Abstract
Cilia and flagella are formed around an evolutionary conserved microtubule-based axoneme and are required for fluid and mucus clearance, tissue homeostasis, cell differentiation and movement. The formation and maintenance of cilia and flagella require bidirectional transit of proteins along the axonemal microtubules, a process called intraflagellar transport (IFT). In humans, IFT defects contribute to a large group of systemic diseases, called ciliopathies, which often display overlapping phenotypes. By performing exome sequencing of a cohort of 167 non-syndromic infertile men displaying multiple morphological abnormalities of the sperm flagellum (MMAF) we identified two unrelated patients carrying a homozygous missense variant adjacent to a splice donor consensus site of IFT74 (c.256G > A;p.Gly86Ser). IFT74 encodes for a core component of the IFT machinery that is essential for the anterograde transport of tubulin. We demonstrate that this missense variant affects IFT74 mRNA splicing and induces the production of at least two distinct mutant proteins with abnormal subcellular localization along the sperm flagellum. Importantly, while IFT74 deficiency was previously implicated in two cases of Bardet–Biedl syndrome, a pleiotropic ciliopathy with variable expressivity, our data indicate that this missense mutation only results in primary male infertility due to MMAF, with no other clinical features. Taken together, our data indicate that the nature of the mutation adds a level of complexity to the clinical manifestations of ciliary dysfunction, thus contributing to the expanding phenotypical spectrum of ciliopathies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The motility of mammalian sperm is provided by the flagellum, an evolutionarily conserved organelle built around a microtubule cytoskeleton, called the axoneme. The axoneme is constituted of nine microtubule doublets surrounding a central pair of microtubules, which is part of the central apparatus. The flagellar axoneme comprises specific components necessary for motility, such as outer and inner dynein arms, nexin–dynein regulatory complex (N-DRC) and Calmodulin and radial spoke-associated complex (CSC) which are tightly connected to the tubulin backbone to coordinate and orchestrate flagellar beating (Ishikawa 2017). This axonemal structure is also found in motile cilia, which for instance are present in mammals at the surface of the respiratory epithelium, fallopian tubes and central nervous ependymal canal, where they facilitate the movements of fluids. In addition, the axoneme is present in primary cilia, which coordinate signaling pathways during development and in tissue homeostasis. Primary cilia are usually immotile solitary organelles extending from the cell surface of most mammalian cells in growth arrest. In these cilia, the structures associated with motility, such as dynein arms and radial spokes are absent, and their tubulin backbone lacks the central pair microtubules. Importantly, the axoneme is present in several ciliated or flagellated unicellular organisms such as Tetrahymena, Chlamydomonas and Trypanosoma, which have proven to constitute useful tools to unravel the molecular mechanisms of assembly and structure of cilia and flagella (Vincensini et al. 2011).
The assembly of cilia and flagella was shown to require a molecular motor-driven process, which ensures the selective transport of proteins along the proximal–distal axis of the growing organelle. This process, called intra flagellar transport (IFT) was mainly studied in protists such as Chlamydomonas reinhardtii cilia and Trypanosoma brucei (Avidor-Reiss and Leroux 2015; Ishikawa and Marshall 2017; Reiter and Leroux 2017). IFT complexes, also called “trains” are multiprotein complexes classified as IFT-A and IFT-B depending on their protein content. The IFT complex A is linked to a kinesin-2 motor to ensure anterograde transport (toward the ciliary and flagellar tips) along axonemal microtubules, whereas the IFT B complex is associated with cytoplasmic dyneins to ensure retrograde transport. IFT trains are specialized for the transport of different sets of protein cargo such as tubulin and are essential to build cilia and flagella. As a result, IFT deficiency is deleterious for cell function and tissue differentiation, and can cause a variety of human diseases and developmental disorders, referred as ciliopathies (Reiter and Leroux 2017).
As an exception, the molecular mechanisms of sperm flagellum assembly in mammals are still poorly defined and the presence of an IFT mechanisms in mature sperm flagella was debated due to the absence of protein turnover in this organelle, in contrast to cilia (Pedersen and Rosenbaum 2008; Rosenbaum and Witman 2002; San Agustin et al. 2015). The recent identification of IFT encoding genes, which mutations in mouse models are associated with sperm flagellum assembly defects, indicated an evolutionary conservation of IFT machinery components and potential similar function in mammalian sperm. In the mouse, Ift88 was the first IFT gene shown to be required for sperm flagella assembly (Pazour et al. 2000), followed by Spef2 (Sironen et al. 2011), Ift20 (Zhang et al. 2016), Ift25 (Liu et al. 2017), Ift27 (Zhang et al. 2017), Ift74 (Shi et al. 2019), Ift81 (Qu et al. 2020), Ift140 (Zhang et al. 2018), Ttc21a (Liu et al. 2019b)and Ttc29 (Lores et al. 2019). Importantly, recent data from genetic analyses of patients showing an isolated phenotype of male sterility due to multiple morphological abnormalities of the sperm flagella (MMAF) also allowed the identification of protein damaging variants affecting IFT genes, such as TTC21A (Liu et al. 2019b), TTC29 (Lores et al. 2019), SPEF2 (Liu et al. 2019a, c) and CFAP69 (Dong et al. 2018; He et al. 2018). In humans, most of those identified IFT-related genes have a predominant expression in the testis and in the case of SPEF2, the mutations were shown to only affect testis-specific isoforms, thus explaining the presence of defects only impacting sperm flagellum structure and leading to an isolated form of male infertility, which is not associated with ciliopathy (Toure et al. 2020). To date, their exact function in sperm flagella and their implication in a canonical IFT process remain unknown.
Here, we report the identification of two individuals presenting a MMAF phenotype and carrying a homozygous missense mutation in IFT74, a gene encoding for an essential IFT component. IFT74 was extensively studied in unicellular organisms such as Chlamydomonas (Bhogaraju et al. 2013; Brown et al. 2015; Kubo et al. 2016) and was shown to associate with IFT81 for tubulin transport within cilia and to be required for ciliogenesis (Kubo et al. 2016). In the mouse, Shi et al. demonstrated that IFT74 deficiency in male germ cells induces male sterility, with immotile and short-tailed sperm cells (Shi et al. 2019). Importantly, in humans, IFT74 deficiency was associated with two cases of Bardet–Biedl syndrome (BBS), a rare multisystemic ciliopathy resulting in retinal dystrophy, obesity, polydactyly, intellectual disability, renal dysfunction, urogenital malformations and male hypogonadism (Kleinendorst et al. 2020; Lindstrand et al. 2016). The work we present here indicates that in humans, mutations in IFT74 can induce isolated male sterility due to impaired flagellogenesis and confirms the importance of IFT machinery for proper assembly and elongation of mammalian sperm flagella.
Results
Identification of an IFT74 bi-allelic missense mutation in two unrelated infertile individuals with MMAF
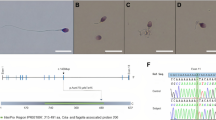
Whole-exome sequencing (WES) data from a cohort of 167 MMAF-affected individuals were analyzed to identify new candidate genes for severe asthenozoospermia due to sperm flagella defects. Previous analyses of this cohort permitted to identify bi-allelic variants in a total of 76 men (45.5%) in 16 confirmed MMAF-associated genes (AK7, ARMC2, CFAP43, CFAP44, CFAP65, CFAP69, CFAP70, CFAP91 (MAATS1), CFAP251 (WDR66), DNAH1, DNAH8, FSIP2, QRICH2, SPEF2, TTC21 and TTC29). In the remaining undiagnosed individuals, we identified a homozygous missense variation c.256G > A; p.Gly86Ser, absent from the genetic variation databases (gnomAD, ClinVar) in IFT74 (Intra Flagellar Transport, Gene ID 80,173). We considered this variant to be a good candidate since two unrelated individuals presented the same homozygous variant and IFT74 was shown to be essential for spermatogenesis and male fertility in mice (Shi et al. 2019). IFT74 is located on 9p21.2 and is subject to alternative splicing resulting in multiple transcripts and protein isoforms. The canonical IFT74 transcript (ENST00000443698; GenBank: NM_025103) contains 19 coding exons (19/20), predicting a 600-amino-acid protein (UniProtKB: Q96LB3) (Fig. 1a). Based on public tissue expression databases (EMBL-EBI Expression Atlas, NCBI, Human protein atlas, and GTEx), IFT74 is highly and preferentially expressed in testes. The protein was also detected by proteomic analyses of human whole sperm (Wang et al. 2013) and human airways ciliary samples (Blackburn et al. 2017; Wang et al. 2013) and comprises an amino-terminal region with basic residues and a unique large coil-coiled domain extending from amino acid 98 to 482 (Fig. 1a). The identified variant, c.256G > A; p.Gly86Ser, impacts the last nucleotide of exon 3 (Fig. 1b). According to this location, next to the splice donor consensus site, the variant is predicted to be deleterious for U2 type splicing; it is also scored as deleterious by both adaptive boosting (ada_score > 0.99999) and random forests (rf_score = 0.998), using dbscSNV tool. At the protein level, the variant is predicted to be probably damaging (0.997) according to PolyPhen2, and disease causing (probability = 1) according to Mutation Taster. We confirmed the presence of the variant at the homozygous state by Sanger sequencing on DNA samples from both MMAF individuals, as illustrated in Fig. 1b.
Identification of IFT74 c.256G > A mutation in two infertile MMAF individuals. a Schematic representation of IFT74 exon–intron structure (top) and predicted protein domains according to SMART webtool and Uniprot (bottom), with position of the c.256G > A mutation identified in the two infertile MMAF individuals. In blue, the Tubulin interacting domain (Tub) and coiled-coil domain spanning from amino acid 98 to 482. The localization of primers used for RT-PCR (F1, R1, R2, blue arrows) and for quantitative RT-PCR (red arrows) is indicated. b Sanger sequencing data of the mutations identified in infertile individuals IFT74_1 and IFT74_2, and pedigree of the two individuals
Clinical and biological features
Individual IFT74_1 (i.e. III.5) originated from Tunisia and his parents were not consanguineous (Fig. 1b). He reported a 3-year primary infertility with his first wife. After they separated, the woman had a child with another partner. After 18 months of infertility with his second wife, the patient approached our reproductive center for medical help. Medical history was free from any medical or surgical previous events. The patient reported no genital, urinary or other particular symptom. A detailed medical interrogation failed to evidence any exposure to potentially gonadotoxic compounds or factors, either professional or in his life mode. The patient displayed no clinical signs of hypogonadism nor hypoandrogenism; secondary sex characteristics were strictly normal. The only clinical feature deserving specific care was excess weight (BMI 27.9, 75 kg for 1.64 m). No diabetes, nor hypertension were reported. The patient had four sisters, all of them with children, and three brothers (Fig. 1c). One of the brothers was also infertile (III.6). The two other brothers were fertile; one of them (III.8) had fathered four children, two healthy daughters and two sons who deceased in the neonatal period; the cause of these deaths was not known. No familial nor genetic pathology were documented in the family.
Semen analyses evidenced a very low progressive sperm motility, contrasting with normal sperm counts and near normal vitality rate (Table 1). The percentage of morphologically normal sperm cells was significantly reduced (14%, lower limit 23%) and detailed analysis of the morphological anomalies indicated sperms with short tails (16%). The pattern of the other abnormalities showed no specificity. In addition, a further analysis performed on a sperm fraction selected through centrifugation on a density gradient showed that instead of being improved as is usual in men with normal semen parameters, motility was markedly decreased after this selection, whereas percentages of vitality and of normal sperms remained close to the initial values.
Individual IFT74_2 originated from Algeria and was recruited in Tunisia at the Polyclinique les Jasmins, Tunis. No history of familial consanguinity has been reported. He has three brothers and two sisters. His three male siblings were married and fertile. His past medical history was marked by a unilateral varicocele treated 6 years before the date of the recruitment. Analyses of its ejaculate evidenced a low sperm count (7 million/ml) associated with a total sperm immotility and 0% of typical forms with 100% of flagellar abnormalities (Table 1). The majority of the flagella presented an irregular caliber (84%) and/or a short length (73%). The vitality was also dramatically reduced (22%). Additional characterization of the sperm defect was not possible for this patient as he did not volunteer to give any additional samples.
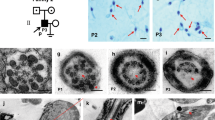
As illustrated for individual IFT74_1 (Fig. 2a), sperm morphological analysis of the patients indicated a typical MMAF phenotype defined by sperm with absent, short and irregular flagella in rates largely the distribution ranges observed in control fertile men (Auger et al. 2016) (Table 1). Ultrastructure analysis of sperm from individual IFT74_1 was performed by transmission electron microscopy (TEM) and showed abnormal mitochondrial sheath and cytoplasmic bags containing unassembled flagellar components, as previously described for the MMAF phenotype (Fig. 2b). Analysis of a total number of 26 transversal sections of sperm cells from individual IFT74_1, evidenced 27% of abnormal axonemal structure, all characterized by the absence of some peripheral doublets; no morphological anomalies were observed on the other transversal sections even at the highest possible magnification and the central pair of the axoneme was always present. Such level of axonemal disorganization appears lower than what was previously reported for MMAF individuals carrying mutations in genes encoding for axonemal or associated structural components such as CFAP43, CFAP44, AK7 and DNAH8 (Coutton et al. 2018; Liu et al. 2020; Lores et al. 2018); the same observation was previously made for patients carrying mutations in TTC29, a gene also encoding for a component of the IFT machinery (Lores et al. 2019).
Characterization of the sperm morphological and ultra-structural phenotype of individual IFT74_1 carrying homozygous IFT74 mutation. a Schorr staining of semen smears from individual IFT74_1, showing sperm with abnormal constituted flagella such as short flagella (arrow) and irregular caliber of the midpiece and principal piece (arrow head). Scale bars represent 5 µm. b TEM analysis of spermatozoa from individual IFT74_1, showing flagella with abnormal midpiece and irregular caliber of the flagella. Ac acrosome, N nucleus, M mitochondria, Ax axoneme, FS fibrous sheath. Scale bars represent 1 µm
Alteration of IFT74 splicing in sperm from patient with homozygous IFT74 mutation
In order to demonstrate the pathogenicity of the identified c.256G > A;p.Gly86Ser variant, we analyzed the transcript and protein levels in semen samples available from individual IFT74_1. We first performed semi-quantitative RT-PCR to amplify IFT74 transcripts in sperm cells from control individual and individual IFT74_1, using primers located in exons 2, 4 and 5 (Fig. 3a). Semi-quantitative analysis of the ubiquitous housekeeping gene HPRT, showed no difference between control individual and individual IFT74_1 (Fig. 3a), indicating that there was no major difference in the quantity and quality of the analyzed mRNA. The signal intensity of IFT74 transcripts appearing slightly decreased in sperm cells from individual IFT74_1, compared with control sperm (Fig. 3a), we performed real time quantitative PCR analysis. This was performed as a single experiment due to limited amount of biological material but the results indicated normal transcript amount in sperm from individual IFT74_1; the ratio of transcript even being slightly increased for the patient (Supplemental Figure S1A). This observation indicates that the mutated transcripts in sperm from individual IFT74_1 were not or only marginally subjected to mRNA decay. However, we note that the PCR amplicons generated in sperm from the control individual formed a sharp band while the amplicons generated with the same primers in sperm from individual IFT74_1 appeared as a smear and were likely heterogeneous. We confirmed the presence of heterogenous transcripts by Sanger sequencing of the amplicons and analysis of the electropherograms; the electropherogram clearly indicated a mixture of at least 2 different transcripts in sample from individual IFT74_1 (Fig. 3b). To elucidate the consequences of the c.256G > A mutation and to define the nature of the different transcripts, we individually cloned and sequenced the heterogeneous PCR products generated from individual IFT74_1. We isolated and sequenced 16 clones and we identified the following different transcripts: (1) type 1: normally spliced transcripts carrying the c.256G > A missense mutation in exon 3 (9/16 clones; 56%), (2) type 2: abnormally spliced transcripts using a cryptic donor site located 30 bp upstream the normal splicing site, which induces an in-frame deletion of 10 amino-acids within exon 3 (5/16 clones, 31%). These two main mutated transcripts isolated in sperm from individual IFT74_1, namely type 1 and type 2, are illustrated in Fig. 3c along with the theoretically encoded proteins. In addition, two minor transcripts were detected as spliced transcripts using cryptic donor sites present in intron 3 (2/16 clones, 13%) (Supplemental Figure S1). Type 3 retains 18 bp of intron 3, and encodes for a mutant protein carrying an in-frame insertion of 6 amino-acids. Type 4 retains 108 bp of intron 3, which induces a frameshift and a truncated protein (p.Gly86Ser*13) (Supplemental Figure S1).
IFT74 splicing and transcript analysis in Sperm from individual carrying IFT74 homozygous mutation. a Semi-quantitative RT-PCR analysis of IFT74 transcripts in sperm cells from control and IFT74_1 individuals, using primers located in exons 2, 4 and 5. The ubiquitous housekeeping gene HPRT is used for quality and quantity control. In sperm from IFT74_1 patient, the absence of a clear and sharp band suggests a heterogenous population of transcripts. b Electrophoregrams of Sanger sequencing of the whole amplicons generated by semiquantitative RT-PCR in sperm from individual IFT74_1. The arrow indicates the presence of at least two distinct superimposed sequences in the RT-PCR products from individual IFT74_1, as compared to control sperm. This indicate a heterogenous population of mRNA in sperm cells from the patient. c Cloning and sequencing of RT-PCR products from individual IFT74_1 identified two main mutated transcripts. Type 1 transcript result from normal splicing of intron 3 and carries a missense mutation (G > A), which predicts a mutated protein p.Gly86Ser. Type 2 transcript carries a deletion of 30 bp due to abnormal splicing of intron 3, which predicts a mutated protein p.Leu77_Gly86del with an in-frame deletion of ten amino acids
Production of IFT74 mutated proteins in sperm from patient with homozygous IFT74 mutation
We next performed western blot experiments using two different antibodies raised against IFT74 epitopes, which are located downstream the mutation site (HPA026684 and HPA020247, Sigma Aldrich®). Using both antibodies, the protein was detected as a single band at the predicted molecular weight of 69 kDa in sperm protein extracts from control individuals. For antibody HPA020247, a shorter exposure of the western blot also clearly indicated a unique band in control sample (data not shown). In contrast, for individual IFT74_1, the protein signal appears as a very tight doublet, which corresponds to the expected sizes of the two principal mutated proteins described above: ENSP00000404122.1:p.Gly86Ser (69.13 kDa; missense mutation) and ENSP00000404122.1:p.Leu77_Gly86del (68.13 kDa; in frame deletion of 10 aa) (Fig. 4a).
Analysis of IFT74 proteins in sperm from individual carrying homozygous IFT74 mutation. a Western blot analysis of spermatozoa from control and IFT74_1 individuals, using two different antibodies raised against IFT74 epitopes located downstream the mutation (antibodies HPA020247 and HPA026684 from Sigma Aldrich®). β-Tubulin hybridization is used for quality and quantity loading control. In sperm from control individual, the protein is detected as a single band at the expected size (69.2 kDa). In sperm from individual IFT74_1, both antibodies detect a doublet, confirming the production of at least 2 principal types of IFT74 mutant proteins. b Immunofluorescence staining on spermatozoa, using IFT74 antibody (HPA026684) (in red) and β-Tubulin antibody (in green). In contrast to control sperm, IFT74 signal is only detected in the proximal part of the flagellum in sperm from individual IFT74_1. Such abnormal pattern is not observed in sperm from MMAF individual carrying mutations in TTC29. Cells were counterstained with DAPI (blue) as nuclei marker. Scale bars represent 5 µm
Alteration of IFT74 protein localization in sperm from patient with homozygous IFT74 mutation
We then performed immunodetection assays using IFT74 antibody HPA026684. In control individuals we observed that IFT74 was homogeneously distributed along the sperm flagellum. However, we noticed that in sperm cells from individual IFT74_1 which achieved to assemble a correct length of flagella (visible by Tubulin staining), IFT74 staining was weaker and its distribution was different (Fig. 4B). When compared to Tubulin staining, the mutated IFT74 proteins appeared concentrated in the proximal part of the flagellum. This feature was observed in 89.4% of normomorphic sperm cells, and was specific for the IFT74 mutant proteins, as sperm from a MMAF individual mutated in TTC29, referred as i.e. TTC29_1, and harboring wild type IFT74 alleles (Lores et al. 2019), did not show such abnormal distribution, although IFT process is likely to be impaired in those cells. Considering that in Chlamydomonas, IFT74 was shown to associate with IFT81 and to form the main module for intraflagellar transport of Tubulin (Bhogaraju et al. 2013; Kubo et al. 2016), we performed IFT81 immunodetection assays and observed that IFT81 protein was also abnormally concentrated in the proximal part of the flagellum, while the staining was homogenously distributed along the sperm flagellum from control individual Supplemental Figure S2. Again, such abnormal localization of IFT81 protein was not observed in sperm from a MMAF individual mutated in TTC29. Interestingly, when analyzing SPAG6, an axonemal component localizing to the central pair, we observed that both sperm from individual IFT74_1 and TTC29_1, displayed a normal distribution pattern along the flagellum Supplemental Figure S3.
Discussion
We report the identification of the IFT74 c.256G > A;p.Gly86Ser bi-allelic missense mutation, in two unrelated infertile men with sperm flagellum assembly defects. We showed that the mutation induces the production of two main mutant IFT74 proteins: one carrying a missense mutation (p.Gly86Ser), the other carrying a 10 amino-acids in-frame deletion (p.Leu77_Gly86del). We suspect that these mutated proteins retain some functional activity, and supporting this hypothesis, we observed a moderate axonemal disorganization in sperm from an IFT74 mutated individual. In particular the central pair was not affected, in contrast to previously investigated MMAF individuals harboring truncating mutations in CFPA43, CFAP44, AK7 and DNAH8, for instance (Coutton et al. 2018; Liu et al. 2020; Lores et al. 2018).
IFT74 is required for ciliogenesis and was shown to associate with IFT81 to form the main module for intraflagellar transport of Tubulin. In vitro, the N-terminus of IFT81 is able to directly bind to tubulin, and IFT74 N-terminus, which is positively charged, enhances the affinity of this interaction with the highly acidic C-terminal tails of β-tubulin (Bhogaraju et al. 2013). Brown et al. (2015) demonstrated that in Chlamydomonas, the N-terminus of IFT74 is required for the recruitment of retrograde IFT-A subcomplexes to IFT-B, for the assembly of new trains at the base of the cilium and their injection into the cilium. Our observation that IFT81 protein is mislocalized and retained in the proximal region of the flagellum, in sperm from individual carrying homozygous IFT74 mutation, suggests that such molecular complex is conserved and functional in mammalian sperm too.
The MMAF phenotype and the male sterility observed in IFT74 homozygous mutated individuals is in line with the phenotype observed in Chlamydomonas reinhardtii, and in mouse invalidated for IFT74. In Chlamydomonas, IFT74 null mutation abrogates flagella assembly (Brown et al. 2015; Kubo et al. 2016), and in the mouse, its specific deletion in male germ cells was associated with infertility due to reduced production of sperm with short or absent tails (Shi et al. 2019). However, in human, individuals carrying biallelic IFT74 loss of function mutations were reported to display Bardet–Biedl syndrome (BBS), a rare developmental genetic disorder linked to severe impairment of primary cilia function (Kleinendorst et al. 2020; Lindstrand et al. 2016). Mutations in IFT81 were also associated with short-Rib polydactyly syndrome, a group of bone malformations characterized by a narrow thorax and polydactyly (Duran et al. 2016). BBS is a genetically heterogeneous disease with considerable phenotypic variability. To date more than 20 genes (BBS1-BBS20) were mapped to the disease; those BBS genes encode for components of the BBSome complex located at ciliary basal body (BBS1, 2, 4, 5, 7, 8, 9), for chaperone and microtubule associated proteins (Khan et al. 2016; Priya et al. 2016). The disease is typified by mixed rod-cone dystrophy, polydactyly associated sometimes with syndactyly and/or brachydactyly. Mental retardation is also present in about 50% of cases, obesity in about 70%, associated with insulin resistance, type 2 diabetes, dyslipidemia, and hypertension (Tsang et al. 2018). So far BBS patients were reported with hypogonadotropic hypogonadism and urogenital malformations with renal anomalies present in about 40% of the patients (Tsang et al. 2018). However, the two BBS patients identified with IFT74 null mutations were suffering from obesity, polydactyly, retinal dystrophy, but no renal abnormality was diagnosed and only one of the patients had intellectual disability (Kleinendorst et al. 2020; Lindstrand et al. 2016). Considering our present data, those patients could also display spermiogenesis defects and male sterility; however, no fertility phenotype was stated in the above studies. While writing this manuscript, a study from Koscinski et al. reported the first characterization of the reproductive functions for a series of 11 adult male BBS patients aged of 18 to 39. Those patients carried mutations in BBS1, 3, 5, 9, 10 and 12 and some of them displayed hypogonadism and urogenital anomalies (unilateral or bilateral cryptorchidism, micropenis, short scrotum, epididymal cysts …). However, these anomalies were not systematically present, which indicated that the reproductive features of BBS patient are also extremely variable (Koscinski et al. 2020). None of those 11 patients were reported by the authors with a typical MMAF phenotype nor with ultra-structural defects of the sperm axoneme, while reduced sperm motility was frequently observed, which the authors assumed to result from an altered spermatogenesis due an abnormal thermoregulation caused by the obesity and genital anomalies of the tested subjects (Dieterich et al. 2009).
Importantly, following a comprehensive clinical examination of the two MMAF individuals we report herein with the IFT74 c. 256G > A mutation, no clinical features indicative of a ciliopathy nor of the BBS were observed. In particular, genital and urogenital malformations, which are part of the BBS phenotype (Tsang et al. 2018) were not detected in the two MMAF patients. The patients did not suffer from intellectual disability neither; one of them (individual IFT74_1) was occupying an engineer position. No diabetes nor hypertension were reported. Only weight excess was reported for individual IFT74_1, which remains limited as compare to the severe obesity usually found in BBS patients. Sperm production was quantitatively normal for individual IFT74_1, which again excluded the presence of hypogonadism, which is so far the only reproductive phenotype described for BBS patients in association with urogenital malformations (Koscinski et al. 2020). Individual IFT74_2 had a significantly reduced sperm count but this patient also suffered from unilateral varicocele, which was treated 6 years earlier. As numerous studies have reported an association between varicocele and altered semen parameters, the reduced sperm count of this patient may be partly caused by the varicocele. Overall, the examinations and clinical signs reported for both patients carrying the IFT74 c. 256G > A mutation, indicated a phenotype of isolated male infertility, with sperm anomalies associated with a MMAF phenotype but no major clinical signs of BBS.
It is of particular interest to understand why spermiogenesis is affected and ciliogenesis preserved in IFT74 mutated patients while IFT74 appears essential for IFT and for both cilia and flagella assembly. We showed that the c. 256G > A missense mutation induces the production of two main mutated IFT74 proteins: one carrying a missense mutation (p.Gly86Ser), the other carrying an in-frame 10 amino-acids deletion, (p.Leu77_Gly86del). Considering the site of these mutations, both types of mutant proteins could be affected in their N-terminal domain, which is known to be required for Tubulin binding. Binding and transport of the Tubulin is therefore expected to be impacted in cells from the patients. However, we suspect that the mutated IFT74 proteins are partly functional and could be sufficient to maintain IFT in most cilia but not in sperm flagella, thus inducing male sterility but not BBS. Such sperm-restricted phenotype was previously observed for a somatic missense mutation in AK7, encoding a Cilia and flagella Adenylate kinase, which only induces male sterility but not Primary Ciliary Dyskinesia in human (Lores et al. 2018). The greater sensibility of sperm flagellum towards these mutations, denotes some specificities in the molecular mechanisms of flagellum assembly and/or a reduced redundancy in functional proteins in this organelle. This point is reinforced by the observation that in mice, the invalidation of several genes demonstrated to be involved in human BBS (Bbs1, Bbs2, Bbs4, Bbs6 and Bbs7), only induced male sterility due to absent or abnormal sperm flagella while the cilia were properly assembled (Koscinski et al. 2020).
Overall the consequences of the IFT74 missense mutation described in this report confirms that the IFT machinery is certainly required for sperm flagellum assembly in mammals, in contrast to previous assumption that were done for both Drosophila and mammalian sperm. Moreover, a large body of studies now shows that despite common molecular alterations (Koscinski et al. 2020; Niederlova et al. 2019), ciliopathies encompass a collection of distinct clinical features. We show here that the nature of the mutation affecting a particular gene required for ciliogenesis may induce different pathologies, adding a level of complexity to the clinical manifestations of ciliary dysfunction.
Materials and methods
Whole exome sequencing and variants filtering
Genomic DNA was isolated from saliva using the Oragen DNA extraction kit (DNAgenotech®, Ottawa, Canada). Coding regions and intron/exon boundaries were sequenced on the Novogen platform based in China (agilent v6, HiSeqX,) after enrichment with Agilent kits (Agilent Technologies, Wokingham, UK).
Bioinformatics analyses: All datasets were reanalyzed using our updated bioinformatics pipeline, following the GATK 3.7 best practice recommendations. Briefly: sequencing reads were aligned against the GRCh38 reference genome using BWA-MEM 0.7.17, resulting BAM files were cleaned and sorted and duplicates were marked using Picard 2.7.1, and variants were called using GATK HaplotypeCaller to produce GVCF files. The GVCF files were then merged with a custom script (https://github.com/bcm-uga/mergeGVCFs) to obtain a single GVCF, and a final VCF file was produced with GATK Genotype GVCFs.
Variants filtering: The most promising candidate variants were identified using an in-house bioinformatics pipeline (Perl scripts available upon request), as follows: low-quality variant calls (DP < 5, GQ < 20, or less than 15% of reads supporting the ALT allele) were removed. We used variant effect predictor (VEP version 92) (McLaren et al. 2016) to annotate the variants and predict their impact. variants with a minor allele frequency greater than 5% in the NHLBI ESP6500 [Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA], greater than 3% in 1000 Genomes Project phase 3 datasets, or greater than 1% in gnomAD v2.0 (https://gnomad.broadinstitute.org/) were discarded. We also compared these variants to an in-house database of over 200 control exomes enriched for North African subjects. All variants present in homozygous state in this control database were excluded. We then only retained variants impacting splice donor or acceptor sites or causing frameshifts, in-frame insertions or deletions, stop gain, stop loss or missense variants except those scored as "tolerated" by SIFT (https://sift.bii.a-star.edu.sg/) and as "benign" by Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/). Gene expression data from the Genotype-Tissue Expression, GTEx project (https://gtexportal.org/home/) were added and used to strengthen the likely implication of the selected candidate variants.
Sanger sequencing
The identified variants in IFT74 were validated by Sanger sequencing performed on ABI 3500XL (Applied Biosystems). Data analyses were performed using SeqScape software (Applied Biosystems). Sequences of primers used and the expected product sizes are summarized in Supplemental Table S1.
Semen analysis
Semen samples were obtained by masturbation after a period of 2 to 7 days of sexual abstinence. Semen samples were incubated at 37 °C for 30 min for liquefaction and semen parameters were evaluated according to World Health Organization (WHO) guidelines (Cooper et al. 2010). Sperm vitality was assessed by eosin staining, and sperm morphology was analyzed on Schorr stained semen smears according to David’s classification (Auger et al. 2016). Moreover, as usual for patients requiring medically assisted procreation, a further analysis was performed on a sperm fraction recovered after selection by centrifugation (300 g, 20 min) on density gradient.
Transmission electron microscopy analysis of sperm cells
Human sperm cells (10 millions) were fixed by incubation in 0.1 M phosphate buffer pH 7 supplemented with 3% glutaraldehyde (Grade I; Sigma-Aldrich Co. Saint-Louis, MO, USA) for 2 h at room temperature. After two washes in PBS, sperm cells were resuspended in 0.2 M sodium cacodylate buffer and the samples were post-fixed by incubation with 1% osmium tetroxide (Electron Microscopy Sciences, Hatfield, UK). Samples were then dehydrated by immersion in a graded series of alcohol solutions and embedded in Epon resin (Polysciences Inc., Warrington, USA). Semi-thin sections were cut and stained with toluidine blue-Azur II. Ultra-thin sections (90 nm) were cut with a Reichert Ultracut S ultramicrotome (Reichert-Jung AG, Wien, Austria) and were then stained with uranyl acetate and lead citrate. Sections were analyzed with a JEOL 1011 microscope and digital images were acquired with a Gatan Erlangshen CCD camera and Digital Micrograph software.
RT-PCR analysis of sperm cells
200–800 ng of total RNA was extracted from 5 to 10 million human spermatozoa using NucleoSpin RNA kit (Macherey Nagel) and subjected to reverse transcription with High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Fisher Scientific) following the manufacturer’s instructions. PCR reactions were performed from 50 ng of retrotranscripted mRNA with GoTaq DNA polymerase (Promega) using IFT74 specific primers with 40 cycles of amplification (95 °C, 30 s; 55 °C, 30 s; 72 °C, 1 min). Amplicons were gel purified and either directly sequenced (Eurofins Genomics), or cloned before sequencing using TA cloning vector (Thermo Fisher Scientific). Sequences of primers used and expected product sizes are summarized in Table S1.
RT-qPCR analysis of sperm cells
cDNA prepared from one semen sample were used as template for real-time fluorescence quantitative PCR with SensiFAST SYBR® No-ROX Kit (Bioline). IFT74 and HPRT qPCR primer pairs were obtained from OriGene. The quantification of mRNA was performed in triplicate with HPRT as an internal control, according to the \(- 2^{{\Delta \Delta C_{{\text{t}}} }}\) method. Sequences of primers used and expected product sizes are summarized in Supplemental Table S1.
Western blot analysis on sperm cells
Denaturized protein samples corresponding to 1–2 million of spermatozoa from control or patient were loaded on SDS-PAGE [8% acrylamide/bisacrylamide (40% 37.5:1)] and transferred onto nitrocellulose membranes. The membranes were blocked in 5% milk in PBS-Tween 0.1%, and immunoblot analysis was performed using the indicated primary antibodies. Details of antibodies and dilutions used for western blot assays are provided in Supplemental Table S2.
Immunofluorescence analysis of sperm cells
10 μL of semen samples were spread onto a Superfrost Plus slide (Menzel Glasbearbeitungswerk, GmbH & Co. KG). Sperm was fixed by incubation with PBS/4% paraformaldehyde for 10 min. The slides were incubated 20 min at 95 °C in citrate buffer (H-3300, VectorLabs). The slides were next treated with 0.2% Triton in PBS for permeabilization and then blocked by incubation in 10% goat serum for 1 h. They were then incubated overnight with primary antibodies at 4 °C and then with secondary antibodies for 1 h at room temperature. The slides were mounted in Vectashield medium (Vector Laboratories) supplemented with 0.5 μg/mL DAPI. Slides were analyzed with a Zeiss Axiophot epifluorescence microscope. Digital images were acquired with a cooled charge-coupled device (CCD) camera (Hamamatsu Co.), under identical instrument settings, with MetaMorph software (Molecular Devices). Details of antibodies and dilutions used for immunofluorescence assays are provided in table S2.
Accession numbers
The IFT74 variant reported in this manuscript is accessible in ClinVar with the submission number SUB8577098.
Web resources
BLAST, (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/
dbscSNV, https://doi.org/10.1093/nar/gku1206
EMBL EBI Expression Atlas, https://www.ebi.ac.uk/gxa/home
gnomAD Browser, http://gnomad.broadinstitute.org
GTEx, https://gtexportal.org
Online Mendelian Inheritance in Man, https://www.omim.org
Smart, http://smart.embl-heidelberg.de/
Uniprot, https://www.uniprot.org/
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Change history
26 March 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00439-021-02278-z
References
Auger J, Jouannet P, Eustache F (2016) Another look at human sperm morphology. Hum Reprod 31:10–23. https://doi.org/10.1093/humrep/dev251
Avidor-Reiss T, Leroux MR (2015) Shared and distinct mechanisms of compartmentalized and cytosolic ciliogenesis. Curr Biol 25:R1143-1150. https://doi.org/10.1016/j.cub.2015.11.001
Bhogaraju S et al (2013) Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science 341:1009–1012. https://doi.org/10.1126/science.1240985
Blackburn K, Bustamante-Marin X, Yin W, Goshe MB, Ostrowski LE (2017) Quantitative proteomic analysis of human airway cilia identifies previously uncharacterized proteins of high abundance. J Proteome Res 16:1579–1592. https://doi.org/10.1021/acs.jproteome.6b00972
Brown JM, Cochran DA, Craige B, Kubo T, Witman GB (2015) Assembly of IFT trains at the ciliary base depends on IFT74. Curr Biol 25:1583–1593. https://doi.org/10.1016/j.cub.2015.04.060
Cooper TG et al (2010) World Health Organization reference values for human semen characteristics. Hum Reprod Update 16:231–245. https://doi.org/10.1093/humupd/dmp048
Coutton C et al (2018) Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat Commun 9:686. https://doi.org/10.1038/s41467-017-02792-7
Dieterich K et al (2009) The Aurora Kinase C c.144delC mutation causes meiosis I arrest in men and is frequent in the North African population. Hum Mol Genet 18:1301–1309. https://doi.org/10.1093/hmg/ddp029
Dong FN et al (2018) Absence of CFAP69 causes male infertility due to multiple morphological abnormalities of the flagella in human and mouse. Am J Hum Genet 102:636–648. https://doi.org/10.1016/j.ajhg.2018.03.007
Duran I et al (2016) Destabilization of the IFT-B cilia core complex due to mutations in IFT81 causes a spectrum of short-rib polydactyly syndrome. Sci Rep 6:34232. https://doi.org/10.1038/srep34232
He X et al (2018) Novel homozygous CFAP69 mutations in humans and mice cause severe asthenoteratospermia with multiple morphological abnormalities of the sperm flagella. J Med Genet. https://doi.org/10.1136/jmedgenet-2018-105486
Ishikawa T (2017) Axoneme structure from motile Cilia. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a028076
Ishikawa H, Marshall WF (2017) Intraflagellar transport and ciliary dynamics. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a021998
Khan SA, Muhammad N, Khan MA, Kamal A, Rehman ZU, Khan S (2016) Genetics of human Bardet–Biedl syndrome, an updates. Clin Genet 90:3–15. https://doi.org/10.1111/cge.12737
Kleinendorst L, Alsters SIM, Abawi O, Waisfisz Q, Boon EMJ, van den Akker ELT, van Haelst MM (2020) Second case of Bardet–Biedl syndrome caused by biallelic variants in IFT74. Eur J Hum Genet 28:943–946. https://doi.org/10.1038/s41431-020-0594-z
Koscinski I et al (2020) Reproduction function in male patients with Bardet Biedl syndrome. J Clin Endocrinol Metab. https://doi.org/10.1210/clinem/dgaa551
Kubo T et al (2016) Together, the IFT81 and IFT74 N-termini form the main module for intraflagellar transport of tubulin. J Cell Sci 129:2106–2119. https://doi.org/10.1242/jcs.187120
Lindstrand A et al (2016) Copy-number variation contributes to the mutational load of Bardet–Biedl syndrome. Am J Hum Genet 99:318–336. https://doi.org/10.1016/j.ajhg.2015.04.023
Liu H et al (2017) IFT25, an intraflagellar transporter protein dispensable for ciliogenesis in somatic cells, is essential for sperm flagella formation. Biol Reprod 96:993–1006. https://doi.org/10.1093/biolre/iox029
Liu C et al (2019a) Homozygous mutations in SPEF2 induce multiple morphological abnormalities of the sperm flagella and male infertility. J Med Genet. https://doi.org/10.1136/jmedgenet-2019-106011
Liu W et al (2019b) Bi-allelic mutations in TTC21A induce asthenoteratospermia in humans and mice. Am J Hum Genet 104:738–748. https://doi.org/10.1016/j.ajhg.2019.02.020
Liu W et al (2019c) Loss-of-function mutations in SPEF2 cause multiple morphological abnormalities of the sperm flagella (MMAF). J Med Genet 56:678–684. https://doi.org/10.1136/jmedgenet-2018-105952
Liu C et al (2020) Bi-allelic DNAH8 variants lead to multiple morphological abnormalities of the sperm flagella and primary male infertility. Am J Hum Genet 107:330–341. https://doi.org/10.1016/j.ajhg.2020.06.004
Lores P et al (2018) Homozygous missense mutation L673P in adenylate kinase 7 (AK7) leads to primary male infertility and multiple morphological anomalies of the flagella but not to primary ciliary dyskinesia. Hum Mol Genet 27:1196–1211. https://doi.org/10.1093/hmg/ddy034
Lores P et al (2019) Mutations in TTC29, encoding an evolutionarily conserved axonemal protein, result in asthenozoospermia and male infertility. Am J Hum Genet 105:1148–1167. https://doi.org/10.1016/j.ajhg.2019.10.007
McLaren W et al (2016) The ensembl variant effect predictor. Genome Biol 17:122. https://doi.org/10.1186/s13059-016-0974-4
Niederlova V, Modrak M, Tsyklauri O, Huranova M, Stepanek O (2019) Meta-analysis of genotype-phenotype associations in Bardet-Biedl syndrome uncovers differences among causative genes. Hum Mutat 40:2068–2087. https://doi.org/10.1002/humu.23862
Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG (2000) Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol 151:709–718. https://doi.org/10.1083/jcb.151.3.709
Pedersen LB, Rosenbaum JL (2008) Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol 85:23–61. https://doi.org/10.1016/S0070-2153(08)00802-8
Priya S, Nampoothiri S, Sen P, Sripriya S (2016) Bardet–Biedl syndrome: genetics, molecular pathophysiology, and disease management. Indian J Ophthalmol 64:620–627. https://doi.org/10.4103/0301-4738.194328
Qu W et al (2020) The essential role of intraflagellar transport protein IFT81 in male mice spermiogenesis and fertility. Am J Physiol Cell Physiol 318:C1092–C1106. https://doi.org/10.1152/ajpcell.00450.2019
Reiter JF, Leroux MR (2017) Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18:533–547. https://doi.org/10.1038/nrm.2017.60
Rosenbaum JL, Witman GB (2002) Intraflagellar transport. Nat Rev Mol Cell Biol 3:813–825. https://doi.org/10.1038/nrm952
San Agustin JT, Pazour GJ, Witman GB (2015) Intraflagellar transport is essential for mammalian spermiogenesis but is absent in mature sperm. Mol Biol Cell 26:4358–4372. https://doi.org/10.1091/mbc.E15-08-0578
Shi L et al (2019) Intraflagellar transport protein 74 is essential for spermatogenesis and male fertility in micedagger. Biol Reprod 101:188–199. https://doi.org/10.1093/biolre/ioz071
Sironen A et al (2011) Loss of SPEF2 function in mice results in spermatogenesis defects and primary ciliary dyskinesia. Biol Reprod 85:690–701. https://doi.org/10.1095/biolreprod.111.091132
Toure A et al (2020) The genetic architecture of morphological abnormalities of the sperm tail. Hum Genet. https://doi.org/10.1007/s00439-020-02113-x
Tsang SH, Aycinena ARP, Sharma T (2018) Ciliopathy: Bardet–Biedl syndrome. Adv Exp Med Biol 1085:171–174. https://doi.org/10.1007/978-3-319-95046-4_33
Vincensini L, Blisnick T, Bastin P (2011) 1001 model organisms to study cilia and flagella biology of the cell/under the auspices. Eur Cell Biol Organ 103:109–130. https://doi.org/10.1042/BC20100104
Wang G et al (2013) In-depth proteomic analysis of the human sperm reveals complex protein compositions. J Proteom 79:114–122. https://doi.org/10.1016/j.jprot.2012.12.008
Zhang Z et al (2016) Intraflagellar transport protein IFT20 is essential for male fertility and spermiogenesis in mice. Mol Biol Cell. https://doi.org/10.1091/mbc.E16-05-0318
Zhang Y et al (2017) Intraflagellar transporter protein (IFT27), an IFT25 binding partner, is essential for male fertility and spermiogenesis in mice. Dev Biol 432:125–139. https://doi.org/10.1016/j.ydbio.2017.09.023
Zhang Y et al (2018) Intraflagellar transporter protein 140 (IFT140), a component of IFT-A complex, is essential for male fertility and spermiogenesis in mice. Cytoskeleton 75:70–84. https://doi.org/10.1002/cm.21427
Acknowledgements
We thank all the individuals and their families for their cooperation, as well as all the referring physicians. We thank all the technicians from the Service de Biologie de la Reproduction at the Hôpital Cochin (Paris) for routine semen sample evaluation (Jacques Bras, Nathalie Chériau, Véronique Hernandorena, Jean-Claude Cambronne and Caroline Villalon). We thank Cellular Imaging Facility of the Institut Cochin for electron microscopy procedures.
Funding
This work was supported by Institut National de la Santé et de la Recherche Médicale (INSERM), Centre National de la Recherche Scientifique (CNRS), Université Paris Descartes, University Grenoble-Alpes and Agence Nationale de la Recherche: Grant MASFLAGELLA ANR-14-CE15 and Grant FLAGEL-OME ANR-19-CE17-0014 to PR, NTM and AT, Grant DIVERCIL-17-CE13-0023 to AT.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. AT designed and supervised the study. M-AL, LF, CP, SFBM, RZ and ED recruited the patients, performed clinical analysis and characterization. NTM, CC and PR performed exome and bioinformatics analysis. PL, Z-EK, AA-Y, AD, CCa and S-HH performed experimental work (Sanger sequencing, RT-PCR, Sequencing, Immunofluorescence, Immunoblotting). AT performed MO and TEM analysis. PL and AT performed data illustrations. PL, MW, LS and EC, ED and AT analyzed the data. PL and AT wrote the manuscript. The first draft of the manuscript was written by PL and AT, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Animal research
Not applicable.
Ethics approval
The study was performed according to the principle of the Declaration of Helsinki. The use of human samples and the genetic studies were approved by local and governmental committees: Comité de Protection des Personnes CPP Ile de France III (authorization 02748), CRB Germethèque (certification under ISO-9001 and NF-S 96-900), Fertithèque collection declared to the French Ministry of Health (DC-2015-2580) and the French Data Protection Authority (DR-2016-392).
Consent to Participate
Informed consent was obtained from all individuals included in the study.
Consent to Publish
Agreement for data publication was obtained from all individuals included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised due to change in the second author's name.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lorès, P., Kherraf, ZE., Amiri-Yekta, A. et al. A missense mutation in IFT74, encoding for an essential component for intraflagellar transport of Tubulin, causes asthenozoospermia and male infertility without clinical signs of Bardet–Biedl syndrome. Hum Genet 140, 1031–1043 (2021). https://doi.org/10.1007/s00439-021-02270-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-021-02270-7