Abstract
Familial mitral valve prolapse in human beings has been associated with several genetic variants; however, in most cases, a known variant has not been identified. Dogs also have a naturally occurring form of familial mitral valve disease (MMVD) with similarities to the human disease. A shared genetic background and clinical phenotype of this disease in some dog breeds has indicated that the disease may share a common genetic cause. We evaluated DNA from 50 affected dogs from five different dog breeds in a whole genome sequencing approach to identify shared variants across and within breeds that could be associated with MMVD. No single causative genetic mutation was found from the 50 dogs with MMVD. Ten variants were identified in 37/50 dogs around and within the MED13L gene. These variants were no longer associated with MMVD when evaluated with a larger cohort including both affected and unaffected dogs. No high/moderate impact variants were identified in 10/10 miniature poodles, one was identified in 10/10 Yorkshire Terriers and 10/10 dachshunds, respectively, 14 were identified in 10/10 Miniature schnauzers, and 19 in 10/10 CKCS. Only one of these could be associated with the cardiac valve (Chr12:36801705, COL12A1; CKCS) but when evaluated in an additional 100 affected CKCS the variant was only identified in 84/100 affected dogs, perhaps indicating genetic heterogeneity in this disease. Our findings indicate that development of MMVD in the dog may be related to a combination of genetic and environmental factors that impact specific molecular pathways rather than a single shared genetic variant across or within breeds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mitral valve prolapse (MVP) is one of the most common cardiac disorders in human beings affecting 2–3% of the general population (Delling et al. 2016). Familial mitral valve prolapse can be isolated or associated with other syndromic conditions, particularly connective tissue disorders (Boudoulas et al. 2020; Parwani et al. 2017). Several genetic variants have been associated with MVP development; however, the majority of patients with MVP do not have one of these known variants and the genetic basis for this disorder is still incompletely understood (Boudoulas et al. 2020; LaHaye et al. 2014).
Dogs have a naturally occurring form of non-syndromic mitral valve disease, myxomatous mitral valve disease (MMVD), with similarities to the human form of MVP particularly Barlow’s syndrome (Pedersen and Haggstrom 2000; Parwani et al. 2017; Markby et al. 2020a; Oyama et al. 2020). It is particularly common in some breeds of dogs that are physically small (small dog breed) including the Yorkshire Terrier, Miniature Poodle, Miniature Schnauzer, Dachshund, Cavalier King Charles Spaniel, Chihuahua and mixed breed dogs among others. (Borgarelli and Buchanan 2012; Han et al. 2013). There is evidence of a familial etiology in some of these breeds and it has been suggested that mitral valve disease may be genetically fixed within some breeds that have a very high incidence (Friedenberg et al. 2016; Lewis et al. 2011; Olsen et al. 1999; Swenson et al. 1996). The shared genetic background and clinical phenotype of this disease in some small dog breeds as well as the high prevalence of disease in those breeds has indicated that the disease may share a common genetic cause (Parker and Kilroy-Glynn 2012). It is possible that MMVD may have been introduced at a very early stage of dog breed development and reached relatively high rates due to reductions in the gene pools of these breeds after their development (Parker and Kilroy-Glynn 2012). However, genome wide association studies have identified associations to different chromosomal regions depending on the dog breed which suggesting more breed specific molecular etiologies (Lee et al 2010; Madsen et al. 2011; Stern et al. 2015). These findings as well as breed specific variations at the transcriptome level could suggest that a more breed specific etiology may be involved (Markby et al. 2020b).
The objective of this study was to utilize DNA from 50 affected dogs from five different dog breeds in a whole genome sequencing approach to identify shared genetic variants across and within breeds that could be associated with the development of MMVD. The increased understanding of the molecular basis for this common canine disease may bring additional insight into our understanding of human MVP as well.
Materials and methods
Phenotyping
This study was conducted in accordance with the guidelines of the North Carolina State University Institutional Animal Care and Use Committee (IACUC, 17-168-0). Ten adult dogs of each of the following five breeds were selected for evaluation including Yorkshire Terrier, Miniature Poodle, Miniature Schnauzer, Dachshund, and Cavalier King Charles Spaniels. These breeds were selected due to the reported increased occurrence in these breeds (Borgarelli and Buchanan 2012; Han et al. 2013). Dogs were evaluated by a board certified veterinary cardiologist or a cardiology resident under the supervision of a boarded cardiologist at North Carolina State University College of Veterinary Medicine. Dogs diagnosed with a systolic left apical heart murmur and had echocardiographic findings consistent with mitral valve disease (valve thickening, prolapse, and/or mitral regurgitation) were selected for DNA collection (Borgarelli and Haggstrom 2010).
DNA collection, isolation, and sequencing
One to two milliliters of blood were collected in an EDTA blood tube from 50 dogs (ten dogs in each of the five breeds) diagnosed with mitral valve disease. DNA was extracted using a Qiagen midiprep kit following the manufacturer's instructions (Qiagen, Hilden, Germany). For each sample, approximately 3 μg of DNA was submitted for library preparation and whole-genome sequencing (https://www.genewiz.com/en). All sequencing experiments were run in one lane of an Illumina HiSeq 4000 high-throughput sequencing system (2 × 150 paired-end reads).
Variant calling
Variant calling from whole-genome sequencing data was performed using a standardized bioinformatics pipeline for all samples as described previously (Friedenberg and Meurs 2016). Briefly, sequence reads were trimmed using Trimmomatic 0.32 to a minimum phred-scaled base quality score of 30 at the start and end of each read with a minimum read length of 70 bp (Bolger, Lohse, and Usadel 2014). Sequences were then aligned to the canFam3 reference sequence using BWA 0.7.13 (Li and Durbin 2009; Lindblad-Toh et al. 2005). Aligned reads were prepared for analysis using Picard Tools 2.5 (http://broadinstitute.github.io/picard/) and GATK 3.7 following best practices for base quality score recalibration and indel realignment as specified by the Broad Institute, Cambridge, MA (DePristo et al. 2011; McKenna et al. 2010; Van der Auwera et al. 2014). Variant calls were made using GATK's HaplotyeCaller walker, and variant quality score recalibration (VQSR) was performed using sites from dbSNP 146 and the Illumina 174 K CanineHD BeadChip as training resources. A VQSR tranche sensitivity cutoff was applied to SNPs at 99.9 and 99% to indels for use in downstream analyses; genotype calls with a phred-scaled quality score < 20 were flagged but not removed from the variant callset.
Variant filtering
Variants present in affected mitral valve disease dogs were selected and filtered against a database of variants derived from whole-genome sequencing of 152 presumed unaffected dogs of 24 breeds (Supplemental Table 1) thought to have a low prevalence for canine mitral valve disease (https://cidd.discoveryspace.ca/). Filtering allowed for variants to be present at an allele frequency of up to 5% within the population of presumed unaffected dogs to prevent exclusion of variants that could infrequently be identified in other breeds. Variants were annotated using Variant Effect Predictor 98 (Mclaren et al. 2016).
Variant evaluation
Data was first filtered to select for variants identified in all 50 affected dogs. Subsequently, the data was filtered for variants found in ≥ 45 of all affected dogs, ≥ 40 of all affected dogs, ≥ 35 of all affected dogs, ≥ 30 of all affected dogs, ≥ 25 of all affected dogs, ≥ 20 of all affected dogs, ≥ 15 of all affected dogs, ≥ 10 of all affected dogs, ≥ 5 affected dogs, and in at least 1 affected dog. Next, with the assumption that a variant might be most common within a specific breed, each breed's whole genome sequencing data was then filtered for variants that were in 10 of 10 dogs within each breed.
Variants were then evaluated for likely pathogenic implications using the Standards and Guidelines for the Interpretation of Sequence Variants (Richards et al. 2015). Variants were separated into four categories: high, moderate, low and modifier impact per Variant Effect Prediction classification. Disease associated low and modifier variants were evaluated using Human splice site finder (HSF) (http://www.umd.be/HSF/), NNsplice (https://www.fruitfly.org/seq_tools/splice.html), NetGene2 (http://www.cbs.dtu.dk/services/NetGene2/), and MaxEntScan (MES) (http://hollywood.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html) to predict the possible genetic importance. Finally genes were evaluated with Ingenuity Pathway Analysis (IPA) to determine gene function and possible association with the development of mitral valve disease (IPA, Qiagen, https://www.qiagen.com/us/) including association with valve development, maintenance and integrity. High and moderate impact variants identified in a gene designated as a novel canine gene were explored more thoroughly by evaluating the gene with Blast (Blast, https://blast.ncbi.nlm.nih.gov/Blast.cgi) for similarity to a known gene in another species that could have valvular relevance.
Two variants (chr26:12745501; chr26:12974780, MED13L gene) (Supplemental Table 2)) with the strongest whole genome association (p < 0.0001) to the development of disease were evaluated with Sanger sequencing in 100 additional affected and 100 apparently unaffected (as determined by lack of a heart murmur by cardiac auscultation at the age of 10 years or older) small breed dogs (Table 2) to validate variant association to the disease. Allele association to MMVD was evaluated with a Fisher’s, p < 0.05. One variant (Chr12:36801705, COL12A1 gene) identified in ten of the ten CKCS was evaluated with Sanger sequencing in 100 additional affected CKCS to validate variant association to the disease in this breed (Supplemental Tables 2 and 3).
Results
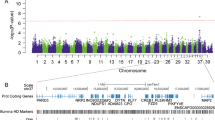
No high, moderate, low or modifier impact variants were found in all 50 dogs. (Fig. 1) One high impact variant (BTNL10, Chr14:507272) was found in ten of ten Yorkshire Terriers, one high impact variant was found in ten of ten Miniature schnauzers (GLYAT, Chr18: 37676090), and one was found in ten of ten CKCS (MFSD13A, Chr6:23283876). No high impact variants were found in all ten Dachshunds or Miniature Poodles. No moderate impact variants were found in all ten Yorkshire Terriers or Miniature Poodles. Four moderate impact variants were found in all ten Schnauzers, one was found in Dachshunds and nine in CKCS (Supplemental Table 4). All of these were only found in these breeds and all but one of these (Chr12:36801705, COL12A1) were in genes that did not have any evident pathophysiological link to valve disease. The one variant in the CKCS in a gene with an association with the heart valve, COL12A1, was subsequently only found in 84 of 100 affected CKCS.
Eight intronic variants in a single gene, MED13L, were found in at least 37 of the affected dogs and two intergenic variants adjacent to MED13L (Chr26:12745501, Chr26:13761780) were found in at least 43 dogs, and were significantly associated with mitral valve disease (p < 0.0001) (Table 1). In silico splice site prediction analysis performed on the MED13L intronic failed to identify alternative splicing sites (Supplemental Table 5).
When the two most common variants (chr26:12745501; chr26:12974780) were evaluated in a population of unaffected small breed dogs they were no longer found to be significantly associated with MMVD (p < 0.6 and p > 0.999). (Table 2).
Discussion
This study demonstrated that there were no shared single high, moderate, low impact, or modifier variants in all 50 MMVD dogs. This suggests that although the disease appears to have many similarities across dogs including clinical presentation, prevalence in small breed dogs and a heritable nature, it does not appear to share a common associated genetic variant. This is consistent with the lack of a single genetic mutation identified in humans with MVP (Boudoulas et al. 2020; LaHaye et al. 2014).
We also did not identify any breed specific variants that were associated with MMVD in the breeds evaluated. Only three of the five breeds had at least one high or moderate impact variant found in ten of ten dogs evaluated but there was no clear association of the variants to valvular development, maintenance, integrity or other aspects of valvular disease. None of these variants were found in the regions previously associated with the development of MMVD by genome wide association in the dog including chromosomes 13q2.2.3 and 14q1.3 (CKCS) (Lee et al 2010; Madsen et al. 2011; Stern et al. 2015). The other two breeds, Dachshunds and miniature poodles, did not have a shared high or moderate impact variant in all ten dogs. These findings could suggest that even within a purebred, closed genetic population that more than one disease associated variant may have been inadvertently selected through breeding practices. This has been previously noted in a few other examples of familial disease in pure bred animals (Meurs et al. 2019; Zeng et al. 2014).
The identification of disease-causing variants is often focused on high and moderate impact variants because of a greater knowledge of how this may impact the gene product; however, there is increasing evidence of the role of intronic variants on the development of disease (Vaz-Drago et al. 2017). Although we did not identify any single variant shared in all 50 dogs, we did find a small number of low impact (intergenic and intronic) variants in a large number of dogs that were located around and within the MED13L (Mediator complex subunit 13-like) gene. MED13L is a subunit of the large Mediator complex that functions as a transcriptional coactivator for most RNA polymerase II transcribed genes and is thought to play an essential role in development (Van Haelst et al. 2015). Mediator complex subunits 10 and 12 have been associated with regulation of cardiac jelly development and atrioventricular valve development in zebrafish (Just et al. 2016; Segert et al. 2018). Mutations in MED13L are associated with MED13L syndrome, which can sometimes cause congenital heart defects, including transposition of the great arteries, ventricular septal defect, and tetralogy of fallot (Asadollahi et al. 2013). Canine mitral valve disease has been assumed to be a middle to later adult-onset disease, since it is most commonly diagnosed at that stage of life; however, an association with MED13L could suggest that that the disease begins as a developmental abnormality with clinical signs developing much later in life (Gordon et al. 2017). We were unable to identify a specific molecular impact of these variants on the gene product using insilico prediction programs and when the most common variants were evaluated in a large number of affected and presumed unaffected dogs, we were no longer able to identify a statistical association with the MMVD phenotype. This could suggest that these variants could simply be associated with other aspects of small breed dog genetic background. Alternatively, it could suggest that these variants are important in the development of MMVD in the dog but are fixed in the population of small breed dogs and that due to incomplete penetrance and late onset were not creating an apparent MMVD phenotype in the unaffected small breed dog cohort. In addition, the unaffected small breed dogs were phenotyped as unaffected by auscultation alone; echocardiography of the presumed unaffected dogs would have provided more rigourous phenotyping and decreased possible incorrect categorization. Further evaluation of the role of variation in this genetic region of MED13l and the development of canine MMVD is warranted.
The lack of a shared common gene variant even within a specific breed with a known familial form of MMVD could suggest that the development of MMVD may be the result of disruptions in shared pathways, rather than a shared genetic variation. It has been suggested that the pathogenesis of MVP in people may be related to multiple factors and that the disease may present a final common pathway for a variety of genetic and molecular factors (Boudoulas et al. 2020). Development of MMVD in the dog may occur in a similar fashion in which even specific breeds develop the disease due to a combination of genetic variants that result in abnormal valvular integrity. Although we did not find a specific shared variant in CKCS with MMVD, the transcriptomic profile of CKCS MMVD mitral valves has been shown to be different than non-CKCS MMVD valves demonstrating some breed specific differences (Markby et al. 2020a). Transcriptomic and pathway analysis work on MMVD valves from 11 different breeds of dogs with severe MMVD identified TGFβ signaling as a dominant pathway which may suggest the role of shared pathways rather than a shared gene (Markby et al. 2020b). This finding could also be consistent with genetic heterogeneity, the development of a clinical phenotype associated with more than one genetic mechanism. Therefore, our data analysis may have been complicated by evaluation for a single shared mutation across a dog breed rather than the evaluation of multiple genetic causes resulting in a similar phenotype within the breed.
As with any study, limitations exist. The number of MMVD dogs evaluated by WGS was relatively small with only ten from each breed. It is possible that contributing genetic variants could have been identified if we evaluated a larger number of animals in each breed. We filtered the MMVD variants in the WGS against a cohort of breeds of dogs that are thought to be of low risk for MMVD. However, large breed dogs can occasionally develop MMVD and it is possible that by doing so we could have missed important variants that were in both populations. To prevent this, we filtered our data so that we allowed variant alleles to exist in up to 5% of the large breed population before we excluded them. It would have been even better to filter the affected population against unaffected small breed dogs; however, given the high prevalence of disease in the small breed dog population, late onset of the disease and possible incomplete penetrance in some dogs we did not feel confident that we could be sure that an unaffected small breed population was truly unaffected. Finally, it is important to note that the annotation of canine reference genes in CanFam3 is still incomplete. Although we attempted to evaluate genes characterized as novel in the genome for their similarity to known reference genes in other species it is likely that future alignments to new versions of the canine genome (CanFam4, CanFam5) will identify some genes that could have a role in the development of MMVD that were not identified here.
In conclusion, no single causative genetic mutation was found in all 50 dogs with MMVD. We did identify a small number of intergenic and intronic variants in a large number of dogs that were located around and within the MED13L gene. However, we were unable to identify a specific molecular impact of these variants on the gene product and the variants were no longer associated with MMVD when evaluated with other small breed dogs that were thought to be unaffected. Given the complexity of the exact phenotyping of this disease which can be impacted by late and variable onset of disease and possibly incomplete penetrance, these variants should not be completely disregarded and further study is warranted. Development of MMVD in the dog may be related to a combination of genetic and environmental factors that impact specific molecular pathways.
Availability of data and material
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Asadollahi R, Oneda B, Sheth F, Azzarello-Burri S, Baldinger R, Joset P, Latal B, Knirsch W, Desai S, Baumer A, Houge G, Andrieux J, Rauch A (2013) Dosage changes of MED13L further delineate its role in congenital heart defects and intellectual disability. Eur J Hum Genet 21:1100–1104
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Borgarelli M, Buchanan JW (2012) Historical review, epidemiology and natural history of degenerative mitral valve disease. J Vet Cardiol 14:93–101
Borgarelli M, Haggstrom J (2010) Canine degenerative myxomatous mitral valve disease: natural history, clinical presentation and therapy. Vet Clin North Am Small Anim Pract 40:651–663
Boudoulas KD, Pitsis AA, Mazzaferri EL, Gumina RJ, Triposkiadis F, Boudoulas H (2020) Floppy mitral valve/mitral valve prolapse: a complex entity with multiple genotypes and phenotypes. Prog Cardiovasc Dis 63:308–326
Delling FN, Rong J, Larson MG, Lehman B, Fuller D, Osypiuk E, Stantchev P, Hackman B, Manning WJ, Benjamin EJ, Levine RA, Vasan RS (2016) Evolution of mitral valve prolapse: insights from the Framingham Heart Study. Circulation 133:1688–1695
DePristo M, Banks E, Poplin RE, Garimella KV, Maguire JR, Hartl C, Philippakis A, del Angel G et al (2011) A framework for variation discovery and genotyping using next- generation DNA sequencing data. Nat Genet 43:491–498
Friedenberg SG, Meurs KM (2016) Genotype imputation in the domestic dog. Mamm Genome 27:485–494
Friedenberg SG, Meurs KM, Mackay TF (2016) Evaluation of artificial selection in Standard Poodles using whole-genome sequencing. Mamm Genome 27:599–609
Gordon SG, Saunders AB, Wesselowski SR (2017) Asymptomatic canine degenerative valve disease. Current and future therapies. Vet Clin North Am Small Anim Pract 47:955–975
Han RI, Clark CH, Black A, French A, Culshaw GJ, Kempson SA, Corcoron BM (2013) Morphological changes to endothelial and interstitial cell and to the extra- cellular matrix in canine myxomatous mitral valve disease. Vet J 197:388–394
Just S, Hirth S, Berger IM, Fishman MC, Rottbauer W (2016) The mediator complex subunit Med10 regulates heart valve formation in zebrafish by controlling Tbx2b-mediated Has2 expression and cardiac jelly formation. Biochem Biophys Res Commun 477:581–588
Lahaye S, Lincoln J, Garg V (2014) Genetics of valvular heart disease. Curr Cardiol Rep 16:487
Lee CM, Song DW, Ro WB, Kang MH, Park HM (2010) Genome-wide association study of degenerative mitral valve disease in Maltese dogs. J Vet Sci 36:63–71
Lewis T, Swift S, Woolliams JA, Blott S (2011) Heritability of premature mitral valve disease in Cavalier King Charles spaniels. Vet J 188:73–76
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, Zody MC (2005) Genomic sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438:803–819
Madsen MB, Olsen LH, Häggström J, Höglund K, Ljungvall I, Falk T, Wess G, Stephenson H, Dukes-McEwan J et al (2011) Identification of 2 Loci associated with development of myxomatous mitral valve disease in Cavalier King Charles spaniels. J Hered 102:S62–S67
Markby GR, Macrae VE, Summers KM, Corcoran BM (2020a) Disease severity-associated gene expression in canine myxomatous mitral valve disease is dominated by TGF β signaling. Front Genet 11:1–13
Markby GR, Macrae VE, Corcoran BM, Summers KM (2020b) Comparative transcriptomic profiling of myxomatous mitral valve disease in the cavalier King Charles spaniel. BMC Vet Res 16:350
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit. Genome Res 20:1297–1303
Mclaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, Flicek P, Cunningham F (2016) The Ensembl variant effect predictor. Genome Biol 17:1–14
Meurs KM, Friedenberg SG, Kolb J, Saripalli C, Tonino P, Woodruff K et al (2019) A missense variant in the titin gene in Doberman pinscher dogs with familial dilated cardiomyopathy and sudden cardiac death. Hum Genet 138:515–524
Olsen LH, Fredholm M, Pedersen HD (1999) Epidemiology and inheritance of mitral valve prolapse in dachshunds. J Vet Intern Med 13:448–456
Oyama MA, Elliott C, Loughran KA, Kossar AP, Castillero E, Levy RJ, Ferrari G (2020) Comparative pathology of human and canine myxomatous mitral valve degeneration: 5HT and TGF-β mechanisms. Cardiovasc Pathol 46:107196
Parker HG, Kilroy-Glynn P (2012) Myxomatous mitral valve disease in dogs: does size matter? J Vet Cardiol 14:19–29
Parwani P, Avierinos JF, Levine RA, Delling FN (2017) Mitral valve prolapse: multimodality imaging and genetic insights. Prog Cardiovasc Dis 60:361–369
Pedersen HD, Haggstrom J (2000) Mitral valve prolapse in the dog: a model of mitral valve prolapse in man. Cardiovasc Res 47:234–243
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424
Segert J, Schneider I, Berger IM, Rottbauer W, Just S (2018) Mediator complex subunit Med12 regulates cardiac jelly development and AV valve formation in zebrafish. Prog Biophys Mol Biol 138:20–31
Stern JA, Hsue W, Song KH, Ontiveros ES, Fuentes VL, Stepien RL (2015) Severity of mitral valve degeneration is associated with chromosome 15 loci in whippet dogs. PLoS ONE 10:1–11
Swenson L, Haggstrom J, Kvart C, Juneja R (1996) Relationship between parental cardiac status in Cavalier King Charles spaniels and prevalence and severity of chronic valvular disease in offspring. J Am Vet Med Assoc 208:2009–2012
Van Der Auwera G, Carneiro MO, Hartl C, Poplin R, Levy-moonshine A, Jordan T, Shakir K et al (2014) From FastQ data to high confidence varant calls: the Genonme Analysis Toolkit best practices pipeline. Curr Protoc Bioinf 43:11.10.1-11.10.33
Van Haelst MM, Monroe GR, Duran K, Van Binsbergen E, Breur JM, Giltay JC, Van Haaften G (2015) Further confirmation of the MED13L haploinsufficiency syndrome. Eur J Hum Genet 23:135–138
Vaz-Drago R, Custódio N, Carmo-Fonseca M (2017) Deep intronic mutations and human disease. Hum Genet 136:1093–1111
Zeng R, Coates JR, Johnson GC, Hansen L et al (2014) Breed distribution of SOD1 alleles previously associated with canine degenerative myelopathy. J Vet Intern Med 28:515–521
Funding
This study was funded by the Mark L. Morris Jr. Investigator Award, D16CA509, Morris Animal Foundation and the American Kennel Club Canine Health Foundation.
Author information
Authors and Affiliations
Contributions
BK, ST and TD phenotyped animals; BW collected, analyzed and interpreted data and assisted with writing the manuscript; KM conceptualized the study, phenotyped animals, collected, analyzed and interpreted data and drafted the manuscript; SF performed and analyzed the bioinformatic work and edited manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
This study was conducted in accordance with the guidelines of the North Carolina State University Institutional Animal Care and Use Committee (17-168-0).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Williams, B., Friedenberg, S.G., Keene, B.W. et al. Use of whole genome analysis to identify shared genomic variants across breeds in canine mitral valve disease. Hum Genet 140, 1563–1568 (2021). https://doi.org/10.1007/s00439-021-02297-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-021-02297-w