis ZM4 culture were harvested after being centrifuged at 12,000 rpm for 1 min. The collected cells were lyophilized and stored at − 80 °C after being disrupted with three cycles of freezing and thawing. Prior to analysis, the freeze‐dried samples were re-suspended and spin‐filtered with 0.22 μm filter.
Data analysis of DNA microarray
DNA microarray data under the stress of phenolic aldehyde inhibitors in Z. mobilis ZM4 was performed in the previous study [3]. In this study, the transporter genes were analyzed according to the threshold of the differently expressed genes (DEGs) with two foldchange and significantly DEGs combining two foldchange with p_ value < 0.05. It analyzed significance correlation of transporter genes following the threshold of | correlation | ≥ 0.95 (Pearson correlation coefficient) and p_ value ≤ 0.05.
HPLC and GC/MS analysis
Phenolic aldehydes, including 4-hydroxybenzaldehyde, syringaldehyde, and vanillin, were determined at 35 °C using reverse-phase HPLC (SPD-20A, Shimadzu, Kyoto, Japan) with the YMC-Pack ODS-A column (Tokyo, Japan). Both 4-hydroxybenzaldehyde and syringaldehyde were measured at 270 nm at 1.0 mL/min using 30% acetonitrile solution as mobile phase, and vanillin was measured at 320 nm using 50% acetonitrile solution at 0.8 mL/min.
The same with the previous study [3], phenolic aldehydes and their intermediates in Z. mobilis ZM4 were determined using GC/MS. Samplings were at 0 and 24 h after inoculation. The intracellular and extracellular phenolic compounds dissolved in ethyl acetate and acetonitrile solution (2:1, v/v) were silylated with NO-bis-trimethylsilyl trifluoro-acetamide after being concentrated by rotary evaporator with vacuum system [18]. The supernatant was analyzed using Agilent 6890 GC/MS fitted from 80 °C for 4 min to 280 °C at 8 °C/min with a HP-5 MS column (30 m × 0.25 mm × 0.25 μm) (Agilent Technologies, Santa Clara, CA, USA). One microliter sample was used to perform splitless GC detection.
Results and discussion
No phenolic compounds accumulated in the intracellular of Z. mobilis ZM4
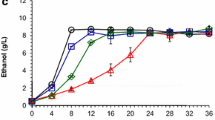
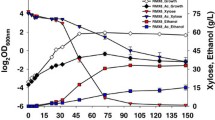
Reduction and transport of biological process were the main molecular mechanism responsible for the degradation of the phenolic aldehydes to the less toxic phenolic alcohols [3]. The phenolic aldehydes tolerance was investigated by assaying the accumulation of the three model phenolic aldehydes in the culture broth of Z. mobilis ZM4 (Fig. 1). Compared with the non-phenolic aldehydes-treated control, the cell growth of Z. mobilis ZM4 was brought down approximately 49.43, 20.10, and 28.03% under the stress of 4-hydroxybenzaldehyde, syringaldehyde, and vanillin, respectively (Fig. 1a). Except for the stress of 4-hydroxybenzaldehyde, Z. mobilis ZM4 completely consumed glucose at 24 h (Fig. 1b). Ethanol productivity was decreased from 0.35 to 0.23, 0.34, and 0.34 g/L/h when separately adding 4-hydroxybenzaldehyde, syringaldehyde, and vanillin (Fig. 1c). Degradation of phenolic aldehydes to the intermediates phenolic alcohols occurred in the extracellular rather than intracellular space of Z. mobilis ZM4 at 24 h (Fig. 1d–e). Furthermore, the decrease of phenolic aldehydes was almost equal to the increase of phenolic alcohols. The results indicated that less accumulation in the intracellular space of Z. mobilis ZM4 was beneficial for the tolerance of Z. mobilis ZM4 to the toxic microenvironment caused by phenolic compounds. The poor water solubility led to the low concentration of phenolic aldehyde inhibitors in extracellular environment and simply not easy to undertake by fermenting strains as the sole carbon source for cell growth on phenolic aldehydes. It is necessary to make the further adjustments (including metabolic engineering and synthetic biology) for phenolic aldehydes degradation in Z. mobilis [3].
The extracellular and intracellular phenolic compounds for ethanol fermentation under the stress of phenolic aldehydes in Z. mobilis ZM4. a Cell growth measured at 600 nm; b glucose consumption; c ethanol production; d extracellular phenolic compounds; e intracellular phenolic compounds. Error bars indicate the standard deviations (SD) of three replicates
Gene transcriptional profiling of transporter proteins under phenolic aldehydes stress
To determine the contribution of transporter proteins to the tolerance of phenolic aldehyde inhibitors in Z. mobilis ZM4, it analyzed the gene transcriptional profiling of transporter proteins in the genome and plasmid of Z. mobilis ZM4, including ABC, MATE, MFS, RND, and SMR. It showed that 15, 8, and 15 genes differentially expressed separately under the stress of 4-hydroxybenzaldehyde, syringaldehyde, and vanillin (Fig. 2). Except for MATE and SMR, other transporter genes covered the DEGs under the stress of the above three model phenolic aldehydes, for example ZMO0799 and ZMO0800 for ABC, ZMO1288 and ZMO1856 for MFS, ZMO0282 and ZMO0798 for RND. GO analysis also showed that the screened DEGs mostly enriched in protein localization, protein transport, and establishment of protein localization of biological process (Table 1). In summary, the transcriptional profiling suggested that the genes encoding transporter proteins were in response to the stress of phenolic aldehydes.
ABC transporters for the tolerance of phenolic aldehydes
ABC transporters contain both uptake and efflux transport systems as the primary active membrane proteins translocating solutes across lipid bilayers [19]. There’re totally 29 genes encoding ABC transporter proteins in the plasmid and genome of Z. mobilis ZM4. Among the genes, ZMO0799 and ZMO0800 were significantly differentially expressed in response to the stress of the three phenolic aldehydes. ZMO0799 was differentially down-regulated by 2.35-, 3.64-, and 2.17-fold, under the stress of 4-hydroxybenzaldehyde, syringaldehyde, and vanillin, respectively, and ZMO0800 was separately differentially up-regulated by 2.23-, 2.19-, and 3.99-fold. Both ZMO0799 and ZMO0800 were the members of the gene cluster ZMO0797-ZMO0798-ZMO0799-ZMO0800-ZMO0801 in respect of phenolic aldehydes degradation according to the previous study [3]. ABC transporters prevented Saccharomyces cerevisiae from HMF damage by exporting excessive HMF [20]. The promotion by ABC transporter was also found for the production of biofuels or chemicals from cellulose in Gram positive strain Ruminiclostridium cellulolyticum [21]. Importantly, ZMO0799 and ZMO0800, especially the gene cluster of ZMO0799-ZMO0800, would be an alternative for genetic engineering of lignocellulosic inhibitor tolerance for fermentation strains in biorefinery fields.
MFS transporters for the tolerance of phenolic aldehydes
MFS, the largest class of secondary active transporters, was found the interaction between protonation and the negative-inside membrane potential [22]. Among thirteen MFS genes in the genome of Z. mobilis ZM4, ZMO1288 was significantly down-regulated by 4.07-, 4.18-, and 3.44-fold under the stress of 4-hydroxybenzaldehyde, syringaldehyde, and vanillin, respectively, and ZMO1856 was separately down-regulated by 4.40, 2.44, and 3.30-fold. It should be noted that the overexpression of ZMO1288 facilitated cell growth, glucose consumption, ethanol productivity, and the conversion of phenolic aldehydes in the previous study [3]. It predicted that the enhanced expression of ZMO1288 would accelerate the transport of phenolic compounds out of intracellular space and no harm was for the intracellular activity of the Z. mobilis ZM4. Membrane transport protein was also involved with the enhancement of phenolic compounds tolerance in Clostridium beijerinckii NCIMB 8052 [23]. Indeed, the gene expression of MFS transporter proteins improved the metabolite production [24, 25]. Therefore, ZMO1288 and ZMO1856 encoding MFS transporters would be the candidates for the genetic strengthening of phenolic aldehydes tolerance in Z. mobilis ZM4.
RND transporters for the tolerance of phenolic aldehydes
RND transporters, the most efficient mechanism of solvent tolerance in Gram-negative bacteria expulsing compounds, are characterized to catalyze substrate efflux via the mechanism of a substrate/H+ antiport [26]. In this study, among 12 genes (ZMO0282, ZMO0285, ZMO0287, ZMO0779, ZMO0780, ZMO0798, ZMO0964, ZMO1429, ZMO1430, ZMO1525, ZMO1529, and ZMO1599) encoding RND efflux pumps in the genome of Z. mobilis ZM4, ZMO0282 was separately significantly differentially up-regulated by 6.05, 2.55, and 7.14 folds under the stress of 4-hydroxybenzaldehyde, syringaldehyde, and vanillin, respectively, and ZMO0798 was separately up-regulated by 2.27, 2.54, and 4.58 folds (Fig. 2). It suggested that RND transporter genes were in response to the stress of phenolic aldehydes. RND was important for the efficient production of ethanol and free fatty acid using genetically engineered cyanobacteria Synechococcus elongatus strain PCC 7942 and Thermoanaerobacter sp. X514 [27, 28]. RND would be a rational choice for the gene engineering of Gram-negative bacteria Z. mobilis ZM4 characteristically surrounded by the additional membrane layer, an outer membrane (OM) serving as the most important function of a selective permeation barrier [29, 30].
MATE transporters for the tolerance of phenolic aldehydes
MATE transporters conserved in biology and functioned in the efflux of cationic and lipophilic substances and xenobiotics with an electrochemical gradient of H+ or Na+ across the cell membrane [31]. In Z. mobilis ZM4, ZMO0214 and ZMO1639 encoding MATE were not differentially expressed under the stress of the three phenolic aldehydes. It showed that more DEGs of the other transporters, especially for ABC, MFS, and RND transporter, than MATE transporter. However, MATE transporter gene worked after co-expressed with biosynthetic gene cluster [32]. This suggested that transporter genes combining with functional genes would be a favorite for inhibitor tolerance enhancement in Z. mobilis ZM4.
SMR transporters for the tolerance of phenolic aldehydes
SMR family of membrane proteins, prominent for its rare dual-topology architecture, exhibited low substrate specificities and was resistant to the large hydrophobic and cationic molecules [33]. Two genes, ZMO0108 and ZMO0697, encoding SMR in the genome of Z. mobilis ZM4, were just differentially down-regulated under the stress of the more toxic 4-hydroxybenzaldehyde and vanillin in Z. mobilis ZM4 (Fig. 2). The expression of the SMR pump potentially enhanced furfural tolerance in ethanologenic Escherichia coli [10]. In few, SMR would be the other synthetic tools for the increase of the lignocellulose-derived inhibitor tolerance in biorefinery field.
Significance correlation analysis of transporter proteins
To determine the expression network in Z. mobilis ZM4, the significance correlation between transporters and their corresponding targets was established (Fig. 3). Among the screened key transporters, ABC transporter (ZMO0799 and ZMO0800), MFS transporter (ZMO1288 and ZMO1856), and RND transporter (ZMO0282 and ZMO0798) just significantly connected with oxidoreductases (ZMO0090, ZMO1116, ZMO1237, and ZMO1885) and transcriptional regulators (ZMO0190, ZMO1574, ZMO1857, and ZMO2030) besides hypothetical proteins (ZMO0020, ZMO0021, ZMO0286, ZMO1215, ZMO1750, ZZM4_0101, and ZZM4_0169). The growing evidence of transporter proteins working as a trigger in transport and regulation to increase transport and stress tolerance had been established, such as transcriptional regulator with ABC transporter [6, 33,34,35], MFS transporter [37, 38], and RND [39, 40].
Significant correlation analysis of transporter proteins under the stress of phenolic aldehydes in Z. mobilis ZM4. Red and blue line separately indicated the positive and negative correlation, and red and green circles were the up- and down-regulated genes, respectively. The transcriptional level of relating genes was listed in the previous study [3]
Specially, ZMO1116 encoding NAD(P)-dependent oxidoreductase was one of the targets of RND transporter (ZMO0798) and ABC transporter (ZMO0799 and ZMO0800), the members of the gene cluster ZMO0797-ZMO0798-ZMO0799-ZMO0800-ZMO0801 [3]. The overexpression of ZMO1116 raised the fermenting parameters including 4-hydroxybenzaldehyde conversion. ZMO0282 for RND transporter was also tightly linked to ZMO1116. The establishment of the significant correlation between transporters and their targets would well elucidate the acceleration mechanism of ethanol fermentability under phenolic aldehydes stress in Z. mobilis ZM4 and provide synthetic tools for the genetic modification of bioethanol production.
Conclusion
Taken together, a globally detailed gene transcriptional profiling for transporter proteins in Gram-negative ethanologenic bacterium Z. mobilis ZM4 was presented under the stress of the model phenolic aldehydes 4-hydroxybenzaldehyde, syringaldehyde, and vanillin using DNA microarray. No phenolic aldehydes and phenolic alcohols were accumulated in the intracellular of Z. mobilis ZM4. Among five bacterial transporter proteins, ABC transporter (ZMO0799 and ZMO0800), MFS transporter (ZMO1288 and ZMO1856), and RND transporter (ZMO0282 and ZMO0798) were identified as the potential candidates for improving phenolic aldehydes tolerance in Z. mobilis ZM4. Meanwhile, it showed that these key transporters were significantly correlated with oxidoreductases and transcriptional regulators. This work would provide the key transporter proteins as synthetic biology tools for the robustness enhancement of biorefinery strains.
Data availability
The data and materials that support the findings of this study are available from the corresponding author upon reasonable request.
References
Jing X, Zhang X, Bao J (2009) Inhibition performance of lignocellulose degradation products on industrial cellulase enzymes during cellulose hydrolysis. Appl Biochem Biotechnol 159:696–707
Klinke HB, Thomsen AB, Ahring BK (2004) Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol 66:10–26
Yi X, Gu H, Gao Q, Liu ZL, Bao J (2015) Transcriptome analysis of Zymomonas mobilis ZM4 reveals mechanisms of tolerance and detoxification of phenolic aldehyde inhibitors from lignocellulose pretreatment. Biotechnol Biofuels 8:153
Gu H, Zhu Y, Peng Y, Liang X, Liu X, Shao L, Xu Y, Xu Z, Liu R, Li J (2019) Physiological mechanism of improved tolerance of Saccharomyces cerevisiae to lignin-derived phenolic acids in lignocellulosic ethanol fermentation by short-term adaptation. Biotechnol Biofuels 12:268
Luo H, Zheng P, Bilal M, Xie F, Zeng Q, Zhu C, Yang R, Wang Z (2020) Efficient bio-butanol production from lignocellulosic waste by elucidating the mechanisms of Clostridium acetobutylicum response to phenolic inhibitors. Sci Total Environ 710:136399
Wang X, Liang Z, Hou J, Shen Y, Bao X (2017) The absence of the transcription factor Yrr1p, identified from comparative genome profiling, increased vanillin tolerance due to enhancements of ABC transporters expressing, rRNA processing and ribosome biogenesis in Saccharomyces cerevisiae. Front Microbiol 8:367
Doshi R, Nguyen T, Chang G (2013) Transporter-mediated biofuel secretion. Proc Natl Acad Sci U S A 110(19):7642–7647
Kumar A, Schweizer HP (2005) Bacterial resistance to antibiotics: active efflux and reduced uptake. Adv Drug Deliv Rev 57(10):1486–1513
Geddes RD, Wang X, Yomano LP, Miller EN, Zheng HB, Shanmugam KT, Ingram LO (2014) Polyamine transporters and polyamines increase furfural tolerance during xylose fermentation with ethanologenic Escherichia coli strain LY180. Appl Environ Microbiol 80(19):5955–5964
Kurgan G, Panyon LA, Rodriguez-Sanchez Y, Pacheco E, Nieves LM, Mann R, Nielsen DR, Wang X (2019) Bioprospecting of native efflux pumps to enhance furfural tolerance in ethanologenic Escherichia coli. Appl Environ Microbiol 85(6):e02985-e3018
Jiménez-Bonilla P, Zhang J, Wang Y, Blersch D, de-Bashan LE, Guo L, Wang Y (2020) Enhancing the tolerance of Clostridium saccharoperbutylacetonicum to lignocellulosic-biomass-derived inhibitors for efficient biobutanol production by overexpressing efflux pumps genes from Pseudomonas putida. Bioresour Technol 312:123532
Franden MA, Pilath HM, Mohagheghi A, Pienkos PT, Zhang M (2013) Inhibition of growth of Zymomonas mobilis by model compounds found in lignocellulosic hydrolysates. Biotechnol Biofuels 6(1):99
Gu H, Zhang J, Bao J (2015) High tolerance and physiological mechanism of Zymomonas mobilis to phenolic inhibitors in ethanol fermentation of corncob residue. Biotechnol Bioeng 112(9):1770–1782
Dunn KL, Rao CV (2014) Expression of a xylose-specific transporter improves ethanol production by metabolically engineered Zymomonas mobilis. Appl Microbiol Biotechnol 98(15):6897–6905
Ren C, Chen T, Zhang J, Liang L, Lin Z (2009) An evolved xylose transporter from Zymomonas mobilis enhances sugar transport in Escherichia coli. Microb Cell Fact 8:66
Weisser P, Krämer R, Sahm H, Sprenger GA (1995) Functional expression of the glucose transporter of Zymomonas mobilis leads to restoration of glucose and fructose uptake in Escherichia coli mutants and provides evidence for its facilitator action. J Bacteriol 177(11):3351–3354
Zhang K, Shao H, Cao Q, He MX, Wu B, Feng H (2015) Transcriptional analysis of adaptation to high glucose concentrations in Zymomonas mobilis. Appl Microbiol Biotechnol 99(4):2009–2022
Klinke HB, Ahring BK, Schmidt AS, Thomsen AB (2002) Characterization of degradation products from alkaline wet oxidation of wheat straw. Bioresour Technol 82:15–26
Locher KP (2016) Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol 23(6):487–493
Ma M, Liu ZL (2010) Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genomics 11:660
Fosses A, Maté M, Franche N, Liu N, Denis Y, Borne R, de Philip P, Fierobe HP, Perret S (2017) A seven-gene cluster in Ruminiclostridium cellulolyticum is essential for signalization, uptake and catabolism of the degradation products of cellulose hydrolysis. Biotechnol Biofuels 10:250
Zhang XC, ZhaoY HJ, Jiang D (2015) Energy coupling mechanisms of MFS transporters. Protein Sci 24(10):1560–1579
Liu J, Lin Q, Chai X, Luo Y, Guo T (2018) Enhanced phenolic compounds tolerance response of Clostridium beijerinckii NCIMB 8052 by inactivation of Cbei_3304. Microb Cell Fact 17(1):35
Chen Z, Huang J, Wu Y, Wu W, Zhang Y, Liu D (2017) Metabolic engineering of Corynebacterium glutamicum for the production of 3-hydroxypropionic acid from glucose and xylose. Metab Eng 39:151–158
Mori K, Niinuma K, Fujita M, Kamimura N, Masai E (2018) DdvK, a novel major facilitator superfamily transporter essential for 5,5’-Dehydrodivanillate uptake by Sphingobium sp. Strain SYK-6. Appl Environ Microbiol 84(20):e01314-e1318
Ramos JL, Duque E, Gallegos MT, Godoy P, Ramos-Gonzalez MI, Rojas A, Teran W, Segura A (2002) Mechanisms of solvent tolerance in gram-negative bacteria. Annu Rev Microbiol 56:743–768
Kato A, Takatani N, Use K, Uesaka K, Ikeda K, Chang Y, Kojima K, Aichi M, Ihara K, Nakahigashi K, Maeda SI, Omata T (2015) Identification of a cyanobacterial RND-type efflux system involved in export of free fatty acids. Plant Cell Physiol 56(12):2467–2477
Lin L, Ji Y, Tu Q, Huang RR, Teng L, Zeng XW, Song HH, Wang K, Zhou Q, Li YF, Cui Q, He ZL, Zhou JZ, Xu J (2013) Microevolution from shock to adaptation revealed strategies improving ethanol tolerance and production in Thermoanaerobacter. Biotechnol Biofuels 6(1):103
Segura A, Molina L, Fillet S, Krell T, Bernal P, Muñoz-Rojas J, Ramos JL (2012) Solvent tolerance in Gram-negative bacteria. Curr Opin Biotechnol 23(3):415–421
Nikaido H (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67(4):593–656
He GX, Kuroda T, Mima T, Morita Y, Mizushima T, Tsuchiya T (2004) An H(+)-coupled multidrug efflux pump, PmpM, a member of the MATE family of transporters, from Pseudomonas aeruginosa. J Bacteriol 186(1):262–265
Darbani B, Motawia MS, Olsen CE, Nour-Eldin HH, Møller BL, Rook F (2016) The biosynthetic gene cluster for the cyanogenic glucoside dhurrin in Sorghum bicolor contains its co-expressed vacuolar MATE transporter. Sci Rep 6:37079
Rapp M, Granseth E, Seppälä S, von Heijne G (2006) Identification and evolution of dual-topology membrane proteins. Nat Struct Mol Biol 13(2):112–116
Poudyal B, Sauer K (2018) The ABC of biofilm drug tolerance: the MerR-like regulator BrlR Is an activator of ABC transport systems, with PA1874-77 contributing to the tolerance of Pseudomonas aeruginosa Biofilms to Tobramycin. J Antimicrob Chemother 62(2):e01981-e2017
Seaton K, Ahn SJ, Sagstetter AM, Burne RA (2011) A transcriptional regulator and ABC transporters link stress tolerance, (p)ppGpp, and genetic competence in Streptococcus mutans. J Bacteriol 193(4):862–874
Görke B (2012) Killing two birds with one stone: an ABC transporter regulates gene expression through sequestration of a transcriptional regulator at the membrane. Mol Microbiol 85(4):597–601
Huang YW, Hu RM, Chu FY, Lin HR, Yang TC (2013) Characterization of a major facilitator superfamily (MFS) tripartite efflux pump EmrCABsm from Stenotrophomonas maltophilia. J Antimicrob Chemother 68(11):2498–2505
Lin HC, Yu PL, Chen LH, Tsai HC, Chung KR (2018) A major facilitator superfamily transporter regulated by the stress-responsive transcription factor Yap1 is required for resistance to fungicides, xenobiotics, and oxidants and full virulence in Alternaria alternata. Front Microbiol 9:2229
Bador J, Neuwirth C, Grangier N, Muniz M, Germé L, Bonnet J, Pillay VG, Llanes C, de Curraize C, Amoureux L (2017) Role of AxyZ transcriptional regulator in overproduction of AxyXY-OprZ multidrug efflux system in Achromobacter Species mutants selected by Tobramycin. Antimicrob Agents Chemother (Bethesda) 61(8):e00290-e317
Cerminati S, Giri GF, Mendoza JI, Soncini FC, Checa SK (2017) The CpxR/CpxA system contributes to Salmonella gold-resistance by controlling the GolS-dependent gesABC transcription. Environ Microbiol 19(10):4035–4044
Funding
This research was supported by Natural Science Foundation of Jiangxi Province (20192BAB204002), Open Funding Project of the State Key Laboratory of Bioreactor Engineering, and Doctor Science Research Foundation of Jiujiang University (8879524).
Author information
Authors and Affiliations
Contributions
XY and WW designed the experiment and drafted the manuscript. XY, JM, and LL carried out the experiment. WW and XY were in charge of the overall project. All authors read and approved to publish the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
All authors are aware of the content and agree with the submission.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yi, X., Lin, L., Mei, J. et al. Transporter proteins in Zymomonas mobilis contribute to the tolerance of lignocellulose-derived phenolic aldehyde inhibitors. Bioprocess Biosyst Eng 44, 1875–1882 (2021). https://doi.org/10.1007/s00449-021-02567-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-021-02567-x