Abstract
Although majority of the genes linked to early-onset cataract exhibit lens fiber cell-enriched expression, our understanding of gene regulation in these cells is limited to function of just eight transcription factors and largely in the context of crystallins. We report on small Maf transcription factors Mafg and Mafk as regulators of several non-crystallin human cataract-associated genes in fiber cells and establish their significance to this disease. We applied a bioinformatics tool for cataract gene discovery iSyTE to identify Mafg and its co-regulators in the lens, and generated various null-allelic combinations of Mafg:Mafk mouse mutants for phenotypic and molecular analysis. By age 4 months, Mafg−/−:Mafk+/− mutants exhibit lens defects that progressively develop into cataract. High-resolution phenotypic characterization of Mafg−/−:Mafk+/− mouse lens reveals severely disorganized fiber cells, while microarray-based expression profiling identifies 97 differentially regulated genes (DRGs). Integrative analysis of Mafg−/−:Mafk+/− lens-DRGs with (1) binding motifs and genomic targets of small Mafs and their regulatory partners, (2) iSyTE lens expression data, and (3) interactions between DRGs in the String database, unravel a detailed small Maf regulatory network in the lens, several nodes of which are linked to cataract. This approach identifies 36 high-priority candidates from the original 97 DRGs. Significantly, 8/36 (22 %) DRGs are associated with cataracts in human (GSTO1, MGST1, SC4MOL, UCHL1) or mouse (Aldh3a1, Crygf, Hspb1, Pcbd1), suggesting a multifactorial etiology that includes oxidative stress and misregulation of sterol synthesis. These data identify Mafg and Mafk as new cataract-associated candidates and define their function in regulating largely non-crystallin genes linked to human cataract.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ocular lens is a transparent tissue that functions to focus light on the retina, and is essential for high-resolution vision (Lachke and Maas 2010; Bassnett et al. 2011). Loss of lens transparency results in an eye defect termed cataract, which is the leading cause of visual impairment worldwide (Shiels et al. 2010). Although cataracts can occur early in life as congenital or pediatric forms of the disease, they are commonly found in aged individuals, and their incidence is expected to rise globally with aging populations (Rao et al. 2011; Churchill and Graw 2011; Shiels and Hejtmancik 2013). Both early- and late-onset cataracts have a genetic basis as revealed by linkage analysis and twin studies (Hammond et al. 2000, 2001; Congdon et al. 2005; Shiels and Hejtmancik 2013), and several modes of inheritance are described for early-onset cataracts that can be presented as an isolated, non-syndromic phenotype or as one of several phenotypes within a syndrome (Shiels et al. 2010; Huang and He 2010; Shiels and Hejtmancik 2013). To understand the pathogenic basis of cataract formation, it is critical to first understand the regulatory events underlying the development and maintenance of lens transparency.
The lens comprises of two principal cell types, the anteriorly localized epithelial cells, which differentiate into posteriorly localized fiber cells that make up the bulk of its tissue. Fiber cells located in the cortical region up-regulate expression of genes encoding structural proteins called crystallins, and as they migrate toward the lens nucleus, undergo terminal differentiation and lose their organelles (Bassnett et al. 2011). These cellular properties are essential for lens transparency. A majority of known mutations that cause pediatric cataract have been identified in genes that exhibit highly enriched expression in fiber cells (Shiels et al. 2010). Therefore, defining the transcriptional basis of gene expression regulation in lens fibers is important for identifying new candidate genes associated with cataract and gaining insights into the etiology of this disease.
In contrast to the detailed understanding of the transcriptional circuitry that functions in early mammalian lens development and in the maintenance of anterior epithelial cells, our understanding of transcription factors that function in lens fiber cells is markedly limited (Lachke and Maas 2010; Cvekl and Ashery-Padan 2014). Besides Pax6, studies on human patients or mouse models have identified only seven other transcription factors—Prox1, Sox1, ATF4/CREB2, Pitx3, Gata3, Hsf4 and the large Maf family member, Maf (also called c-Maf)—that function to regulate gene expression in differentiating fiber cells (Donner et al. 2007; Tanaka et al. 1998; Kim et al. 1999; Kawauchi et al. 1999; Ring et al. 2000; Jamieson et al. 2002; Nishiguchi et al. 1998; Wigle et al. 1999; Bu et al. 2002; Fujimoto et al. 2004; Maeda et al. 2009; Shaham et al. 2009; Sorokina et al. 2011). Moreover, studies on their regulation of fiber cell gene expression have primarily focused on crystallin-encoding genes. Thus, there is a need to identify new transcription factors that function in lens fibers to comprehensively understand gene expression in these cells, specifically in regard to the control of non-crystallin-coding genes that are linked to human cataract.
The musculoaponeurotic fibrosarcoma (MAF) gene family encodes basic leucine zipper transcription regulators that are classified into “large” and “small” MAF subgroups (Motohashi et al. 2002; Kannan et al. 2012). The small Maf subgroup members Maff, Mafg, and Mafk are low molecular weight (18 kDa) proteins that share high homology. Similar to large Maf proteins, small Mafs exhibit a modular structure, containing a basic domain that mediates DNA binding, and a leucine zipper region (bZIP) that facilitates dimerization. Both subgroups of proteins also share an extended homology region (EHR), which is important for DNA binding. However, the major difference between these subgroups is that small Maf proteins lack the histidine/glycine repeat region and a P/S/T-rich acidic domain that is present in large Maf proteins. Thus, small Maf proteins lack domains with known transactivation function, implying that they likely function as obligate repressors when bound to target DNA as homodimers. On the other hand, their heterodimeric pairing with trans-activator proteins allows them to function as activators. Studies on mouse mutants carrying different small Maf allelic combinations have revealed that these proteins are critical regulators of various cellular processes including hematopoiesis, neuronal homeostasis, and stress signaling (Onodera et al. 2000; Katsuoka et al. 2003; Motohashi et al. 2004; Yamazaki et al. 2012). However, while mutations in the large Maf family gene MAF (c-MAF) are associated with human juvenile cataracts (Jamieson et al. 2002; Vanita et al. 2006; Hansen et al. 2007) and other large Maf proteins [Mafa (L-Maf), Mafb, and Nrl] are implicated in vertebrate lens development (Kawauchi et al. 1999; Cvekl et al. 1994; Ogino and Yasuda 1998; Yoshida and Yasuda 2002), the function of small Maf subgroup proteins in the lens and their significance to cataract remain unaddressed.
Here, we identify a new function for the small Maf proteins Mafg and Mafk in regulation of gene expression in lens fiber cells. We find that Mafg−/−:Mafk+/− mouse mutants exhibit profound lens defects, including abnormalities in fiber cell morphology and organization, and develop cataract. Whole genome expression profiling analysis of mutant lens indicates altered expression of fiber cell-expressed genes, several of which are linked to mouse and human cataract and are associated with response to stress. Collectively, these data unravel a new small Maf transcriptional regulatory circuitry that controls fiber cell expression of several non-crystallin genes essential to maintain lens transparency.
Methods
Gene expression analysis by iSyTE and RT-qPCR
To determine the expression of Mafg, Mafk, and Maff in the lens during embryonic and postnatal stages, previously generated and publically available mouse wild-type lens microarray datasets on Affymetrix Mouse Genome 430 2.0 array chip were obtained from GEO (GSE32334, GSE47694, GSE16533, GSE31643, GSE9711) and analyzed for expression of small Mafs. Full details of the bioinformatics analysis will be published elsewhere (Kakrana and Lachke unpublished). In brief, probe-binding fluorescent intensity signals for Mafg, Mafk, and Maff were measured for lens and whole body embryonic tissue without eyes (WB) samples to determine the expression (as well as enrichment) of these genes in the lens expression as described (Lachke et al. 2012b). Expression of all three small Maf genes was further tested by reverse transcriptase quantitative PCR (RT-qPCR) for select postnatal stages.
Generation of mouse mutants
Previously generated Mafg+/−:Mafk+/− double heterozygous germ line mouse mutants (Shavit et al. 1998; Onodera et al. 2000) on a mixed background, with contributions from the 129Sv/J, C57BL/6J, and ICR strains, were housed in the animal facility at the University of Delaware. All animal experiments were conducted in accordance with the Association of Research in Vision and Ophthalmology (ARVO) statement for the use of animals in ophthalmic and vision research and protocols were approved by the Institutional Animal Care and Use Committee (IACUC). The double heterozygote mice (Mafg+/−:Mafk+/−) were cross-bred to generate various combinations of mutant allele progeny. Genotyping was performed as previously described (Onodera et al. 2000). In brief, genomic DNA was extracted from mouse tails and genotyped for wild-type and mutant alleles of Mafg and Mafk. Mafg−/−:Mafk+/− mutants, which appeared to be smaller in size and developed hind limb paralysis evident at the time of weaning, were placed in separate cages and provided with supplemental food gels. Mice containing all combinations of Mafg:Mafk mutant alleles were examined. As previously reported, Mafg−/−:Mafk+/− mutant animals were not born in the expected Mendelian ratios, and were incapable of reproducing with each other. Mutant mice were physically evaluated for the presence of lens defects and cataracts, euthanized, and processed as required. Because these mice were of mixed background with contribution from 129Sv/J that is known to carry mutant CP49 locus, they were genotyped for CP49 as previously described (Alizadeh et al. 2004), and only animals that did not carry the mutant copy of the gene were used for further analysis.
Dark field microscopy, grid imaging and histology
Eyes were dissected from mutant and control animals and carefully cleaned in 1× PBS solution. After the eyes were imaged on a light microscope (Zeiss Stemi SV dissecting microscope), the lens was extracted, cleaned, placed in media 199 (1×, with Earle’s salts and l-glutamine) (Cellgro Mediatech, Inc., Manassas, VA, USA) pre-warmed to 37 °C, and imaged immediately against a 200-mesh electron microscopy grid (Electron Microscopy Sciences, Hatfield, PA, USA; Catalog G300H-Cu) to observe the refractory property of the tissue as described (Shiels et al. 2007). For hematoxylin and eosin (H&E) staining, whole eyes (postnatal mice) or heads (embryos) were collected from control and mutants at appropriate stages. Samples were fixed overnight in Pen-Fix (Richard Allan Scientific, Kalamazoo, MI, USA), dehydrated with ethanol, and paraffin-embedded for microtome sectioning. Serial sagittal paraffin sections (5 µm) were H&E stained according a standard protocol and visualized using light microscopy (Zeiss Axiophot) and a Nikon digital camera.
Scanning electron microscopy
Scanning electron microscopy (SEM) was performed on 2- to 7-month-old Mafg+/+:Mafk+/+ and Mafg+/−:Mafk+/− controls and Mafg−/−:Mafk+/−, Mafg+/−:Mafk−/− mutant lenses as previously described (Scheiblin et al. 2014). In brief, eyes were enucleated from mice and lenses were removed and transferred to a fixative (0.08 M sodium cacodylate pH 7.4, 1.25 % glutaraldehyde, 1 % paraformaldehyde) (Electron Microscopy Sciences) for 48 h. After fixation, lenses were washed in 1× PBS and the lens capsule and initial fiber cell layers were removed to view cortical fiber cells (~150–450 µm from the capsule). Then, lenses were dehydrated through an alcohol dilution series. After overnight incubation in 100 % ethanol, lenses were washed in ethanol two times for 2.5 h each. Lenses were then dried in 1:2 hexamethyldisilazane (HMDS, Sigma, St. Louis, MO, USA)/ethanol for 1 h followed by 2:1 ratio of ethanol to HDMS for 1 h, and 100 % HMDS for 30 min. Lenses were sputter coated for 2.5 min with gold/palladium and imaged using Hitachi S-4700 field emission scanning electron microscope (Tokyo, Japan). The analysis was performed on three biological replicates.
In situ hybridization
Mouse embryonic head tissues at E12.5 and E14.5 were dissected and fixed overnight in 4 % PFA, dehydrated, and embedded in tissue freezing media, OCT (Tissue Tek, Torrance, CA, USA) and coronal frozen sections were prepared at 16 µm thickness. RNA probes for in situ hybridization analysis were generated as described using primers that incorporated the T7 or SP6 promoter sequences upstream of Mafg cDNA sequence (Lachke et al. 2012b). The amplified cDNA was purified and used as a template for in vitro transcription to prepare sense and antisense digoxigenin-labeled RNA probes. In situ hybridization was performed as described, and imaged using a light microscope (Zeiss Axiophot) with a Nikon digital camera (Lachke et al. 2012b).
Immunostaining
Mouse embryonic head tissues at E14.5 and E16.5 or eyes from adults were embedded in tissue freezing media, OCT (Tissue Tek, Torrance California) and frozen sections were prepared at 16 µm thickness (Reed et al. 2001). Sections were fixed with 4 % paraformaldehyde in 1× PBS for 20 min at room temperature followed by two 1× PBS washes. Then they were blocked for 1 h at room temperature in 5 % chicken serum, 0.1 % tween and 1 % BSA in 1× PBS (for Maff/g/k and Foxe3 antibody) or in 5 % chicken serum, 0.3 % triton and 1 % BSA in 1× PBS (for E-cadherin and gamma-crystallin antibody). The tissue was then covered with either of the following primary antibodies: Maff/g/k (SC-22831, Santa Cruz Biotech, Santa Cruz, CA, USA), Foxe3 (SC-134536, Santa Cruz Biotech, Santa Cruz, CA, USA), E-cadherin (4065, Cell signaling) and gamma-crystallin (SC-22415, Santa Cruz Biotech, Santa Cruz, CA, USA; detects γA-, γB-, γC-, γD-, γE- and γF-crystallins, and to a lower extent, γS-crystallin) at 1:100 for overnight at 4 °C. Slides were washed three times in 1× PBS and incubated for 1 h at room temperature with chicken anti-rabbit IgG conjugated to Alexa Fluor 594 secondary antibody (Life Technologies, Carlsbad, CA, USA) at 1:200 dilution mixed with 1:2000 dilution of DRAQ5 (Biostatus Limited, Leicestershire, United Kingdom). Slides were washed again three times with 1× PBS and slides were mounted with cover slip, and stored at −20 °C until imaged. Samples were imaged using Zeiss LSM 780 confocal configured with argon/krypton laser (488 and 561 nm excitation lines) and helium/neon laser (633 nm excitation line) (Carl Zeiss Inc., Göttingen, Germany). Optimal adjustment of brightness/contract was performed in Adobe Photoshop and applied consistently for all images.
Microarray analysis
Mafg+/−:Mafk+/− (control) and Mafg−/−:Mafk+/− (test) mouse mutant lenses at 2 month of age were collected in biological replicates for gene expression profiling by microarrays. Lens total RNA was isolated using the RNeasy Mini Kit (Qiagen). BeadChip MouseWG-6 v2.0 expression arrays (Illumina) were used to perform microarrays using standard procedures for hybridization and scanning by the Illumina BeadArray reader. Analysis of microarray datasets was performed under ‘R’ statistical environment (http://www.r-project.org/). Raw files were imported and background corrected using lumi package (Du et al. 2008) available at Bioconductor (www.bioconductor.org), followed by normalization using rank-invariant method. Present-absent calls were generated using lumi inbuilt function. Probe sets present with detection p value ≤0.05 in at least two samples were considered significantly present and were used to reduce probe-level to the gene-level experiment by selecting the probe with highest median expression for a gene. Differentially regulated genes (DRGs) were identified in Mafg−/−:Mafk+/− mutant lens samples at a fold change (FC) cut-off of ±1.5 for up-regulation and down-regulation. Initial analysis to gain biological significance was performed using a bioinformatics tool, database for annotation, visualization and integrated discovery (DAVID), which provides functional interpretation of genes (Huang et al. 2009). Up-regulated (n = 42) and down-regulated (n = 55) DRGs were separately analyzed in DAVID. All the microarray data reported in this article have been deposited at NCBI and the Gene Expression Omnibus accession number is GSE65500.
Quantitative RT-PCR
RNA from Mafg+/−:Mafk+/− control and Mafg−/−:Mafk+/− mutants was isolated as described above and was used to synthesize cDNA using the RT-qPCR Primer Assay (SABiosciences, Valencia, CA, USA) according to the manufacturer’s instructions. All samples were prepared in a 96-well reaction plate with three biological replicates (independent samples from those used for microarray analysis) and at least two technical triplicates. RT-qPCR was performed on the ABI 7500 fast real-time PCR system and Software v2.0.3 (Applied Biosystems, Carlsbad, CA, USA). Mean FC was calculated using log (base 10)-transformed data in a nested ANOVA by determining the mean and standard deviations. These values were then back transformed to obtain the final FC values.
Integrated analysis of Mafg−/−:Mafk+/−DRGs
To determine biological significance of the Mafg−/−:Mafk+/− lens-DRGs, we took the following approaches. Previous reports suggest that small Mafs (Mafg, Mafk and Maff) heterodimerize with cap and collar (CNC) family transcription factors and recognize specific cis-DNA-binding motifs (Toki et al. 1997; Kataoka 2007; Li et al. 2008). Therefore, we first examined the expression of small Maf-binding partners and co-regulatory molecules in lens microarray datasets and iSyTE. We next examined DRGs that overlap with candidates that are recognized by small Maf or their co-regulatory proteins in vivo by chromatin immune-precipitation (ChIP) assays, albeit in non-lens cells. Studies considered for this step of the analysis were: (1) Nrf2-binding regions identified in human lymphoblastoid cells (Chorley et al. 2012), (2) Nrf2-and Keap1-binding regions identified in mouse embryonic fibroblasts (Malhotra et al. 2010), and (3) Nrf2–Mafg-binding regions identified in hepatoma cell lines (Hirotsu et al. 2012). Next, we used our recently developed bioinformatics tool iSyTE (Lachke et al. 2012b) to analyze the expression of up-regulated and down-regulated DRGs to identify genes that are expressed in the lens. DRGs that were lens enriched or lens expressed, as well as genes that had biological relevance based on the literature were further analyzed to gain insights into their role in lens biology. Finally, the DRGs were also analyzed for the presence of DNA-binding motifs recognized by small Mafs and their co-regulatory proteins. Specifically, the ARE core, NF-E2, and MARE motifs were identified in the 2.5 kb upstream region of DRGs as well as in the genomic regions identified in the above ChIP studies through sequence matching of the position weight matrices (PWMs) by implementing MotifDb R package (version 1.6.0). Of the 528 PWMs for mouse transcription factors, 113 were from JASPAR database (Portales-Casamar et al. 2010), 133 were from the Jolma and coworkers study (Jolma et al. 2013), and 282 were from UniPROBE (Newburger and Bulyk 2009; Robasky and Bulyk 2011). Using the matchPWM algorithm (Wasserman and Sandelin 2004), overrepresented motifs were identified at a significant score threshold of 80–95 %.
Derivation of the small Maf regulatory network
To extend the above analysis by deriving further insights into the relationships of the identified DRGs in Mafg−/−:Mafk+/− lens with their known interactors, we sought to identify and represent molecular interactions in the form of a regulatory network. An in-house Python script was applied to fetch out statistically significant interactions between the DRGs from the String database (http://string-db.org). Next, we overlaid lens expression and enrichment information from iSyTE along with the small Maf-binding motif information identified for DRGs. The resultant small Maf interaction network file was visualized using an open-source tool, Cytoscape.
Results
iSyTE identifies small Maf transcription factors Mafg and Mafk in the lens
We recently developed a novel system-based strategy termed integrated Systems Tool for Eye gene discovery (iSyTE) that has been successful in identifying genes associated with cataract (Lachke et al. 2012b). Here, we utilized iSyTE to identify new regulators in the lens. Specifically, we analyzed the human genome using iSyTE tracks for genes encoding transcription factors and identified the small Maf family member MAFG with an exceptional lens enrichment score strongly indicating it as a promising candidate associated with lens biology and cataract (Fig. 1a). Candidate genes are considered to have an exceptional lens enrichment score if they are placed among the top 1 % of all lens-enriched genes as determined by a t-statistic-based comparative analysis between the E10.5, E11.5 and E12.5 lens microarray datasets and the whole body embryonic tissue (WB) microarray dataset.
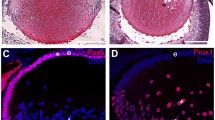
Expression of small Maf transcription factors in the mouse lens. a iSyTE identifies MAFG as a highly lens-enriched gene, among the top 1 % of lens-enriched genes. iSyTE is based on microarray expression datasets of genes that are scored for their differential regulation in the lens when compared to a reference dataset of whole body embryonic tissue (WB) allowing for t-statistic-based estimation of “lens enrichment”. Based on the t-statistic values, lens-enriched genes can be viewed through user-friendly “iSyTE” tracks in the UCSC Genome browser to aid prioritization of genes with potential lens function. Genes with high lens enrichment are represented by intense red color while genes that are not lens enriched are represented by intense blue color. b Analysis of lens microarrays from mouse embryonic and postnatal stages indicates that while Maff is largely absent and Mafk is expressed at low levels, Mafg exhibits highly enriched expression in the embryonic and early postnatal mouse lens. Probe-binding fluorescent signal intensity values for all three small Maf genes, which are reflective of their expression, are plotted on the Y-axis for different lens stages and the WB reference dataset described in iSyTE. c Real-time quantitative RT-PCR confirms that Mafg and Mafk, but not Maff are expressed in postnatal mouse lens, and Mafg expression, although always lens enriched, is progressively reduced in postnatal development. d In situ hybridization demonstrates the presence of Mafg transcripts in transition zone (tz) cells and fiber cells (f) but not in epithelium (e) of E12.5 mouse lens. d′ High-magnification image of area indicated by dotted box in d. Mafg expression is indicated by white asterisks. e At E14.5, in situ hybridization demonstrates the continued presence of Mafg transcripts in mouse lens transition zone (tz) cells and fiber cells (f) but not in epithelium (e). e′ High-magnification image of area indicated by dotted box in e, in which white asterisk indicates Mafg expression. f Immunostaining with antibody that recognizes Mafg, Mafk, Maff demonstrates the presence of small Maf proteins (sMaf) in the nuclei and cytoplasm of transition zone (tz) cells and fiber cells (f) but not in epithelium (e) of E14.5 mouse lens. f′ High-magnification image of area indicated by dotted box in f. sMaf expression is indicated by white arrowheads. g At E16.5, the above antibody demonstrates the continued presence of small Maf proteins in the nuclei and cytoplasm of mouse lens transition zone (tz) cells and fiber cells (f) but not in epithelium (e). g′ High-magnification image of area indicated by dotted box in g, in which white arrowheads indicate the expression of sMaf proteins. Statistical significance in b, c, is as follows: one asterisk indicates p value of 0.05, two asterisks indicate p value of 0.005, three asterisks indicate p value of 0.001. Scale bar in f is 50 μm, f′ is 15 μm; g is 100 μm and g′ is 15 μm
Expression of Mafg and Mafk in the lens
Because redundancy between the three small Maf proteins (Maff, Mafg, Mafk) is known in context of other tissues (Onodera et al. 2000; Katsuoka et al. 2003; Motohashi et al. 2004), we first sought to investigate the expression of all three genes in the mouse lens. We analyzed previously generated, publically available Affymetrix-based microarray gene expression profiles of the mouse lens, as it transitions from the stage of placode invagination to adult, and evaluated the expression of small Maf genes in the lens at these different stages and in the WB reference dataset described in iSyTE. Probe-binding fluorescence signal intensity values for Maff, Mafg, and Mafk were compared in each of these datasets. This analysis indicated that while expression of Mafk and Maff is low or absent, respectively, Mafg is highly enriched in the lens (Fig. 1b). Interestingly, as the lens develops postnatally, Mafg lens expression and enrichment—although remain highest among the three small Mafs at all stages tested—are progressively reduced, while Mafk expression is unaltered and Maff expression remains undetected (Fig. 1b). To validate the differences in expression in individual small Maf members as indicated by microarray analysis, we performed qRT-PCR on different postnatal mouse lens stages and confirmed that while Mafg is enriched and Mafk is expressed at lower levels, Maff is undetectable in the mouse lens at all stages (Fig. 1c). These analyses also confirmed that Mafg expression is progressively down-regulated in postnatal lens (Fig. 1c).
We next performed in situ RNA hybridization at stages E12.5 and E14.5 with Mafg-specific probe and confirmed embryonic lens-enriched expression of Mafg (Fig. 1d–e′). Moreover, this analysis indicated that Mafg transcripts are highly expressed in differentiating lens fiber cells at E12.5 and get progressively restricted to the fiber cells of the equatorial zone at E14.5. This finding is supported by an earlier study that suggests Mafg to be expressed in the lens at E14.5, based on lacZ reporter assay (Shavit et al. 1998). Immunostaining experiments using an antibody that recognizes all three small Mafs confirmed that proteins in this family exhibit highly enriched expression in the embryonic lens at E14.5 and E16.5, specifically in fiber cells (Fig. 1f–g′). Moreover, their presence is detected in both cytoplasm and the nuclei of fiber cells. Together, these data offer support for further investigation of Mafg and Mafk in the lens.
Mafg−/−:Mafk+/− mutants exhibit cataracts
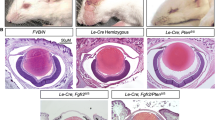
Integrated systems tool for eye gene discovery analysis and expression data suggest that Mafg and Mafk may potentially function in the lens. To test this prediction, we chose to focus on analyzing mouse mutants for these two small Maf genes expressed in the lens. Given the functional redundancy between small Maf proteins in other organs (Yamazaki et al. 2012; Kannan et al. 2012), we analyzed various combinations of Mafg and Mafk mutant alleles. From the different combinations tested, we find that beginning at 4-month age Mafg−/−:Mafk+/− mutants exhibit lens defects including lens opacities that are visible by gross examination of live animals (Fig. 2a). Although the onset is variable, the lens defect phenotype progresses with age and is eventually fully penetrant in Mafg−/−:Mafk+/− animals (Fig. 2b). In contrast, no opacity is visible in Mafg+/+:Mafk+/+ wild-type (WT) and Mafg+/−:Mafk+/− heterozygous animals used as controls in this study, as well as in Mafg+/−:Mafk−/−, or Mafg−/−or Mafk−/− individual mutants. This indicates that Mafg and Mafk have overlapping functions in the maintenance of lens transparency.
Investigation of lens defects in Mafg:Mafk mouse mutants. a Imaging of various Mafg:Mafk mutants at age 4 months revealed that Mafg−/−:Mafk+/− mice exhibit distinct lens opacity (indicated by arrowhead). Dark field imaging of dissected eyes from Mafg−/−:Mafk+/− mice demonstrates the presence of an overt cataract phenotype (back asterisk). Bright field imaging of dissected lens on metal grid indicates a complete lack of visibility of underlying hexagonal patterns in turn demonstrating the severe nature of lens abnormality in Mafg−/−:Mafk+/− mice. Comparative analysis was performed with mouse mutants including Mafg+/+:Mafk+/+, Mafg+/−:Mafk+/−, Mafg+/−:Mafk−/−, and Mafg−/−:Mafk+/+, all of which lacked lens defects. Mafg+/−:Mafk+/+, Mafg+/+:Mafk+/− and Mafg+/+:Mafk−/− mutants were also tested and lacked lens defects (data not shown). b Progression of lens defects in Mafg−/−:Mafk+/− mouse mutants. Lens defects were analyzed in ages 2 through 8 months Mafg−/−:Mafk+/− mouse mutants (represented by closed circles) and Mafg+/−:Mafk+/− controls (represented by open circles). Lenses were scored as clear, hazy, or opaque as indicated by the images on right. All Mafg+/−:Mafk+/− control double mutants exhibited normal eye and lens through all stages tested. At age 3 months, hazy eyes were observed in Mafg−/−:Mafk+/− mutants, while from age 4 months onwards, lenses with severe opacity and cataract were detected. By age 8 months, all Mafg−/−:Mafk+/− mutants tested exhibit severe lens opacities. Scale bar represents 1 mm
Mafg−/−:Mafk+/− mutants exhibit severe lens fiber cell defects
To characterize the morphology of fiber cell defects in 4-month-old Mafg−/−:Mafk+/− mutant mice exhibiting cataracts, we performed histological analysis of their eye tissue and compared these to Mafg+/+:Mafk+/+ WT, Mafg+/−:Mafk+/− double heterozygous, and Mafg+/−:Mafk−/− mutant mice. As expected, both the WT and the Mafg+/−:Mafk+/− animals did not exhibit any lens defects (Fig. 3). Interestingly, fiber cell organization appears normal and Mafg+/−:Mafk−/− mutant mice did not exhibit any observable lens phenotype. However, in Mafg−/−:Mafk+/− mutants, the fiber cell region exhibits large vacuoles in the periphery as well as in the lens nucleus, and a posterior capsular rupture may also be evident (~42 % penetrance) (Fig. 3).
Mafg−/−:Mafk+/− mouse mutant exhibits fiber cell defects. Histological analysis using hematoxylin and eosin staining was performed on eye sections from various Mafg:Mafk mutants at age 4 months. Severely defective Mafg−/−:Mafk+/− mutant lens exhibits large cortical vacuoles (arrowheads) in the fiber cell compartment, while eyes and lens of Mafg+/+:Mafk+/+, Mafg+/−:Mafk+/−, and Mafg+/−:Mafk−/− appear normal. High-resolution scanning electron microscopy of severely affected Mafg−/−:Mafk+/− mutant mouse lens shows disorganization of fiber cell packing, lack of membrane protrusions, and overall severe disruption of the cortical fibers. In contrast, cortical fiber cells of Mafg+/+:Mafk+/+, Mafg+/−:Mafk+/−, and Mafg+/−:Mafk−/− mouse lens at age 4 months appear normal. For both analyses, high-magnification images of specific areas are indicated by dotted box in lower panels. Histology scare bars: top panel, 100 µm; bottom panel, scale bar, 50 µm. Scanning electron microscopy scale bars: top panel 10 µm; bottom panel 5 µm
Next, we performed a high-resolution analysis of lens cortical fiber cells in these animals by scanning electron microscopy (SEM). This analysis reveals that 4-month-old Mafg−/−:Mafk+/− mutant lens fiber cells are severely disrupted exhibiting abnormal membrane protrusions compared to the controls (Fig. 3). These severe fiber cell defects were observed in Mafg−/−:Mafk+/− mutants regardless of whether the lenses had capsule rupture. Similar to the histological analysis, SEM revealed no significant differences between control and Mafg+/−:Mafk−/− mutant fiber cells. Together, these findings indicate that Mafg homozygous null in Mafk heterozygous background, but not the converse, leads to lens fiber cell defects, in turn suggesting that Mafg has an important function in the lens that may be partially compensated by Mafk.
Mafg−/−:Mafk−/− double knockout mouse mutants exhibit lens defects
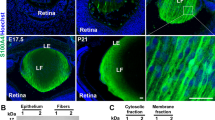
Although Mafg−/−:Mafk−/− double null mutants are perinatal lethal and could not be analyzed for late-onset cataract, we sought to evaluate if they exhibited lens defects. Therefore, we performed histological analysis of E16.5 Mafg−/−:Mafk−/− double null mutants and controls. We find that the Mafg−/−:Mafk−/− double null mutants exhibit an abnormality in newly differentiating fiber cells that suggest a defect in the lens fulcrum, although fiber elongation is not affected (Fig. 4). The lens fulcrum is the region near the lens equator where the epithelial cells, before beginning differentiation into elongating fiber cells, form an anchor point (Cheng et al. 2013). Histological analysis indicates no presence of a “focused” fulcrum and as a consequence the apical ends of newly differentiated fiber cells do not appear to form proper contact with epithelium. These data suggest that in addition to their role in the postnatal lens, Mafg and Mafk function in embryonic lens development.
Mafg−/−:Mafk−/− double knockout mouse mutants exhibit defects in embryonic lens development. Histological analysis using hematoxylin and eosin staining was performed on embryonic head sections from E16.5 Mafg−/−:Mafk−/− mutant or Mafg+/−:Mafk+/− control mice. The white broken line box in the image on left indicates area that is shown at high magnification on the right. While the lens appears normal in control, Mafg−/−:Mafk−/− mutant lens exhibits abnormalities (indicated by white arrowhead) near the lens fulcrum and beyond the transition zone (tz) where cells of the epithelium (e) exit the cell cycle and begin differentiating into fiber cells (f)
Microarray-based gene expression profiling of Mafg−/−:Mafk+/− lens
Examination of adult Mafg−/−:Mafk+/− mutant lens for markers of epithelium or fiber cells did not reveal any changes in the expression of Foxe3, E-cadherin or gamma-crystallin (Fig. S1). To gain a global perspective of gene expression changes in the Mafg−/−:Mafk+/− mutant lens, we performed microarray-based gene expression analysis on lenses at a postnatal stage prior to the onset of overt lens defects. This approach increases the likelihood for detecting primary gene expression alterations, while lowering the number of late-onset secondary changes. At age 2 months, Mafg−/−:Mafk+/− mutant lenses exhibited no opacities and therefore this stage was selected for gene expression profiling analysis. Microarray analysis identified 97 genes that are differentially expressed at ±1.5-FC at p values ≤0.05 in the Mafg−/−:Mafk+/− mutant lens (Fig. 5a, b) compared to control (Mafg+/−:Mafk+/−) lens. While these candidates include several non-crystallin-encoding genes linked to cataract, strikingly there is only one crystallin-encoding gene (Crygf) that is misregulated in the Mafg−/−:Mafk+/− mutant lens (Table S1). These candidates will be discussed in greater detail in later sections.
Mafg−/−:Mafk+/− mouse mutants exhibit defects in lens gene expression. a Analysis of genes that are up-regulated in Mafg−/−:Mafk+/− lens. Column on left (mutant vs. control) is a heatmap that is indicative of genes up-regulated in Mafg−/−:Mafk+/− lens compared to control (Mafg+/−:Mafk+/−) lens. Increased expression in fold change is indicated by intensity of red color. Column on right (lens enrichment) is a heatmap that is indicative of lens enrichment of each candidate gene as per the iSyTE approach. Increased lens enrichment in fold change is indicated by intensity of red color. Decreased lens enrichment in fold change is indicated by intensity of green color. b Analysis of genes that are down-regulated in Mafg−/−:Mafk+/− lens. Column on left (mutant vs. control) is a heatmap that is indicative of genes down-regulated in Mafg−/−:Mafk+/− lens compared to control lens. Increased expression in fold change is indicated by intensity of green color. Column on right (lens enrichment) is a heatmap that is indicative lens enrichment of each candidate gene as per the iSyTE approach as described above. c Genes down-regulated in Mafg−/−:Mafk+/− mutants are identified as significantly lens enriched by iSyTE. Candidate genes from microarray analysis are plotted on the X-axis based on their differential regulation (up-regulated genes are represented by triangles; down-regulated genes are represented by circles) in Mafg−/−:Mafk+/− mutant lens and on the Y-axis based on their lens enrichment (lens enrichment represented by red intensity; non-enrichment in lens represented by green intensity) as per the iSyTE approach. While 46 of the 55 down-regulated genes in mutant lens are lens enriched, only 15 of 42 up-regulated genes are identified as such. Chi-square calculated for differences between lens-enriched and non-enriched genes between these datasets equals 425.92 at two-tailed p value less than 0.0001, and therefore is statistically significant. d Real-time quantitative RT-PCR analysis validates the differential regulation in Mafg−/−:Mafk+/− mutant lens of select candidate genes identified by microarray analysis. Fold change over Mafg+/−:Mafk+/− control lens is indicated on Y-axis. Statistical significance is indicated by asterisk as p value of <0.05
We next sought to investigate whether the differentially regulated genes (DRGs) are also lens enriched according to the iSyTE approach, which would help to identify and prioritize Mafg/k targets as potential new candidates important to lens biology. Comparative analysis of the Mafg−/−:Mafk+/− lens microarray datasets with whole embryonic body (WB) microarray data as per the iSyTE approach (Lachke et al. 2012b) reveals that only 36 % (n = 42) of up-regulated genes are lens enriched, while 84 % (n = 55) of down-regulated genes are lens enriched (Fig. 5c). Furthermore, the difference between lens-enriched and non-enriched down-regulated genes in Mafg−/−:Mafk+/− lens is statistically highly significant, compared to the difference between lens-enriched and non-enriched up-regulated genes (Fig. 5c). These data indicate that genes preferentially expressed in the lens are specifically down-regulated in Mafg−/−:Mafk+/− mutants, indicative of their potential importance in the tissue.
Validation and gene ontology investigation of Mafg−/−:Mafk+/− DRGs
To validate the microarray data, we tested the misregulation of select up- and down-regulated DRGs in the Mafg−/−:Mafk+/− lens by performing RT-qPCR. This analysis confirmed the microarray-predicted up-regulation of several genes, namely Dctn4, Ddit3 and Hmox1, as well as down-regulation of genes Lims2, Aldh3a1, and Hspb1 (Fig. 5d).
Next, to test if the DRGs are enriched for candidates that function in specific pathways or cellular events, we performed traditional cluster analysis following functional enrichment analysis with the bioinformatics tool DAVID. This analysis led to clustering of several candidate genes into specific categories for DRGs, among which is “oxidative stress induced gene expression via Nrf2” (a known heterodimeric partner of small Maf proteins) (Fig. S2, Table S2). While these were interesting findings, we noted that for many of these clusters, less than five genes were identified, and therefore, were unsure of their full significance toward interpretation of the lens phenotype. Thus, we sought to take a more integrated bioinformatics approach (below) toward understanding the function of Mafg and Mafk in the lens.
Integrated analysis to derive a small Maf regulatory network in the lens
To extract biological meaning from the identified DRGs, select promising candidates, and gain insight into the molecular circuitry controlled by small Maf proteins in the lens, we applied the following in silico analysis-based integrated approach. First we sought to identify the small Maf-binding partners and co-regulatory molecules that are of relevance to lens biology. Therefore, we tested the expression of genes that encode well-established small Maf heterodimeric partner proteins (Motohashi et al. 2004; Kannan et al. 2012) by analyzing iSyTE and other lens microarray datasets available in the GEO database. While Bach1, Nfe2l3 (Nrf3) and Nfe2 exhibit low expression or are absent in the lens at all stages examined, Nfe2l1 (Nrf1), Nfe2l2 (Nrf2) and Bach2 are enriched in the lens, indicating their candidacy as potential co-regulators of small Maf proteins in the lens (Fig. S3).
Next, we investigated the known genomic targets of small Mafs and their partner proteins. Interestingly, previous studies have described in vivo DNA-binding cis-regions of Mafg as well as Nrf2 based on chromatin immunoprecipitation-sequencing (ChIP-seq) experiments, albeit in the context of different cell types (Hirotsu et al. 2012; Chorley et al. 2012; Malhotra et al. 2010). Nrf2 represents a good co-regulatory candidate in the lens, because in addition to its expression in the lens tissue, the GO analysis of DRGs suggests its involvement in small Maf function in the lens (Fig. S2). We investigated if these known direct targets overlapped with lens-DRGs. Therefore, we first compared candidates from published ChIP-seq experiments on Nrf2 and Mafg with DRGs identified in Mafg−/−:Mafk+/− mouse lens. We analyzed the genomic regions of these candidate genes for the presence of DNA-binding motifs that have been described for small Maf proteins and their binding partners, including the antioxidant response element (ARE) motif, the Nrf-binding motif, and the MARE (Maf recognition element) motif (Fig. S4). We then examined if these candidates were of relevance to lens biology according to iSyTE. Further, a comprehensive literature-based analysis to identify function in the lens was considered for each candidate. Based on this strategy, from the initial list of 97 DRGs, we could identify 36 high promising candidate genes that were found to be direct targets of Mafg or Nrf2 in vivo and were either expressed in the lens and/or had known biological role in the eye, or contained a small Maf-binding motif (Table 1). These include several genes associated with lens defects and cataract (Table 1).
We next sought to investigate the regulatory relationships between these DRGs. Therefore, to derive a regulatory network for small Maf proteins in the mouse lens, we proceeded to further integrate known interactions between Mafg−/−:Mafk+/− DRGs that are present in the String database (Franceschini et al. 2013), and analyzed the resulting network for enrichment of GO categories (Fig. 6). We draw the circuitry using Mafg as the central node since its requirement, compared to Mafk, seems to be more critical to the lens. The analysis was performed for both mouse (Fig. 6) and human (Fig. 7), and the emerging Mafg network in both is largely overlapping. Based on these various parameters, the above analysis and network representation, together serve to identify 8 genes (Gsto1, Mgst1, Sc4mol, Uchl1, Pcbd1, Aldh3a1, Crygf, Hspb1) that are linked to mammalian cataract (Figs. 6, 7; Table 1). In addition, the regulatory network serves to also identify relationships between genes and their function in common pathways, in turn enabling the prioritization of important candidates. For example, the network identifies the genes Mapk14, Ubr5, and Hspb1 that are down-regulated in Mafg−/−:Mafk+/− lens and are associated with cellular response to stress. Furthermore, it serves to identify a gene cluster associated with lipid and sterol synthesis, several members of which are excellent candidates for direct regulation by small Maf proteins in the lens due to the presence of a binding motif (Figs. 6, 7, Fig. S4; Table 1). Thus, the integrated approach leads to the identification and prioritization of several new candidate genes in the small Maf regulatory network that are associated with lens physiology, the misregulation of which results in cataract. Further, it serves to highlight the contribution of various non-crystallin-encoding genes that contribute to the lens phenotype.
Integrated analysis-derived small Maf regulatory network in the mouse lens. Based on the integration of various datasets—including differentially regulated genes (DRGs) in Mafg−/−:Mafk+/− mutant lens and their interactions in String database, in vivo cis-binding evidence for small Maf or their co-regulatory proteins in DRGs, presence of small Maf-binding motifs in DRGs, as well as lens-relevant expression in iSyTE—a model for the small Maf functional regulatory network in the lens is proposed. Since mouse genetics-based analysis indicates the importance of Mafg in the lens, the circuitry is featured around it. This regulatory network suggests that lens defects in the small Maf mutant Mafg−/−:Mafk+/− are caused by altered regulation of genes largely encoding non-crystallin proteins that function in diverse pathways critical to various aspects of lens biology. Key to nodes, edges, and color schemes is provided and more details are discussed in the study. In GO terminology, “Cataract-associated gene” represents a custom-assigned category
Integrated analysis-derived small MAF regulatory network predicted in the human lens. Similar to the analysis performed using mouse datasets, a model for the small MAF functional regulatory network in the human lens is proposed. This analysis is based on the integration of various datasets—including DRGs in Mafg−/−:Mafk+/− mutant lens and their specific interactions in the String database for human, in vivo cis-binding evidence for small MAF or their co-regulatory proteins in DRGs, presence of small MAF-binding motifs in DRGs, and expression in the lens according to the iSyTE approach. Key to nodes, edges, and color schemes is provided
Discussion
Elucidating the regulatory network that controls the formation and maintenance of lens transparency is essential for understanding the pathophysiology of cataracts (Lachke and Maas 2010). Function of the large Maf transcription factor MAF (c-Maf) in lens fiber cells and its association with inherited juvenile cataract are well established (Kim et al. 1999; Kawauchi et al. 1999; Ring et al. 2000; Jamieson et al. 2002; Vanita et al. 2006; Hansen et al. 2007). On the other hand, although investigated in several tissues and implicated in diverse diseases such as thrombocytopenia, cancer, and neuronal disorders (Onodera et al. 2000; Katsuoka et al. 2003; Motohashi et al. 2004; Yamazaki et al. 2012), the function of small Mafs in the lens or their association with cataract remains undefined.
In this study, we apply an effective bioinformatics-based cataract gene discovery tool iSyTE (Lachke et al. 2011; Kasaikina et al. 2011; Lachke et al. 2012a, b) to identify a new function for the small Maf transcriptional regulators Mafg and Mafk in regulating a network of human and mouse cataract genes in lens fiber cells. We find that Mafg−/−:Mafk+/− mutants develop fully penetrant lens defects that get severe with age and develop cataracts. In addition, analysis of Mafg−/−:Mafk−/− double knockout mutants indicates a defect in the organization of fiber cells near the lens fulcrum region, where epithelial cells anchor before beginning differentiation into fiber cells (Cheng et al. 2013). Because these germ line knockout mutants are perinatal lethal, it will be intriguing to analyze the function of these regulators in future analysis of conditional null mutants. Interestingly, although these data indicate that Mafg and Mafk may have overlapping functions, they also suggest that the requirement of Mafg is more critical in the lens. Indeed, in previous studies on double or triple mutant analyses of small Mafs, Mafg has been identified as the most critical regulator among the three genes (Onodera et al. 2000; Katsuoka et al. 2003; Motohashi et al. 2004; Yamazaki et al. 2012). Histology and scanning electron microscopy of Mafg−/−:Mafk+/− mutants demonstrate abnormalities in lens fiber cell organization, while gene expression profiling by microarrays identifies 97 DRGs in mutant lens. Genome-wide expression profiling is now increasingly applied to characterize mutant tissues on the molecular level. However, these experiments generate numerous candidates that are identified as differentially regulated in the tissue. Moreover, traditional approaches like DAVID (Fig. S2) are not always effective in prioritizing candidates. Therefore, a formidable challenge is to select for the most promising high-priority candidate genes within these datasets. To further investigate the molecular basis of the cataract, we took an integrated approach in analyzing these DRGs. Our approach of using ChIP-seq data from non-lens cells is effective because we select only those candidate target genes that are also expressed or have a described function in the lens (which we can determine because of the iSyTE tool or through literature-based analysis). Therefore, although the ChIP data are not from lens cells, we can generate specific hypotheses about TF-targets in the lens for future in-depth analysis, as well as prioritize promising candidate genes. Based on these analyses, we have derived a comprehensive gene association network—which we term as “integrated analysis-derived regulatory network”—for small Maf function in the mouse and human lens.
From the original list of 97 DRGs identified in the microarrays, the integrated approach enables the recognition of 36 high-priority candidates for further investigation. Importantly, 8 of these 36 (~22 %) high-priority candidates have already been associated with human or mouse cataracts, indicating the effectiveness of the approach. However, the regulation of these 8 cataract genes had never been addressed, and the findings presented here indicate them to be either direct or indirect targets of Mafg and Mafk in the lens. Furthermore, because only 1 of the 97 DRGs encodes a crystallin protein (Crygf), this work provides a critical advance on regulation of non-crystallin genes associated with cataract—an under-addressed area in lens research. Below, we discuss several high-priority genes and their significance to the manifestation of the lens phenotype in Mafg−/−:Mafk+/− mutants.
Mgst1, a DRG identified in the Mafg−/−:Mafk+/− mutant lens, encodes microsomal glutathione S-transferase that is considered to function in the protective response for lipid peroxidation. In human patients with pseudoexfoliation syndrome (PEX) and cataract, MGST1 mRNA levels are found to be elevated in anterior lens capsules, while in PEX and glaucoma patients these are down-regulated in anterior segment tissues (Zenkel et al. 2007; Strzalka-Mrozik et al. 2013). However, we did not find evidence for optic nerve damage in Mafg−/−:Mafk+/− mutants. Nevertheless, both findings are suggestive of alterations in oxidative stress response in these tissues. Another DRG linked to cataract and oxidative stress is Gsto1, which encodes a glutathione S-transferase (GST) omega 1 that is considered to function in oxidative stress response by mediating regeneration of ascorbate. Interestingly, homozygous GSTO1Ala140Asp/GSTO2Asn142Asp haplotype carriers are associated with an elevated risk of cataracts in humans (Stamenkovic et al. 2014). Gsto1 is identified as a direct target of Nrf2 in ChIP assays and is down-regulated in Mafg−/−:Mafk+/− mutant lens.
Two other DRGs associated with stress, Aldh3a1 and Hspb1, are of significance to the lens and cataractogenesis as indicated by studies in mouse mutants. Aldh3a1 exhibits high expression in mammalian corneal epithelial cells where it has been considered to function in resistance to UV-induced oxidative damage (Lassen et al. 2007). We find Aldh3a1 to be expressed in the lens (this study, and Kakrana and Lachke, unpublished) and Aldh3a1−/− mouse mutants have been shown to develop cataracts (Lassen et al. 2007). Our analysis demonstrates Aldh3a1 to be down-regulated in Mafg−/−:Mafk+/− mutant lens and suggests it to be a potential direct target of small Mafs based on the presence of ARE core motif, as well as its recognition as a Mafg/Nrf2 target in multiple ChIP experiments (Hirotsu et al. 2012; Chorley et al. 2012; Malhotra et al. 2010). Another candidate down-regulated in Mafg−/−:Mafk+/− mutant lens is Hspb1, which encodes a stress response heat-shock protein that directly interacts with and stabilizes lens crystallin proteins CryαA and αB—mutations in both of which cause congenital cataracts in humans (Litt et al. 1998; Berry et al. 2001). In addition, similar to Mafg−/−:Mafk+/− mutants, Hspb1 is down-regulated in the lens of Tdrd7 and Hsf4 mouse mutants, both of which are directly linked to human cataract (Bu et al. 2002; Fujimoto et al. 2004; Lachke et al. 2011).
Thus, from the above misregulated genes, it is likely that the oxidant–antioxidant balance is affected in the Mafg−/−:Mafk+/− mutant lens. Oxidative stress can cause damage to various cellular components (lipids, proteins, DNA) and has been linked to cataracts (Berthoud and Beyer 2009). Small Mafs’ function has also been linked to oxidative stress response through heterodimeric regulatory interactions with the CNC proteins. Indeed, in addition to the stress-associated candidates discussed above (Mgst1, Gsto1, Aldh3a1, Hspb1), GO analysis identifies three other genes (Hmox1, Ddit3, Psmb5) among the DRGs that are classified into “response to oxidative stress” and are up-regulated in the Mafg−/−:Mafk+/− mutant lens. Indeed, Hmox1 has been identified as a direct target of Mafg and Nrf2 and is found to be elevated in small Maf mutants (Katsuoka et al. 2003; Yamazaki et al. 2012; Hirotsu et al. 2012). These findings may reflect a possible contribution of oxidative stress in the Mafg−/−:Mafk+/− mutant lens cataract. Interestingly, neither the known oxidative damage response targets of small Mafs in other tissues, nor the known antioxidant genes of the lens tissue are found to be misregulated in the Mafg−/−:Mafk+/− mutant lens (Berthoud and Beyer 2009; Yamazaki et al. 2012). However, our initial analysis of the oxidative stress status of aged lenses suggests that Mafg−/−:Mafk+/− mutants have higher levels of GSSG compared to control (Mafg+/−:Mafk+/−), indicative of elevated oxidative stress (data not shown). These findings will be pursued further in a comprehensive developmental time-course analysis of these mutants.
Our network analysis identifies a DRG cluster enriched for lipid and sterol synthesis genes and the presence of the gene Sc4mol within this cluster. Human mutations in SC4MOL (MSMO1), which codes for a methyl oxidase enzyme that is involved in cholesterol biosynthesis, cause an autosomal recessive syndrome that is presented with cataract (He et al. 2011). Indeed, misregulation of cholesterol synthesis has been associated with cataract in the Shumiya rat model (Mori et al. 2006). These data suggest a function for Mafg and Mafk in fine-tuning expression of the sterol pathway genes in lens cells. In addition to potentially affecting fiber cell membrane structure, which may provide a partial explanation for the fiber cell disorganization observed in Mafg−/−:Mafk+/− mutants, increased lipid/sterol production due to misregulation of this pathway may contribute to the overall elevation of stress in the mutant lens.
Another gene down-regulated in small Maf mutant lens, Uchl1, encodes a peptidase that removes C’ glycine residue of ubiquitin. Examination of UCHL1 Ser18Tyr polymorphisms in human patients indicates a positive association with cataract (Rudolph et al. 2011). Moreover, a second proteasomal pathway-associated gene Ubr5, which encodes a E3 ubiquitin ligase that functions in response to DNA damage, is down-regulated in the Mafg−/−:Mafk+/− mutant lens. Another non-crystallin cataract gene down-regulated in small Maf mutant lens is involved in two separate functions in cells—as a co-factor for HNF1 in transcription and in the synthesis of tetrahydrobiopterin. Pcbd1 null mouse mutants exhibit cataracts (Bayle et al. 2002).
Finally, the only crystallin gene that is down-regulated in Mafg−/−:Mafk+/− mutant lens is Crygf. Although a missense mutation in Crygf causes cataract in mice, its ortholog in humans has not been identified (Graw et al. 2002). It is possible that Crygf down-regulation may contribute to the phenotype in Mafg−/−:Mafk+/− lenses. However, Crygf is just one of the seven highly expressed gamma-crystallin genes found in the mouse, and none of the other six Cryg-genes are found altered by microarrays in these mutants. Furthermore, Cryg protein staining by immunofluorescence with a pan-Cryg antibody indicates no change in expression of the entire gamma-crystallin repertoire in the adult mutant lens (Fig. S1). Therefore, this potential redundancy makes it unclear if the modest reduction in Crygf levels, although may contribute to the phenotype, represents the principle mechanism of cataract formation in Mafg−/−:Mafk+/− lenses.
Together, these findings indicate that Mafg and Mafk are required for normal expression of functionally diverse genes in lens fiber cells. Therefore, the pathogenesis of cataracts in Mafg−/−:Mafk+/− mutant mice is likely to be multifactorial. Indeed, in addition to the cataract-linked genes discussed above, GO analysis identifies several DRGs to be classified into “extracellular matrix” (Metrn, Bgn, C1qtnf2, Gp2, Hdgf, Tfpi, Wfdc1, Serping1, Mdk, Igfbp5), which may provide an explanation for the posterior capsule rupture observed in Mafg−/−:Mafk+/− mutant lens. Furthermore, mutations in ALDH1A3, which is down-regulated in Mafg−/−:Mafk+/− mutant lens, cause recessive microphthalmia and anophthalmia in humans, and therefore may potentially contribute toward the lens phenotype (Fares-Taie et al. 2013; Yahyavi et al. 2013).
It will be interesting to investigate the significance and potential overlap of regulation by small Maf proteins through MARE motif, which is also the target of other large Mafs in the lens, such as Mafb, Nrl, and importantly, the human cataract gene MAF (Cvekl et al. 1994; Yang et al. 2004). It is plausible that Mafg and Mafk homo- or hetero-dimers occupy specific MARE sites and prevent other Maf proteins from binding and activation, thus serving to fine-tune gene expression. It is also likely that some of the targets are regulated by both small and large Maf proteins. Although whole genome expression profiles have not been generated for c-Maf−/− mouse mutant lens, some differences can be noted regarding its targets and those in Mafg−/−:Mafk+/− mutants. For example, the down-regulation of Cryaa in c-Maf−/− mouse lens is also not observed in Mafg−/−:Mafk+/− mutant lens. Notably, while the c-Maf−/− mouse mutant lens exhibits a reduction of several gamma-crystallin genes, specifically Crygb, Crygd, Cryge, and Crygf, our findings demonstrate that only Crygf is down-regulated in Mafg−/−:Mafk+/− mutant lens. We have identified a core ARE motif in the upstream region of Crygf (Fig. S2) that potentially supports binding by small Maf proteins. These findings suggest the Mafg and Mafk may function in rendering specificity to the transcriptional regulation of individual gamma-crystallin genes. It can be speculated that the balance and specificity of small and large Maf proteins in fiber cell transcriptional regulation may be achieved by binding with specific co-regulatory partner proteins. AP-1 (Jun), which is previously implicated in gene regulation in the lens, is downstream of Fgf signaling, and is also identified in the small Maf integrated network, has a putative binding site embedded in MARE sequence in the rat Crygb promoter (Cvekl et al. 1994; Cvekl and Duncan 2007). It will be interesting to investigate the dynamics of small Maf regulation in the context of these regulatory proteins that can bind near MARE sequences.
Although present analysis of the small Maf-binding protein Nrf2 data serves to potentially explain control of a subset of the 97 DRGs identified in the Mafg−/−:Mafk+/− mutant lens, it is possible that the other small Maf-binding proteins expressed in the lens (e.g., Nrf1, Bach2, and to a lower extent Bach1), are also functionally important in the lens. Regulation through these partners may explain the remainder of 97 DRGs detected in Mafg−/−:Mafk+/− mutant lens. Thus, the characterization of the lens in select Nrf and Bach family gene deletion mouse mutants will allow a comprehensive derivation of the small Maf regulatory network in the lens.
Collectively, in addition to identifying and characterizing two new transcriptional regulators in the lens that are necessary for normal expression of established non-crystallin human cataract genes, these findings serve to highlight the general utility of an integrated approach in gaining new insights into human disease genes. Finally, the small Maf regulatory network inferred from the integrated analysis in this study represents a critical first step toward assembly of a comprehensive fiber cell gene regulatory network, and provides several promising candidates for future investigations on cataract using resequencing analysis or association studies.
References
Alizadeh A, Clark J, Seeberger T et al (2004) Characterization of a mutation in the lens-specific CP49 in the 129 strain of mouse. Invest Ophthalmol Vis Sci 45:884–891
Bassnett S, Shi Y, Vrensen GFJM (2011) Biological glass: structural determinants of eye lens transparency. Philos Trans R Soc Lond B Biol Sci 366:1250–1264. doi:10.1098/rstb.2010.0302
Bayle JH, Randazzo F, Johnen G et al (2002) Hyperphenylalaninemia and impaired glucose tolerance in mice lacking the bifunctional DCoH gene. J Biol Chem 277:28884–28891. doi:10.1074/jbc.M201983200
Berry V, Francis P, Reddy MA et al (2001) Alpha-B crystallin gene (CRYAB) mutation causes dominant congenital posterior polar cataract in humans. Am J Hum Genet 69:1141–1145. doi:10.1086/324158
Berthoud VM, Beyer EC (2009) Oxidative stress, lens gap junctions, and cataracts. Antioxid Redox Signal 11:339–353. doi:10.1089/ars.2008.2119
Bu L, Jin Y, Shi Y et al (2002) Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat Genet 31:276–278. doi:10.1038/ng921
Cheng C, Ansari MM, Cooper JA, Gong X (2013) EphA2 and Src regulate equatorial cell morphogenesis during lens development. Development 140:4237–4245. doi:10.1242/dev.100727
Chorley BN, Campbell MR, Wang X et al (2012) Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res 40:7416–7429. doi:10.1093/nar/gks409
Churchill A, Graw J (2011) Clinical and experimental advances in congenital and paediatric cataracts. Philos Trans R Soc Lond B Biol Sci 366:1234–1249. doi:10.1098/rstb.2010.0227
Congdon N, Broman KW, Lai H et al (2005) Cortical, but not posterior subcapsular, cataract shows significant familial aggregation in an older population after adjustment for possible shared environmental factors. Ophthalmology 112:73–77. doi:10.1016/j.ophtha.2004.07.012
Cvekl A, Ashery-Padan R (2014) The cellular and molecular mechanisms of vertebrate lens development. Development 141:4432–4447. doi:10.1242/dev.107953
Cvekl A, Duncan MK (2007) Genetic and epigenetic mechanisms of gene regulation during lens development. Prog Retin Eye Res 26:555–597. doi:10.1016/j.preteyeres.2007.07.002
Cvekl A, Sax CM, Bresnick EH, Piatigorsky J (1994) A complex array of positive and negative elements regulates the chicken alpha A-crystallin gene: involvement of Pax-6, USF, CREB and/or CREM, and AP-1 proteins. Mol Cell Biol 14:7363–7376
Donner AL, Episkopou V, Maas RL (2007) Sox2 and Pou2f1 interact to control lens and olfactory placode development. Dev Biol 303:784–799. doi:10.1016/j.ydbio.2006.10.047
Du P, Kibbe WA, Lin SM (2008) lumi: a pipeline for processing Illumina microarray. Bioinformatics 24:1547–1548. doi:10.1093/bioinformatics/btn224
Fares-Taie L, Gerber S, Chassaing N et al (2013) ALDH1A3 mutations cause recessive anophthalmia and microphthalmia. Am J Hum Genet 92:265–270. doi:10.1016/j.ajhg.2012.12.003
Franceschini A, Szklarczyk D, Frankild S et al (2013) STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41:D808–D815. doi:10.1093/nar/gks1094
Fu L, Liang JJ-N (2002) Detection of protein–protein interactions among lens crystallins in a mammalian two-hybrid system assay. J Biol Chem 277:4255–4260. doi:10.1074/jbc.M110027200
Fujimoto M, Izu H, Seki K et al (2004) HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J 23:4297–4306. doi:10.1038/sj.emboj.7600435
Graw J, Klopp N, Neuhäuser-Klaus A et al (2002) Crygf(Rop): the first mutation in the Crygf gene causing a unique radial lens opacity. Invest Ophthalmol Vis Sci 43:2998–3002
Hammond CJ, Snieder H, Spector TD, Gilbert CE (2000) Genetic and environmental factors in age-related nuclear cataracts in monozygotic and dizygotic twins. N Engl J Med 342:1786–1790. doi:10.1056/NEJM200006153422404
Hammond CJ, Duncan DD, Snieder H et al (2001) The heritability of age-related cortical cataract: the twin eye study. Invest Ophthalmol Vis Sci 42:601–605
Hansen L, Eiberg H, Rosenberg T (2007) Novel MAF mutation in a family with congenital cataract–microcornea syndrome. Mol Vis 13:2019–2022
He M, Kratz LE, Michel JJ et al (2011) Mutations in the human SC4MOL gene encoding a methyl sterol oxidase cause psoriasiform dermatitis, microcephaly, and developmental delay. J Clin Invest 121:976–984. doi:10.1172/JCI42650
Hirotsu Y, Katsuoka F, Funayama R et al (2012) Nrf2–MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res 40:10228–10239. doi:10.1093/nar/gks827
Huang B, He W (2010) Molecular characteristics of inherited congenital cataracts. Eur J Med Genet 53:347–357. doi:10.1016/j.ejmg.2010.07.001
Huang DW, Sherman BT, Zheng X et al (2009) Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinform. doi:10.1002/0471250953.bi1311s27 (Chapter 13:Unit 13.11)
Jamieson RV, Perveen R, Kerr B et al (2002) Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum Mol Genet 11:33–42
Jolma A, Yan J, Whitington T et al (2013) DNA-binding specificities of human transcription factors. Cell 152:327–339. doi:10.1016/j.cell.2012.12.009
Kannan MB, Solovieva V, Blank V (2012) The small MAF transcription factors MAFF, MAFG and MAFK: current knowledge and perspectives. Biochim Biophys Acta 1823:1841–1846. doi:10.1016/j.bbamcr.2012.06.012
Kasaikina MV, Fomenko DE, Labunskyy VM et al (2011) Roles of the 15-kDa selenoprotein (Sep15) in redox homeostasis and cataract development revealed by the analysis of Sep 15 knockout mice. J Biol Chem 286:33203–33212. doi:10.1074/jbc.M111.259218
Kataoka K (2007) Multiple mechanisms and functions of maf transcription factors in the regulation of tissue-specific genes. J Biochem 141:775–781. doi:10.1093/jb/mvm105
Katsuoka F, Motohashi H, Tamagawa Y et al (2003) Small Maf compound mutants display central nervous system neuronal degeneration, aberrant transcription, and Bach protein mislocalization coincident with myoclonus and abnormal startle response. Mol Cell Biol 23:1163–1174
Kawauchi S, Takahashi S, Nakajima O et al (1999) Regulation of lens fiber cell differentiation by transcription factor c-Maf. J Biol Chem 274:19254–19260
Kim JI, Li T, Ho IC et al (1999) Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci 96:3781–3785
Lachke SA, Maas RL (2010) Building the developmental oculome: systems biology in vertebrate eye development and disease. Wiley Interdiscip Rev Syst Biol Med 2:305–323. doi:10.1002/wsbm.59
Lachke SA, Alkuraya FS, Kneeland SC et al (2011) Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science 331:1571–1576. doi:10.1126/science.1195970
Lachke SA, Higgins AW, Inagaki M et al (2012a) The cell adhesion gene PVRL3 is associated with congenital ocular defects. Hum Genet 131:235–250. doi:10.1007/s00439-011-1064-z
Lachke SA, Ho JWK, Kryukov GV et al (2012b) iSyTE: integrated systems tool for eye gene discovery. Invest Ophthalmol Vis Sci 53:1617–1627. doi:10.1167/iovs.11-8839
Lassen N, Bateman JB, Estey T et al (2007) Multiple and additive functions of ALDH3A1 and ALDH1A1: cataract phenotype and ocular oxidative damage in Aldh3a1(−/−)/Aldh1a1(−/−) knock-out mice. J Biol Chem 282:25668–25676. doi:10.1074/jbc.M702076200
Li W, Yu S, Liu T et al (2008) Heterodimerization with small Maf proteins enhances nuclear retention of Nrf2 via masking the NESzip motif. Biochim Biophys Acta 1783:1847–1856. doi:10.1016/j.bbamcr.2008.05.024
Litt M, Kramer P, LaMorticella DM et al (1998) Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet 7:471–474
Liu Y, Liu X, Zhang T et al (2007) Cytoprotective effects of proteasome beta5 subunit overexpression in lens epithelial cells. Mol Vis 13:31–38
Long Y, Li Q, Li J, Cui Z (2011) Molecular analysis, developmental function and heavy metal-induced expression of ABCC5 in zebrafish. Comp Biochem Physiol B Biochem Mol Biol 158:46–55. doi:10.1016/j.cbpb.2010.09.005
Maeda A, Moriguchi T, Hamada M et al (2009) Transcription factor GATA-3 is essential for lens development. Dev Dyn 238:2280–2291. doi:10.1002/dvdy.22035
Malhotra D, Portales-Casamar E, Singh A et al (2010) Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res 38:5718–5734. doi:10.1093/nar/gkq212
Min J-N, Zhang Y, Moskophidis D, Mivechi NF (2004) Unique contribution of heat shock transcription factor 4 in ocular lens development and fiber cell differentiation. Genesis 40:205–217. doi:10.1002/gene.20087
Mori M, Li G, Abe I et al (2006) Lanosterol synthase mutations cause cholesterol deficiency-associated cataracts in the Shumiya cataract rat. J Clin Invest 116:395–404. doi:10.1172/JCI20797
Motohashi H, O’Connor T, Katsuoka F et al (2002) Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294:1–12
Motohashi H, Katsuoka F, Engel JD, Yamamoto M (2004) Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1–Nrf2 regulatory pathway. Proc Natl Acad Sci 101:6379–6384. doi:10.1073/pnas.0305902101
Newburger DE, Bulyk ML (2009) UniPROBE: an online database of protein binding microarray data on protein–DNA interactions. Nucleic Acids Res 37:D77–D82. doi:10.1093/nar/gkn660
Nishiguchi S, Wood H, Kondoh H et al (1998) Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev 12:776–781
Nongpiur ME, Khor CC, Jia H et al (2014) ABCC5, a gene that influences the anterior chamber depth, is associated with primary angle closure glaucoma. PLoS Genet 10:e1004089. doi:10.1371/journal.pgen.1004089
Ogino H, Yasuda K (1998) Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science 280:115–118
Onodera K, Shavit JA, Motohashi H et al (2000) Perinatal synthetic lethality and hematopoietic defects in compound mafG:mafK mutant mice. EMBO J 19:1335–1345. doi:10.1093/emboj/19.6.1335
Portales-Casamar E, Thongjuea S, Kwon AT et al (2010) JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res 38:D105–D110. doi:10.1093/nar/gkp950
Rao GN, Khanna R, Payal A (2011) The global burden of cataract. Curr Opin Ophthalmol 22:4–9. doi:10.1097/ICU.0b013e3283414fc8
Reed NA, Oh DJ, Czymmek KJ, Duncan MK (2001) An immunohistochemical method for the detection of proteins in the vertebrate lens. J Immunol Methods 253:243–252
Ring BZ, Cordes SP, Overbeek PA, Barsh GS (2000) Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development 127:307–317
Robasky K, Bulyk ML (2011) UniPROBE, update 2011: expanded content and search tools in the online database of protein-binding microarray data on protein–DNA interactions. Nucleic Acids Res 39:D124–D128. doi:10.1093/nar/gkq992
Rudolph T, Sjölander A, Palmér MS et al (2011) Ubiquitin carboxyl-terminal esterase LI (UCHLI) SI8Y polymorphism in patients with cataracts. Ophthalmic Genet 32:75–79. doi:10.3109/13816810.2010.544360
Scheiblin DA, Gao J, Caplan JL et al (2014) Beta-1 integrin is important for the structural maintenance and homeostasis of differentiating fiber cells. Int J Biochem Cell Biol 50:132–145. doi:10.1016/j.biocel.2014.02.021
Shaham O, Smith AN, Robinson ML et al (2009) Pax6 is essential for lens fiber cell differentiation. Development 136:2567–2578. doi:10.1242/dev.032888
Shavit JA, Motohashi H, Onodera K et al (1998) Impaired megakaryopoiesis and behavioral defects in mafG-null mutant mice. Genes Dev 12:2164–2174
Shiels A, Hejtmancik JF (2013) Genetics of human cataract. Clin Genet 84:120–127. doi:10.1111/cge.12182
Shiels A, King JM, Mackay DS, Bassnett S (2007) Refractive defects and cataracts in mice lacking lens intrinsic membrane protein-2. Invest Ophthalmol Vis Sci 48:500–508. doi:10.1167/iovs.06-0947
Shiels A, Bennett TM, Hejtmancik JF (2010) Cat-map: putting cataract on the map. Mol Vis 16:2007–2015
Sorokina EA, Muheisen S, Mlodik N, Semina EV (2011) MIP/Aquaporin 0 represents a direct transcriptional target of PITX3 in the developing lens. PLoS One 6:e21122. doi:10.1371/journal.pone.0021122
Stamenkovic M, Radic T, Stefanovic I et al (2014) Glutathione S-transferase omega-2 polymorphism Asn142Asp modifies the risk of age-related cataract in smokers and subjects exposed to ultraviolet irradiation. Clin Exp Ophthalmol 42:277–283. doi:10.1111/ceo.12180
Strzalka-Mrozik B, Prudlo L, Kimsa MW et al (2013) Quantitative analysis of SOD2, ALDH1A1 and MGST1 messenger ribonucleic acid in anterior lens epithelium of patients with pseudoexfoliation syndrome. Mol Vis 19:1341–1349
Tanaka T, Tsujimura T, Takeda K et al (1998) Targeted disruption of ATF4 discloses its essential role in the formation of eye lens fibres. Genes Cells 3:801–810
Toki T, Itoh J, Kitazawa J et al (1997) Human small Maf proteins form heterodimers with CNC family transcription factors and recognize the NF-E2 motif. Oncogene 14:1901–1910. doi:10.1038/sj.onc.1201024
Vanita V, Singh D, Robinson PN et al (2006) A novel mutation in the DNA-binding domain of MAF at 16q23.1 associated with autosomal dominant “cerulean cataract” in an Indian family. Am J Med Genet A 140:558–566. doi:10.1002/ajmg.a.31126
Wasserman WW, Sandelin A (2004) Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet 5:276–287. doi:10.1038/nrg1315
Wigle JT, Chowdhury K, Gruss P, Oliver G (1999) Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet 21:318–322. doi:10.1038/6844
Yahyavi M, Abouzeid H, Gawdat G et al (2013) ALDH1A3 loss of function causes bilateral anophthalmia/microphthalmia and hypoplasia of the optic nerve and optic chiasm. Hum Mol Genet 22:3250–3258. doi:10.1093/hmg/ddt179
Yamazaki H, Katsuoka F, Motohashi H et al (2012) Embryonic lethality and fetal liver apoptosis in mice lacking all three small Maf proteins. Mol Cell Biol 32:808–816. doi:10.1128/MCB.06543-11
Yang Y, Chauhan BK, Cveklova K, Cvekl A (2004) Transcriptional regulation of mouse alphaB- and gammaF-crystallin genes in lens: opposite promoter-specific interactions between Pax6 and large Maf transcription factors. J Mol Biol 344:351–368. doi:10.1016/j.jmb.2004.07.102
Yoshida T, Yasuda K (2002) Characterization of the chicken L-Maf, MafB and c-Maf in crystallin gene regulation and lens differentiation. Genes Cells 7:693–706
Zenkel M, Kruse FE, Naumann GOH, Schlötzer-Schrehardt U (2007) Impaired cytoprotective mechanisms in eyes with pseudoexfoliation syndrome/glaucoma. Invest Ophthalmol Vis Sci 48:5558–5566. doi:10.1167/iovs.07-0750
Acknowledgments
We thank Drs. Melinda Duncan and Irfan Saadi for helpful comments and discussions. This work was supported by National Institutes of Health [R01 EY021505 to S.L.]; and Fight for Sight Grant-in-Aid award to S.L. S.L. is a Pew Scholar in Biomedical Sciences.
Conflict of interest
None declared.
Author information
Authors and Affiliations
Corresponding author
Additional information
S. A. Agrawal, D. Anand and A. D. Siddam contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Agrawal, S.A., Anand, D., Siddam, A.D. et al. Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non-crystallin genes associated with cataract. Hum Genet 134, 717–735 (2015). https://doi.org/10.1007/s00439-015-1554-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-015-1554-5