Abstract
Spermatozoa are polarized cells with a head and a flagellum joined together by the connecting piece. Flagellum integrity is critical for normal sperm function, and flagellum defects consistently lead to male infertility. Multiple morphological abnormalities of the flagella (MMAF) is a distinct sperm phenotype consistently leading to male infertility due to a reduced or absent sperm motility associated with severe morphological and ultrastructural flagellum defects. Despite numerous genes recently described to be recurrently associated with MMAF, more than half of the cases analyzed remain unresolved, suggesting that many yet uncharacterized gene defects account for this phenotype. By performing a retrospective exome analysis of the unsolved cases from our initial cohort of 167 infertile men with a MMAF phenotype, we identified one individual carrying a homozygous frameshift variant in CFAP206, a gene encoding a microtubule-docking adapter for radial spoke and inner dynein arm. Immunostaining experiments in the patient’s sperm cells demonstrated the absence of WDR66 and RSPH1 proteins suggesting severe radial spokes and calmodulin and spoke-associated complex defects. Using the CRISPR–Cas9 technique, we generated homozygous Cfap206 knockout (KO) mice which presented with male infertility due to functional, structural and ultrastructural sperm flagellum defects associated with a very low rate of embryo development using ICSI. Overall, we showed that CFAP206 is essential for normal sperm flagellum structure and function in human and mouse and that bi-allelic mutations in CFAP206 cause male infertility in man and mouse by inducing morphological and functional defects of the sperm flagellum that may also cause ICSI failures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Male infertility is a major and contemporary public health concern which regroups a wide range of sperm phenotypes often caused by severe gene defects. Recent approaches for genetic investigation allowed to significantly increase our knowledge about the genetic causes involved in the most severe phenotypes of male infertility (Krausz and Riera-Escamilla 2018). Besides the mere identification of the genetic cause, these findings also allowed to better understand the physiopathological mechanisms associated with a specific sperm phenotype and to adapt the best clinical management. This assumption is particularly true for the “MMAF phenotype”, for Multiple Morphological Anomalies of the Flagella which is defined by a mosaic of sperm cells with absent, short, irregular and coiled flagellum. This condition is systematically associated with an extreme asthenozoospermia with near zero progressive sperm motility leading to male infertility (Touré et al. 2021). The sperm flagellum is a highly specialized organelle responsible for sperm motility and its migration in the female reproductive tract. The mammalian sperm has a central “9 + 2” conformation of microtubule doublets associated with hundreds of accessory proteins that together constitute an axoneme (Lindemann and Lesich 2016). The sperm flagellum axoneme shares key structural similarities with the axoneme of other motile cilia (Brown and Witman 2014). However, the sperm flagellum is unique by its specific functions and mode of assembly (Inaba 2007) and differs from other cilia by the presence of numerous and specific peri‐axonemal structures including the outer dense fibers (ODFs), and the fibrous and mitochondrial sheaths. At least twenty genes, encoding different proteins located the sperm flagellum, have so far been shown to be associated with the MMAF phenotype (Touré et al. 2021). A large majority of these genes encodes for axonemal protein highlighting the critical role of this structure in sperm flagellum integrity. Moreover, it seems that many of these MMAF-related proteins are closely located or related to the calmodulin- and spoke-associated complex (CSC) suggesting a key and specific role of this structure in the sperm flagellum assembly, stability and function (Touré et al. 2021). A large part of the genetic causes of MMAF remains to be identified and might further support this hypothesis. Here, we report that bi-allelic truncating mutations in CFAP206, a gene encoding an axonemal protein essential for the CSC stability, are associated with male infertility in human and mouse.
Materials and methods
Patients
In our previously established cohort of 167 MMAF patients (Coutton et al. 2019), we previously identified 77 patients with harmful variants in known MMAF-related genes (Lorès et al. 2021). To identify some additional causes associated with human asthenozoospermia due to MMAF, we retrospectively analyzed the exomes of the remaining 90 unsolved cases. All patients of the cohort presented a typical MMAF phenotype characterized by severe asthenozoospermia (total sperm motility below 10%) with at least three of the following flagellar abnormalities present in > 5% of the spermatozoa: short, absent, coiled, bent or irregular flagella (Coutton et al. 2019). All patients had a normal somatic karyotype (46, XY) with normal bilateral testicular size, normal hormone levels (FSH, testosterone and prolactin) and secondary sexual characteristics. All these patients presented only with non-syndromic infertility without any other clinical features. Sperm analysis was carried out in the source laboratories during routine biological examination of the patients according to World Health Organization (WHO) guidelines (Wang et al. 2014). One patient (P1), who was identified to carry a homozygous CFAP206 variant, is described in this study. P1 sperm morphology assessed with Papanicolaou staining and detailed semen parameters were performed. The patient originated from the Middle East (Iran) and was recruited at the Royan Institute (Reproductive Biomedicine Research Center) for primary infertility in 2016 and was born to first-cousin parents. Informed consent was obtained from the patient and controls participating in the study according to local protocols and the principles of the Declaration of Helsinki. The study was approved by local ethics committees, and samples were stored in the Fertithèque collection declared to the French Ministry of health (DC-2015-2580) and the French Data Protection Authority (DR-2016-392).
Exome sequencing and bioinformatic analysis
Data processing of the whole cohort of 167 MMAF patients was performed according to our previously described protocol (Coutton et al. 2019). In brief, the enrichment of coding regions together with intron/exon boundaries was performed with the Exon V6 kit (Agilent Technologies, Wokingham, UK). Sequencing was performed with Illumina HiSeq X by a service provider (Novogene, Cambridge, UK). Exomes data were analyzed using a bioinformatics pipeline developed in-house using two modules, both distributed under the GNU General Public License v3.0 and available on GitHub: https://github.com/ntm/grexome-TIMC-Primary and https://github.com/ntm/grexome-TIMC-Secondary and as described in part in Martinez et al. (2020). Variants with a minor allele frequency greater than 1% in gnomAD v2.0, 3% in 1000 Genomes Project phase 3, or 5% in NHLBI ESP6500 were filtered out and only variants predicted to have high-impact (e.g., stop-gain or frameshift variants) by variant Effect Predictor v92 (McLaren et al. 2016) were scrutinized.
Sanger sequencing
The identified variant in CFAP206 was validated by Sanger sequencing performed on ABI 3500XL (Applied Biosystems). Data analyses were performed using SeqScape software (Applied Biosystems). Sequences of primers used are reported in Table S1. Unfortunately, no parental DNA was available to perform the segregation analysis.
Quantitative real-time RT-PCR (RT-qPCR) analysis
RT-qPCR was performed with cDNAs from a panel of 6 different human tissues and one human cDNAs reference (pooled tissues) purchased from Life Technologies®. Each sample was assayed in triplicate for each gene on a StepOnePlus (LifeTechnologies®) with Power SYBR®Green PCR Master Mix (Life Technologies®). The PCR cycle was as follows: 10 min at 95 °C, 1 cycle for enzyme activation; 15 s at 95 °C, 60 s at 58 °C with fluorescence acquisition, 40 cycles for the PCR. Primer sequences and RT-qPCR conditions are indicated in Table S2. The efficacy of primers was checked using a standard curve. Melting curve analysis was used to confirm the presence of a single PCR product. RT-qPCR data were normalized using the reference housekeeping gene GAPDH for human with the − ΔΔCt method (Livak and Schmittgen 2001). The 2−ΔΔCt value was set at 0 in the cDNAs pooled tissues, resulting in an arbitrary expression of 1. Statistics were performed using a two-tailed t-test on Prism 4.0 software (GraphPad, San Diego, CA) to compare the relative expression of CFAP206 transcripts in several organs. Statistical tests with a two-tailed P value ≤ 0.05 were considered significant.
Immunostaining in human sperm cells
Immunofluorescence (IF) experiments were performed using sperm cells from control individuals and from the individual carrying the CFAP206 variant. Sperm cells were fixed in phosphate-buffered saline (PBS)/4% paraformaldehyde for 1 min at room temperature. After washing in 1 ml PBS, the sperm suspension was spotted onto 0.1% poly L-lysine pre-coated slides (Thermo Scientific). After attachment, sperm were permeabilized with 0.1% (v/v) Triton X-100–DPBS (Triton X-100; Sigma-Aldrich) for 5 min at RT. Slides were then blocked in 5% normal serum–DPBS (normal goat or donkey serum; GIBCO, Invitrogen) and incubated overnight at 4 °C with the following primary antibodies: rabbit polyclonal anti-WDR66 (HPA040005, Sigma-Aldrich, rabbit, 1:50, green), rabbit polyclonal anti-RSPH1 (HPA017382, Sigma-Aldrich, 1:100) and monoclonal mouse anti-acetylated-α-tubulin (T7451, Sigma-Aldrich, 1:2000). Washes were performed with 0.1% (v/v) Tween-20–DPBS, followed by 1-h incubation at room temperature with secondary antibodies. Highly cross-adsorbed secondary antibodies (Dylight 488 and Dylight 549, 1:1000) were from Jackson Immunoresearch®. Appropriate controls were performed, omitting the primary antibodies. Samples were counterstained with 5 mg/ml Hoechst 33,342 (Sigma-Aldrich) and mounted with DAKO mounting media (Life Technology). Fluorescence images were captured with a confocal microscope (Zeiss LSM 710). Two hundred sperm cells were manually analyzed by two different experienced operators and the IF staining intensity and pattern were compared with a fertile control. The specificity of the anti-WDR66 and anti-RSPH1 antibodies was previously validated (Kott et al. 2013; Auguste et al. 2018).
CRISPR/Cas9 KO mice
Cfap206 knockout mouse model
The Cfap206 knockout mouse model (C57BL/6) was created by CRISPR/Cas-mediated genome engineering. Briefly, the single-guide RNAs (sgRNAs) were designed against Cfap206 exon 3–11 (Table S3). Cas9 mRNA and sgRNA were prepared according to the reference (Yang et al. 2014). One-cell-stage embryos were collected and injected with prepared Cas9 mRNA and sgRNA. Then the injected embryos were further cultured in KSOM medium (Millipore, Cat. #MR-106-D) at 37 °C under 5% CO2 until blastocyst stage and transferred into pseudopregnant female mice. A frameshift variant in Cfap206 was detected by Sanger sequencing in the founder mouse and its offspring, and the primer sequences are available in Table S4. Male mice (aged 8–12 weeks) were used for subsequent experiments in this study. All animal experiments were carried out in accordance with the recommendation in the Guide for the Care and Use of Laboratory Animals of the National Institutes of health. The study was approved by the animal ethics committee at Anhui Medical University.
Immunoblotting of mouse testis
Mouse testis samples were homogenized in radioimmunoprecipitation assay (RIPA) buffer (Beyotime) via an ultrasonic homogenizer and then heated at 100 °C for 10 min. The lysates were separated on 10% polyacrylamide gel by SDS-PAGE and transferred to PVDF (polyvinylidene fluoride) membrane. Then, the membrane was sealed for 1 h at 25 °C with 5% milk diluted with TBST (TBS-0.1% Tween-20, Sangon Biotech). Anti-CFAP206 antibody (HPA044891, Atlas Antibodies) was diluted in TBST at 1:1,000 and incubated with the membranes at 4 °C overnight. The membranes were then washed in TBST three times and incubated with HRP-conjugated anti-Rabbit IgG antibody (M21002, Abmart,1:10,000) in blocking solution for 1 h at room temperature. Enhanced chemiluminescence (ECL) (BL520A, Biosharp) was used for visualization. β-tubulin was used as a loading control.
Histological analysis of mouse tissues
Fresh mouse testes were gently fixed with modified Davidson’s solution (50% diluted water, 30% formaldehyde, 15% ethanol, and 5% glacial acetic acid) for over 48 h. After fixation, the tissue was dehydrated in gradient ethanol (70% ethanol for 24 h, 80% ethanol for 2 h, 90% ethanol for 2 h, and 100% ethanol for 1 h). Then, tissues were placed in xylene for 1 h and embedded in paraffin wax. Finally, sections were cut at a 3 µm thickness. For H&E staining, sections were deparaffinated in xylene at 65 °C overnight. After deparaffinating, slides were stained with H&E, dehydrated, and mounted.
Mating test
Fertility was investigated in wild-type and Cfap206 knockout adult male mice. At least three male mice that were 8–12 weeks of age were analyzed in each group. Each male mouse and three wild-type C57BL/6 females (8–12 weeks of age) were caged. Vaginal plugs were checked every morning. Once a vaginal plug was identified, the male mouse was allowed to rest for 2 days before another two females were placed in the cage. After mating, the female mice were separated and fed in a single cage and the pregnancy results and number of pups were recorded.
Mouse semen parameters and sperm morphological analysis
For sperm morphology and parameters analyses of the mouse, spermatozoa were extracted from the cauda epididymis through dissection of adult male mice and diluted in 1 mL human tubal fluid (HTF, 90,126, Millipore) for 15 min at 37 °C. Sperm count, progressive sperm rate and motile sperm rate were further analyzed by a computer-assisted analysis system (IVOSII, Hamilton). At least three C57BL/6 male mice aged 8–10 weeks were analyzed in each group. Sperm from cauda epididymis were fixed and stained by H&E staining for the morphology analysis.
Electron microscopy evaluation
For scanning electron microscope (SEM) analysis of mouse sperm cells, cauda epididymis samples were prepared as previously described (Liu et al. 2019b). In brief, mouse sperm specimens were deposited on poly-L-lysine-coated coverslips, fixed in 2.5% glutaraldehyde, washed in 0.1 mol/L phosphate buffer, and post-fixed in osmic acid. The specimens were then progressively dehydrated with ethanol and isoamyl acetate gradient, then dried with a CO2 critical-point dryer (Eiko HCP-2, Hitachi). Next, the specimens were mounted on aluminum stubs, sputter-coated using of an ionic sprayer meter (Eiko E-1020, Hitachi), and analyzed via SEM (Stereoscan 260) under an accelerating voltage of 20 kV.
For transmission electron microscope (TEM) analysis of mouse sperm cells, mouse semen samples were rinsed and were progressively dehydrated with graded ethanol (50%, 70%, 90%, and 100%) and 100% acetone, followed by infiltration with 1:1 acetone and SPI-Chem resin overnight at 37 °C. After being embedded in Epon 812, the specimens were sliced with ultra-microtome, stained with uranyl acetate and lead citrate, and observed and photographed via TEM (Tecnai G2, FEI) with an accelerating voltage of 120 kV.
Intracytoplasmic sperm injection with the sperm of wild-type and Cfap206 knockout mice
Mice intracytoplasmic sperm injection (ICSI) was conducted as previously described (Liu et al. 2019a). Briefly, MII oocytes were collected from the oviduct of superovulated wild-type C57BL/6 female mice. Sperm were collected from wild-type and Cfap206 knockout mice. Sperm heads only were injected into mouse oocytes by a Piezo driven pipette according to the previously reference (Ron-El et al. 1995).Then, the injected oocytes were cultured in KSOM medium at 37 °C under 5% CO2. Cleavage and blastocyst rates were further recorded around 24 h and 96 h, respectively.
Results
Exome sequencing identified bi-allelic variants in CFAP206 in an MMAF patient
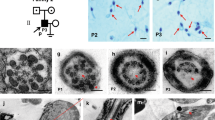
In the whole cohort of 167 MMAF patients, we previously identified 77 patients (46%) with deleterious variants in known MMAF-related genes (Lorès et al. 2021). After reanalysis of the remaining exomes, we identified one additional subject with a homozygous variant in CFAP206 (MIM *609910), a gene not previously associated with any pathology in human. Patient’s sperm morphology and his detailed semen parameters are presented in Fig. 1A–D and Table 1, respectively. The patient had a frameshift variant c.1430dupA; p.Asn477LysfsTer15 (NM_001031743.2) (Fig. 1E; Table 1). The variant c.1430dupA is a single-nucleotide duplication predicted to induce a translational frameshift and a premature stop codon (p.Asn477LysfsTer15) expected to lead to the complete absence of the protein or the production of a truncated protein. The variant is present in the Genome Aggregation Database (gnomADv3.1) with a minor allele frequency (MAF) of 6.63 × 10−6. The presence of this homozygous variant in the patient was also consistent with the known consanguinity of his parents (cousins). The variant identified by exome sequencing was validated by Sanger sequencing as illustrated in Fig. 1F. This CFAP206 variant is deposited in ClinVar under reference SUB9294549. No other candidate variants reported to be associated with cilia, flagella or male fertility were otherwise detected.
Morphology of normal and CFAP206 mutant spermatozoa and presentation of the patient’s variant. A–D Light microscopy analysis of spermatozoa from fertile control individuals (A) and the CFAP206 patient (B–D). Most spermatozoa from the CFAP206 patient have flagella that are short, absent, coiled or of irregular caliber. Scale bars: 10 µm. E Structure of the canonical transcript of CFAP206 showing the position of the observed variant. The functional structure of the encoded protein is shown in the lower panel. CFAP206-domain (IPR021897) is highlighted in green. F Electropherograms from Sanger sequencing indicating the homozygous state of the identified variant c.1430dupA; p.Asn477LysfsTer15 (NM_001031743.2) in CFAP206. Variants are annotated following HGVS recommendations
CFAP206 (also known as c6orf165) is located on chromosome 6 and contains 14 exons encoding cilia- and flagella-associated protein 206 (CFAP206), a predicted 622-amino acid protein (Q8IYR0). CFAP206 is predominantly expressed in the testis according to data from GTEx (https://gtexportal.org) and described to be associated with cilia and flagella (Vasudevan et al. 2015; Beckers et al. 2020). In addition, the encoded CFAP206 protein was detected in human sperm proteome (Wang et al. 2013) whereas it was found at a low level in human airway cilia (Blackburn et al. 2017). RT-qPCR experiments performed with a panel of six human tissues including other ciliated tissues such as trachea confirmed these results, showing that CFAP206 is largely overexpressed in the testis compared to all the other tested tissues (Fig. S1).
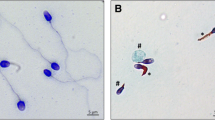
To explore the ultrastructural defects induced by the c.1430dupA CFAP206 variant in human sperm, we subsequently studied the presence of different proteins belonging to various axonemal substructures by immunofluorescence (IF). Taking into account the expected role of CFAP206 in RS and CSC stability (Vasudevan et al. 2015; Beckers et al. 2020), the presence of the following proteins was first investigated: RSPH1 as a marker of the radial spokes (RS) and WDR66, a protein localized in the CSC at the base of radial spoke 3 in Tetrahymena and Chlamydomonas (Urbanska et al. 2015). Due to the limited amount of sperm cells, no further IF experiments could be performed. In the patient’s sperm cells, RSPH1 staining was totally absent, dramatically reduced or displayed an abnormal dotted and irregular pattern (Fig. 2A). In control sperm, WDR66 immunostaining decorated the full-length flagella, but in the CFAP206 patient, WDR66 staining was completely absent whereas tubulin staining remained detectable (Fig. 2B). These results demonstrate that RS and the CSC are strongly disorganized and support that CFAP206 is a key axonemal component ensuring the stability of the CSC and the anchoring of the RS. Unfortunately, Transmission Electron Microscopy (TEM), which might provide evidence of the resulting defect, could not be performed due to the very low number of sperm cells available.
Radial spokes and the calmodulin- and spoke-associated complex are affected in the CFAP206 patient. A Sperm cells from a fertile control individual and the CFAP206 patient stained with anti-RSPH1 (HPA017382, Sigma-Aldrich, rabbit, 1:100, green), a protein located at the head of the radial spokes, and anti-acetylated tubulin (32–2500, ThermoFisher, mouse, 1: 1000, red) antibodies. DNA was counterstained with DAPI II. RSPH1 immunostaining is present throughout the flagellum in control sperm cells but is mainly absent or strongly reduced in CFAP206 patients. In a few sperm cells the RSPH1 staining is present but displays an abnormal pattern with an irregular signal. Scale bars: 10 µm. B Sperm cells from a fertile control individual and the CFAP206 patient stained with anti-WDR66 (HPA040005, Sigma-Aldrich, rabbit, 1:50, green) and anti-acetylated tubulin (32–2500, ThermoFisher, mouse, 1: 1000, red) antibodies. DNA was counterstained with DAPI II. Contrary to the control, the WDR66 immunostaining is not detectable in the sperm flagellum from the CFAP206 patient. Scale bars: 10 µm
CRISPR/Cas 9 KO mice
Then, we assessed the impact of the absence of CFAP206 on mouse by generating KO mice using the CRISPR-Cas9 technology. We obtained a strain with a deletion of nine exons (exons 3–11) (Fig. S2A). RT-PCR and Western-blot analyses performed on testes from KO mouse Cfap206−/− confirmed the deleterious effect of the CRISPR/Cas9 induced deletion with the production of abnormal transcripts leading to the total absence of the protein (Fig. S2B, C). Reproductive phenotypes were then studied. KO females were fully fertile and gave litters of normal size, contrary to KO males, which exhibited complete infertility when mated with females (Fig. 3A). The testis to body weight ratio was comparable between Cfap206+/+ and Cfap206−/− male mice (Fig. 3B, C). As well, hematoxylin and eosin staining of testicular tissues showed no significant inter-group difference in the overall morphology of germ cells (Fig. S3). Total sperm counts obtained from the epididymes of Cfap206−/− male mice were significantly lower than those from Cfap206+/+ male mice (3.92 × 106 ± 0.64 versus 22.63 × 106 ± 2.47, respectively; ***P < 0.001) (Fig. 3D; Table S5), and sperm from Cfap206−/− animals showed a significant decrease in total and progressive motility (Fig. 3E, F; Table S5). Consistently, these functional flagellum defects were associated with morphological defects (Figs. 3G and S4). Sperm from Cfap206−/− males mainly had a flagellum of normal length (Table S5) but most of them showed abnormal forms, sperm exhibiting bent and coiled flagella (Fig. 3G and S4; Table S5). We also observed a significant increase of sperm cells with absent or short flagella compared to the WT mice (Table S5). To visualize the impact of the absence of the CFAP206 protein on the flagellum ultrastructure, sperm from Cfap206−/− males were analyzed by TEM (Fig. S5). Some longitudinal sections of Cfap206−/− sperm showed a distorted axoneme (Fig. S5) which may be the cause or the consequence of the bent and coiled flagella. In addition, observations of transversal sections often revealed a deep disorganization with several defects such as lack of peripheral doublets, absence of the central pair or abnormal distribution of the DMTs or/and of the ODFs (Fig. S5E and F). No extra-reproductive features (including hydrocephaly) were observed in Cfap206−/− mice.
Cfap206 deficiency induces sperm flagellar abnormalities and infertility in male mice. A The mean number of the pups per litter was 6.83 ± 0.40 in Cfap206+/+ male mice, whereas all the four Cfap206−/− male mice were completely infertile. ***P < 0.001. B–C The ratio of testes to body weight was comparable between Cfap206+/+ and Cfap206−/− male mice. D Sperm counts (× 106) of Cfap206−/− male mice was significantly lower compared to Cfap206+/+ male mice (3.92 ± 0.64 versus 22.63 ± 2.47, respectively; ***P < 0.001). E Sperm motility rate in Cfap206+/+ male mice was 56.33 ± 5.67%, whereas the motility decreased to 37.33 ± 2.94% in Cfap206−/− male mice. *P < 0.05. F The progressive motility was significantly reduced in Cfap206−/− male mice compared to the wild-type (31.00 ± 2.75% and 49.17 ± 5.17% respectively, *P < 0.05). G Sperm morphology using scanning electron microscopy of the Cfap206+/+ (i) and Cfap206−/− male mice (ii–v). Most spermatozoa of Cfap206−/− male mice presented with coiled and bent flagellum. Scale bars: 10 μm. n.s. not significant
ICSI experiments performed with CFAP206 deficient sperm cells showed a significant lower rate of 2-cell embryos (56.18% vs 91.11%, respectively) and a dramatic decrease of blastocyst rate (7% vs 31.7%, respectively) compared to WT mice (Fig. 4; Table S6).
Discussion
In the present work, we show that the presence of bi-allelic truncating CFAP206 variants induces male infertility in human and mouse associated with morphological and motility defects demonstrating that CFAP206 is critical for sperm flagellum structure and function. Ultrastructure of the sperm flagellum was found to be deeply affected by the absence of CFAP206’s protein. In Cfap206−/− mouse, TEM experiments revealed severe axoneme disorganization and peri-axonemal defects (Fig. S5). In human, immunofluorescent analyses showed that the CFAP251/WDR66 was totally absent from sperm cells from our patient, supporting a role of CFAP206 in the assembly and/or the stability the CSC in mammalian sperm flagella (Fig. 2B). The CSC is a complex multi-protein structure located within the region that interconnects the bases of RS2, RS3, and the N-DRC (Urbanska et al. 2015) and is expected to mediate the regulatory signals between the radial spokes and the dynein arms, therefore regulating flagellar motility (Urbanska et al. 2015). Based on the localization of the CSC complex, at the base of the radial spokes, it was expected that the absence of CSC’s members would result in radial spoke assembly defects (Dymek and Smith 2007). This assumption was confirmed by IF analyses which showed that the RS were severely and consistently impacted, although few sperm cells showed residual RSPH1 staining (Fig. 2A). Such results are consistent with the known localization of CFAP206 in the axoneme. Inactivation of the conserved ciliary protein FAP206 in the ciliate Tetrahymena resulted in slow cell motility. In addition, cryo–electron tomography showed that the 96 nm repeats along the axoneme, lacked the RS2 and the dynein c, suggesting that FAP206 is essential for docking the RS2 and dynein c to the microtubule. Interestingly, it was also demonstrated that the assembly of the CSC component FAP91/CaM-IP2 into the axoneme was dependent on FAP206 which was shown to be indirectly associated with the CSC through the RSP3 protein (Vasudevan et al. 2015). Consistently, we previously demonstrated that deleterious variants in MAATS1 encoding CFAP91, the human FAP91 ortholog, led to a MMAF phenotype with similar axonemal defects (lack of WDR66 and RSPH1) to what has been observed here in the CFAP206 affected patient (Martinez et al. 2020). Altogether, these observations strongly reinforce the assumption that CFAP206 is critical for sperm flagellum axoneme assembly and stability through its essential role within the CSC–RS complex and its interaction with other axonemal partners such as CFAP251/WDR66 or CFAP91 (Vasudevan et al. 2015; Beckers et al. 2020).
These data further reinforce the hypothesis that the RS3–CSC complex is critical for the assembly and stability of the sperm flagellum axoneme and its alteration constantly lead to the MMAF phenotype. In our cohort, variants in CSC-related genes (CFAP251/WDR66, MAATS1 and now CFAP206) are found in about 11% of the tested subjects suggesting that the CSC is the main structure affected in MMAF patients (Kherraf et al. 2018; Martinez et al. 2020). Interestingly, although the axoneme is a common structure shared between cilia and flagellum, we observed that MMAF patients only present with an isolated infertility without any other clinical features or ciliopathies suggesting that, in other ciliated cells, these axonemal MMAF-related proteins are dispensable for the structure and function of cilia and that axonemal biogenesis/structure of sperm flagella and cilia may require different proteins and mechanisms in particular regarding the RS3–CSC complex (Touré et al. 2021). Interestingly, another team recently published another mouse model inactivated for the Cfap206 gene and evidenced a similar sperm phenotype. Electron tomography on cryo-conserved sperm flagella revealed defects in the repetitive pattern of radial spokes with only one RS per 96 nm repeat confirming that CFAP206 is needed for the establishment of radial spokes in mammalian sperm flagella. In addition, the authors reported that about 80% of KO mice developed externally visible enlarged cranial vaults, suggesting ventricular dilatation and hydrocephalus. In addition, these mutant mice presented mucus accumulation in nasal cavities and a significant increase in ciliary beat frequency compared to wild-type (Beckers et al. 2020). These extra-reproductive observations are in opposition with the clinical features observed in our mouse model and in our patient carrying the bi-allelic truncating variations in CFAP206 who presented only isolated infertility without any other clinical features. We do not believe that this phenotypic discrepancy can be caused by a residual CFAP206 activity in our patient nor in our KO mice, as all carried a clear cut loss of function variant similar to that induced in Beckers et al.’s mice. Such phenotypic discrepancies between patients with MMAF phenotype or two different mouse models inactivated for the same gene have been described previously (Ben Khelifa et al. 2014; Coutton et al. 2018; Morimoto et al. 2019; Rachev et al. 2020; Touré et al. 2021). These observations might be explained by different mouse genetic background, various genome-editing method or unknown off-target effect. Alternatively, we cannot exclude a slight effect on ciliary beating in MMAF patients without pathological consequences, or at least none that have been noticed by the affected men themselves. Nasal brushings or curette biopsies from affected MMAF individuals could be useful to formally explore this possibility.
It was generally accepted that MMAF affected individuals have a good prognosis following ICSI, in particular for patients carrying mutations in genes encoding axonemal proteins (Wambergue et al. 2016; Touré et al. 2021). In contrast to what was observed for most other MMAF patients, the patient carrying the CFAP206 mutation did not achieve any pregnancy when his sperm was used for ICSI. Furthermore, ICSI experiments performed in Cfap206−/− mouse showed a dramatic decrease in early division and blastocyst rate compared to WT, suggesting that CFAP206 deficiency may also affect early embryonic development (Fig. 4; Table S6). This effect on early embryonic development following IVF was also observed in the previously published Cfap206−/− mouse line (Beckers et al. 2020). This very low rate of embryo development is a clear additional factor compromising the fertility of CFAP206-mutated patients as it was observed in the Cfap206−/− mouse. Beckers et al. (2020) previously demonstrated that CFAP206 also localized to the basal body/centrosome of motile cilia. Interestingly, unsuccessful ICSI were already reported in MMAF patients with mutations impacting centrosomal proteins (Touré et al. 2021). These data provide further support to the assumption that a poor ICSI prognosis is expected for MMAF patients with mutation in genes encoding centrosomal proteins which persist after fertilization and are required for embryo development conversely to axonemal proteins. However, no basal body defects could be evidenced in Cfap206−/− sperm cells using IF and TEM (data not shown). These results should be confirmed with further cases and experimentation but suggest that CFAP206 mutations may constitute an adverse factor to obtain pregnancies with ICSI and may modify the clinical management of CFAP206-mutated MMAF patient.
Overall, these data demonstrate that CFAP206 is essential for normal sperm flagellum structure and function in human and mouse, and that variants in CFAP206 lead to severe flagellum malformations and may also cause poor early embryo development resulting in primary male infertility.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. Accession numbers: The CFAP206 variant reported in this manuscript is accessible in ClinVar with the submission number SUB9294549.
References
Auguste Y, Delague V, Desvignes J-P et al (2018) Loss of calmodulin- and radial-spoke-associated complex protein CFAP251 leads to immotile spermatozoa lacking mitochondria and infertility in men. Am J Hum Genet 103:413–420. https://doi.org/10.1016/j.ajhg.2018.07.013
Beckers A, Adis C, Schuster-Gossler K et al (2020) The FOXJ1 target Cfap206 is required for sperm motility, mucociliary clearance of the airways and brain development. Development. https://doi.org/10.1242/dev.188052
Ben Khelifa M, Coutton C, Zouari R et al (2014) Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet 94:95–104. https://doi.org/10.1016/j.ajhg.2013.11.017
Blackburn K, Bustamante-Marin X, Yin W et al (2017) Quantitative proteomic analysis of human airway cilia identifies previously uncharacterized proteins of high abundance. J Proteome Res 16:1579–1592. https://doi.org/10.1021/acs.jproteome.6b00972
Brown JM, Witman GB (2014) Cilia and diseases. Bioscience 64:1126–1137. https://doi.org/10.1093/biosci/biu174
Coutton C, Vargas AS, Amiri-Yekta A et al (2018) Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in trypanosoma and human. Nat Commun 9:686. https://doi.org/10.1038/s41467-017-02792-7
Coutton C, Martinez G, Kherraf Z-E et al (2019) Bi-allelic mutations in ARMC2 lead to severe astheno-teratozoospermia due to sperm flagellum malformations in humans and mice. Am J Hum Genet 104:331–340. https://doi.org/10.1016/j.ajhg.2018.12.013
Dymek EE, Smith EF (2007) A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. J Cell Biol 179:515–526. https://doi.org/10.1083/jcb.200703107
Inaba K (2007) Molecular basis of sperm flagellar axonemes: structural and evolutionary aspects. Ann N Y Acad Sci 1101:506–526. https://doi.org/10.1196/annals.1389.017
Kherraf Z-E, Amiri-Yekta A, Dacheux D et al (2018) A Homozygous ancestral SVA-insertion-mediated deletion in WDR66 induces multiple morphological abnormalities of the sperm flagellum and male infertility. Am J Hum Genet 103:400–412. https://doi.org/10.1016/j.ajhg.2018.07.014
Kott E, Legendre M, Copin B et al (2013) Loss-of-function mutations in RSPH1 cause primary ciliary dyskinesia with central-complex and radial-spoke defects. Am J Hum Genet 93:561–570. https://doi.org/10.1016/j.ajhg.2013.07.013
Krausz C, Riera-Escamilla A (2018) Genetics of male infertility. Nat Rev Urol 15:369–384. https://doi.org/10.1038/s41585-018-0003-3
Lindemann CB, Lesich KA (2016) Functional anatomy of the mammalian sperm flagellum. Cytoskeleton (hoboken) 73:652–669. https://doi.org/10.1002/cm.21338
Liu C, He X, Liu W et al (2019a) Bi-allelic mutations in TTC29 cause male subfertility with asthenoteratospermia in humans and mice. Am J Hum Genet 105:1168–1181. https://doi.org/10.1016/j.ajhg.2019.10.010
Liu W, He X, Yang S et al (2019b) Bi-allelic mutations in TTC21A induce asthenoteratospermia in humans and mice. Am J Hum Genet 104:738–748. https://doi.org/10.1016/j.ajhg.2019.02.020
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lorès P, Kheraff Z-E, Amiri-Yekta A et al (2021) A missense mutation in IFT74, encoding for an essential component for intraflagellar transport of Tubulin, causes asthenozoospermia and male infertility without clinical signs of Bardet-Biedl syndrome. Hum Genet. https://doi.org/10.1007/s00439-021-02270-7
Martinez G, Beurois J, Dacheux D et al (2020) Biallelic variants in MAATS1 encoding CFAP91, a calmodulin-associated and spoke-associated complex protein, cause severe astheno-teratozoospermia and male infertility. J Med Genet 57:708–716. https://doi.org/10.1136/jmedgenet-2019-106775
McLaren W, Gil L, Hunt SE et al (2016) The ensembl variant effect predictor. Genome Biol 17:122. https://doi.org/10.1186/s13059-016-0974-4
Morimoto Y, Yoshida S, Kinoshita A et al (2019) Nonsense mutation in CFAP43 causes normal-pressure hydrocephalus with ciliary abnormalities. Neurology 92:e2364–e2374. https://doi.org/10.1212/WNL.0000000000007505
Rachev E, Schuster-Gossler K, Fuhl F et al (2020) CFAP43 modulates ciliary beating in mouse and Xenopus. Dev Biol 459:109–125. https://doi.org/10.1016/j.ydbio.2019.12.010
Ron-El R, Liu J, Nagy Z et al (1995) Intracytoplasmic sperm injection in the mouse. Hum Reprod 10:2831–2834. https://doi.org/10.1093/oxfordjournals.humrep.a135802
Touré A, Martinez G, Kherraf Z-E et al (2021) The genetic architecture of morphological abnormalities of the sperm tail. Hum Genet 140:21–42. https://doi.org/10.1007/s00439-020-02113-x
Urbanska P, Song K, Joachimiak E et al (2015) The CSC proteins FAP61 and FAP251 build the basal substructures of radial spoke 3 in cilia. Mol Biol Cell 26:1463–1475. https://doi.org/10.1091/mbc.E14-11-1545
Vasudevan KK, Song K, Alford LM et al (2015) FAP206 is a microtubule-docking adapter for ciliary radial spoke 2 and dynein c. Mol Biol Cell 26:696–710. https://doi.org/10.1091/mbc.E14-11-1506
Wambergue C, Zouari R, Fourati Ben Mustapha S et al (2016) Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum Reprod 31:1164–1172. https://doi.org/10.1093/humrep/dew083
Wang G, Guo Y, Zhou T et al (2013) In-depth proteomic analysis of the human sperm reveals complex protein compositions. J Proteomics 79:114–122. https://doi.org/10.1016/j.jprot.2012.12.008
Wang Y, Yang J, Jia Y et al (2014) Variability in the morphologic assessment of human sperm: use of the strict criteria recommended by the World Health Organization in 2010. Fertil Steril. https://doi.org/10.1016/j.fertnstert.2013.12.047
Yang H, Wang H, Jaenisch R (2014) Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc 9:1956–1968. https://doi.org/10.1038/nprot.2014.134
Acknowledgements
We thank the patient for their participation in this study as well as all the referring physicians.
Funding
This work was mainly supported by the following grants by the Agence Nationale de la Recherche (ANR) MASFLAGELLA (ANR-14-CE15-0002) and FLAGEL-OME (ANR-19-CE17-0014). This work was also supported by the National Key R&D Program of China [Grant number 2019YFC1005106]; Supported by the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2019PT310002; the National Natural Science Foundation of China [Grant numbers 81901541 and 81971441].
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. CCo, YXC, PFR, FZ, XJH and XA designed and supervised the study. AA-Y, VS recruited the patients, performed clinical analysis and characterization. NTM, ZEK, PFR, TC and CCo performed exome and bioinformatics analysis. GM, JB, CCa, MB and TC performed patient’s experimental work. HBL and QSS provided Cfap206 knockout mice. BS, MRL and QSS performed animal’s experimental work. QSS and DL performed ICSI of mice. GM, CCA, KKL, FBT and XJH performed data illustrations. XA, FZ, PFR, AT, MW, CA, HW and CCo analyzed the data. XA, GM, QSS and CCo wrote the manuscript. The first draft of the manuscript was written by XA, GM and CCo, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethical approval
The study was approved by local ethics committees, and samples were then stored in the CRB Germethèque (certification under ISO-9001 and NF-S 96-900) following a standardized procedure or were part of the Fertithèque collection declared to the French Ministry of health (DC-2015-2580) and the French Data Protection Authority (DR-2016-392).
Animal research
All animal procedures were run according to the Chinese guidelines on the use of animals in scientific investigations with the approval of the animal Ethics Committee at the First Affiliated Hospital of Anhui Medical University (P2020-12-36).
Consent to participate
Informed consent was obtained from all individuals included in the study.
Consent to publish
Agreement for data publication was obtained from all individuals included in the study.
Web resources
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/
gnomAD Browser, http://gnomad.broadinstitute.org
GTEx, https://gtexportal.org
Uniprot, https://www.uniprot.org/
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shen, Q., Martinez, G., Liu, H. et al. Bi-allelic truncating variants in CFAP206 cause male infertility in human and mouse. Hum Genet 140, 1367–1377 (2021). https://doi.org/10.1007/s00439-021-02313-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-021-02313-z