Abstract
Soybean [Glycine max (L.) Merr.] is an important legume crop worldwide, which provides abundant plant protein and oil for human beings. Soybean mosaic virus (SMV) can cause serious damage to the yield and quality of soybean, but it is difficult to control SMV with chemicals, breeding SMV-resistant varieties has become the most effective way to control the disease. Therefore, it is important to identify SMV resistance genes from soybean resources and apply them to soybean breeding. In this study, the disease rates (DRs) of 219 soybean accessions to SMV strain SC7 in two environments were investigated. A high-density NJAU 355 K SoySNP array was used for genome-wide association study (GWAS) of DR. A 274 kb region on chromosome 15 (1,110,567 bp to 1,384,173 bp) was repeatedly detected in two environments. Six new significant single nucleotide polymorphisms (SNPs) on chromosome 15 were identified. Four of these six SNPs were located within two candidate genes, Glyma.15G015700 and Glyma.15G015800. The elite haplotype Glyma.15G015700Hap I with low DR exhibited strong resistance to SC7. The expression of Glyma.15G015700 in the SMV-resistant accession increased significantly after inoculation with SC7. Furthermore, most of the proteins predicted to interact with Glyma.15G015700 are heat shock proteins, which have been shown to be related to disease resistance. In summary, new SMV resistance loci and a new candidate gene, Glyma.15G015700, were identified and might be utilized in further soybean disease resistance breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean [Glycine max (L.) Merr.] originated from China and is the most important legume crop in the world, serving as a primary source of edible vegetable oil and protein (Cheng et al. 2022). Soybean plants are susceptible to various diseases and environmental stresses during growth. Among all soybean diseases, soybean mosaic virus (SMV) can cause serious damage to the yield and quality of soybean. SMV is a single-stranded sense RNA virus (Jayaram et al. 1992), which is primarily transmitted by viral vectors and aphids (Yang and Gai 2011). The soybean yield loss caused by SMV is generally approximately 10%, but can reach 35–50% or even complete loss in some years when SMV is seriously harmful (Li et al. 2013). Breeding and planting SMV-resistant soybean varieties is the most economical and effective way for controlling SMV due to its wide distribution, severity, and resistance to chemicals. It is particularly important to identify SMV resistance genes and apply them to soybean disease resistance breeding. However, most SMV resistance genes identified from SMV-resistant varieties are R genes with leucine-rich repeats (LRRs) (Widyasari et al. 2020). R genes usually exert strong selection pressure on pathogens, but pathogens can easily overcome resistance by developing escape mutations in their effector genes (Dodds and Rathjen 2010). R genes generally do not provide broad-spectrum resistance (Widyasari et al. 2020), thus SMV-resistant soybean varieties bred with the R genes are not suitable for widespread cultivation. Therefore, the identification of new SMV resistance genes from soybean with rich genetic resources is important for the practical application of soybean disease resistance breeding.
During long-term interactions with the host and environment, SMV can produce various pathogenic differentiations, resulting in different pathogenic strains. The 98 SMV isolates collected in the U.S. were divided into seven main strains (G1–G7) by Cho and Goodman (1979). Four major SMV resistance loci (Rsv1, Rsv3, Rsv4 and Rsv5) have been identified from different SMV-resistant germplasm resources (Widyasari et al. 2020). These resistance loci are located on different chromosomes and resistant to different SMV strains. Rsv1 has been mapped on chromosome 13, and more than 10 genes encoding nucleotide-binding leucine-rich repeat (NLR) proteins have been found to be resistant to G1–G6 (Hayes et al. 2004). Rsv3 has been mapped on chromosome 14, and Glyma.14G204700 is a candidate gene of Rsv3, encoding coiled-coil NLR, which confers resistance to G5–G7 (Tran et al. 2018). Rsv4 is located on chromosome 2 and encodes an RNase H family protein that inhibits Potyvirus multiplication by degrading dsRNA and confers resistance to G1–G7 (Ishibashi et al. 2019). Rsv5 has been mapped on chromosome 13 and is resistant to G1 (Zheng et al. 2005), but the mechanism underlying Rsv5-mediated resistance is still unknown.
In China, researchers have classified thousands of SMV isolates into 22 strains, named SC1–SC22 (Li and Zhi 2016). The SMV resistance locus is named RSC (resistance of SMV in China), mainly includes RSC3Q, RSC3, RSC4, RSC5, RSC7, RSC8, RSC12, RSC14Q, RSC18 and RSC20 (Widyasari et al. 2020). The R plus strain indicates loci with resistance to a specific strain; for instance, RSC5 represents locus with resistance to SC5. Similar to Rsv, the reported RSC resistance loci are also mainly detected on chromosomes 2, 13 and 14. RSC5, RSC8 and RSC18 are all located on chromosome 2 and were identified from the SMV-resistant accession Kefeng No. 1 (Widyasari et al. 2020). Among them, Glyma02g13495 is considered the most likely candidate gene of RSC5 (Karthikeyan et al. 2017). Glyma.02G121500 and Glyma.02G121600 are candidate genes of RSC8 (Zhao et al. 2016). Glyma.02G127800, Glyma.02G128200 and Glyma.02G128300 are candidate genes of RSC18 (Li et al. 2015). Additionally, a coding gene GmMLRK1 of malectin-like receptor kinase located on chromosome 2 may be candidate gene for RSC7 (Che et al. 2023). RSC3Q, RSC3, RSC14Q and RSC20 are all located on chromosome 13 and were identified from the SMV-resistant accession Qihuang-1 (Widyasari et al. 2020). Glyma13g25730, Glyma13g25750, Glyma13g25950, Glyma13g25970 and Glyma13g26000 are considered candidate genes of RSC3Q (Zheng et al. 2014). Glyma13g25920, Glyma13g25950, Glyma13g25970 and Glyma13g26000 are candidate genes of RSC3 (Li et al. 2017). RSC14Q (Ma et al. 2011) represents the resistance locus of SC14 strain identified from Qihuang-1, but the candidate gene of RSC14Q is unknown at present. Glyma.13G194700 and Glyma.13G195100 are candidate genes of RSC20 (Karthikeyan et al. 2018). RSC12 is also located on chromosome 13 and has been identified from the SMV-resistant accession Qihuang-22 (Ma et al. 2010). RSC4 is located on chromosome 14 and was cloned from the SMV-resistant accession Dabaima. Glyma.14G205000 is candidate gene of RSC4, which encodes a cell wall-localized NLR-type resistant protein (Yin et al. 2021).
The SMV strain SC7 utilized in this study is a prevalent strain in the Huang-Huai soybean planting region in China (Yang et al. 2014), it can infect most soybean varieties and can cause severe yield loss. Regarding this SMV strain, a quantitative trait locus (QTL) near Rsv4 on chromosome 2 was found to be repeatedly detected by both QTL mapping and GWAS (Yan et al. 2015). Che et al. (2023) identified 20 SNPs by GWAS that were significantly associated with resistance to SC7 in soybean population. These SNPs were located on chromosomes 2, 13, and 19, and the candidate gene for resistance to SC7 was the malectin-like receptor kinase-encoding gene GmMLRK1. To date, the SMV resistance loci have been identified primarily by QTL mapping (Hayes et al. 2004; Zheng et al. 2005; Tran et al. 2018; Ishibashi et al. 2019; Yin et al. 2021) and partly by GWAS (Yan et al. 2015; Che et al. 2023). Therefore, the purpose of this study was to identify new SMV resistance genes that differ from the known R genes by performing a GWAS on 219 soybean accessions. These findings of this study will benefit to SMV resistance breeding.
Materials and methods
Materials and planting conditions
In this study, 219 soybean accessions (39 cultivars, 180 landraces) from all over the world (Wang et al. 2016) provided by the National Center for Soybean Improvement (NCSI, Nanjing Agricultural University, Nanjing, China) were utilized as soybean natural population accessions with extensive genetic variation. The accessions were mainly from the northern region of China (NR, 20 soybean accessions), Huang-Huai region of China (HR, 57 soybean accessions) and southern region of China (SR, 122 soybean accessions) (Supplementary Table 1). The SMV inoculation experiments were conducted at the Jiangpu Experimental Station (32° 1′ 53″ N and 118° 37′ 50″ E) and Baima Experimental Station (31° 36′ 50″ N and 119° 10′ 50″ E) of Nanjing Agricultural University in two different environments (E1, September 2018, the Jiangpu Experimental Station; E2, May 2019, the Baima Experimental Station). The SMV strain selected in this study was the virulent strain SC7, which is prevalent in the Huang-Huai region, and preserved the virus in the SMV-susceptible accession Nannong 1138-2.
Approximately 30 seedlings of each soybean accession were planted in a pot for inoculation. A random complete block design with 3 replicates for each soybean accession was used. After the real leaves had fully expanded, they were manually and evenly rubbed with prefabricated solution for inoculation. The solution was made by grinding virus-infected leaves of SC7 in 0.01 mol/L phosphate buffer (PB) (pH 7.2–7.4). The soybean leaves infected by SC7 were stored in −80 °C refrigerator. Prior to the inoculation of the 219 soybean accessions, Nannong 1138-2 was inoculated with the original SC7-infected leaves, which were subsequently used for the inoculation of the 219 soybean accessions.
Based on the standard survey method described in a previous report (Pu et al. 1983), the disease rate (DR) was defined as the percentage of plants showing mosaic symptoms out of the total number of inoculated plants. Accessions with DRs > 10% were considered susceptible, while those with DRs < 10% were considered resistant. The DRs of 219 soybean accessions were observed from the first week to the third week after inoculation, and the DRs were investigated at the fourth week after inoculation. Symptoms of soybean infection appeared several weeks after inoculation, and the DRs remained stable in the fourth week. Thus, the stable DRs of the fourth week were chosen as the trait for GWAS.
Analysis of phenotypic data
A statistical analysis of phenotypic data, including descriptive statistics, analysis of variance (ANOVA) and broad-sense heritability (the ratio of genetic variance to phenotypic variance), was performed with R software (Team 2010) and SPSS software (Arkkelin 2014). The formula for calculating the broad-sense heritability (h2) for DR was as follows Kim et al. (2014): h2 = σ2g/(σ2g + σ2ge/n + σ2e/nr), where σ2g represents the genotype variance, σ2ge represents the interaction variance between genotype and environment, σ2e represents the error variance, n represents the number of environments, r represents the number of replications in each environment. To assess the consistency of the phenotypic data, the Cronbach’s alpha coefficient of the DRs of two environments was calculated with SPSS software (Sun et al. 2007).
GWAS and haplotype analyses
The 219 soybean accessions were genotyped by the NJAU 355 K SoySNP array as previously reported (Wang et al. 2016). A total of 292,053 single nucleotide polymorphisms (SNPs) were identified on 20 chromosomes of soybean using Williams 82 as the reference genome (a2 version). In addition, 207,608 SNPs with a minor allele frequency (MAF) ≥ 0.05 were screened for association analysis. A mixed linear model (MLM) (Lipka et al. 2012) in the GAPIT package (Zhang et al. 2010) was used to perform GWAS for the DRs of two environments. The threshold for significant association was set at 1/n (n represents the number of markers), with a threshold of P ≤ (1/207608) = 4.82e−06 or −log10(P) ≥ 5.3. Manhattan plots and quantile‒quantile (Q‒Q) plots were generated by the qqman package in R software (Turner 2014). The linkage disequilibrium (LD) decay distance was 130 kb (Wang et al. 2016). LD block analysis and haplotype analysis were performed by the LD plot package and haplotype package of Haploview 4.2 software (Barrett et al. 2005).
Prediction and expression analysis of candidate genes
For the GWAS results, we extracted all genes within the LD decay distance (130 kb) of significant SNPs that were repeatedly detected in two environments. Subsequently, their functions were annotated based on the soybean reference genome of Williams 82 (a2 version).
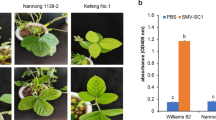
To clarify whether the candidate genes associated with disease resistance were induced by SC7, the SMV-resistant accession Kefeng No. 1 and SMV-susceptible accession Nannong1138-2 were inoculated with SMV strain SC7 and PB to detect the expression of the candidate genes. The real leaves at 0 h, 2 h, 4 h, 8 h, 12 h, 24 h and 48 h after inoculation were collected and rapidly frozen in liquid nitrogen. Total RNA from the leaves was extracted by a High-purity Total RNA Extraction Kit (Proteins Sci Biotech), and complementary DNA (cDNA) was synthesized using HiScript® II Q RT Super Mix for qPCR (+ gDNA wiper) (Vazyme) following the manufacturer’s instructions. The ChamQ SYBR qPCR Master Mix (without ROX) (Vazyme) was used for quantitative real-time polymerase chain reaction (qRT‒PCR) to detect the expression of candidate genes in the leaves. Three biological replicates were conducted for each sample, and tubulin (Glyma.05G157300) was used as the reference gene. The primer sequences for the candidate genes and internal reference gene are shown in Supplementary Table 2.
To analyze the expression patterns of the candidate genes, their expression levels in different tissues of soybean were examined using the SoyBase database (https://www.soybase.org/soyseq/). To visualize the expression data, heatmaps were constructed with HemI software (Deng et al. 2014).
Bioinformatic analysis of candidate genes
The amino acid sequences of the candidate genes were subjected to BLAST-P analysis to screen for Arabidopsis homologous genes and predict the conserved structural domains of the candidate genes. The protein sequence of a candidate gene was used to predict possible interacting proteins through the STRING database (https://cn.string-db.org/).
Results
Phenotypic variation in the disease rates of the soybean natural population
A total of 219 soybean accessions were inoculated with SMV strain SC7 to investigate the DRs in two environments (E1, September 2018; E2, May 2019) (Table 1). The mean values of the DRs in two environments were 0.50 and 0.17, ranging from 0 to 1. The descriptive statistics showed that the standard deviations of the DRs in two environments were 0.24 and 0.13, and the median values were 0.52 and 0.13. The ANOVA revealed that genotype of soybean accession significantly affected the DR of soybean to SC7. The DRs of 219 soybean accessions in two environments were approximately normal distribution (Supplementary Fig. 1), indicating the wide variation in DRs among different soybean accessions. The broad-sense heritability for two environments was 58.58%, indicating that the genetic effect was dominant in phenotypic variation, and the DR trait was stably inherited in the soybean natural population. The Cronbach's alpha coefficient of the DRs across two environments was 0.67, indicating that the DRs of the 219 soybean accessions in two environments were relatively consistent. These results indicated that the soybean natural population has high levels of variation diversity and heritability for DR, which is a complex quantitative trait controlled by multiple genes. Therefore, DR is suitable for genome-wide association study (GWAS).
GWAS revealed significant SNPs conferring resistance to SMV strain SC7
This study utilized the DRs of 219 soybean accessions to conduct a GWAS aimed at identifying significant loci associated with resistance to SMV strain SC7.The GWAS results are shown in Fig. 1a. The Q‒Q plot was used to represent the difference between the observed value and the predicted value. The X coordinate represents the predicted value, and the Y coordinate represents the observed value. The Q‒Q plots showed that the first half of the observed and predicted values fit well (Fig. 1a), indicating that the MLM was suitable for the trait of DR. The Manhattan plots revealed SNP clusters above the significance threshold [−log10(P) ≥ 5.3] on chromosomes 13 and 15 (Fig. 1a). Six significant SNPs (AX-93833468, AX-93833469, AX-93833470, AX-93833471, AX-93833472 and AX-94130556), which were located on chromosome 15, were repeatedly detected in two environments (Fig. 1a; Table 2).
Significant loci related to soybean resistance to SMV strain SC7. a Manhattan plots and Q‒Q plots of GWAS for DR in the soybean natural population across two environments. The horizontal line indicates the significance threshold −log10(P) ≥ 5.3. b GWAS signal of a 0.3 Mb region on chromosome 15 in E1 and E2. The arrow denotes a significant SNP identified in the GWAS results of E1 and E2. c LD heatmap and three haplotype blocks (block 1, block 2 and block 3) of the 0.3 Mb region on chromosome 15
Some loci on chromosome 15 were also identified in previous studies as being significantly associated with SMV (Yan et al. 2015). Currently, no candidate gene for SMV resistance on chromosome 15 has been reported, and there may be new SMV resistance genes. Therefore, the six newly identified significant SNPs [AX-93833468 (Fig. 1b), AX-93833469, AX-93833470, AX-93833471, AX-93833472 and AX-94130556] located on chromosome 15 were chosen for further study.
Haplotype analysis of significant SNPs and prediction of candidate genes conferring resistance to SMV strain SC7
A 274 kb region centered on the six significant SNPs (AX-93833468, AX-93833469, AX-93833470, AX-93833471, AX-93833472 and AX-94130556) on chromosome 15 (1,110,567 bp to 1,384,173 bp) was identified as the candidate region by LD decay (Wang et al. 2016). Subsequently, all the genes in the candidate region were functionally annotated (Supplementary Table 3). LD block analysis was performed on the candidate region. All SNPs in the 274 kb region were divided into three haplotype blocks (block 1, block 2 and block 3) (Fig. 1c), and the six significant SNPs were located within block 2. Notably, four of these six SNPs were located within two genes. AX-93833468 and AX-93833469 were located within Glyma.15G015700, while AX-93833470 and AX-93833471 were located within Glyma.15G015800. Haplotype analysis was performed on 219 soybean accessions using these four SNPs (Fig. 2a), and the results revealed two haplotypes (Hap 1: AGTA; Hap 2: GACG). Hap 1 and Hap 2 included 166 accessions and 40 accessions from the soybean natural population, respectively (Supplementary Table 4). The mean DR of Hap 1 and Hap 2 in two environments (E1 and E2) was 0.30 and 0.55, respectively. The ANOVA revealed that the DR of Hap 1 was significantly lower than that of Hap 2 (P < 0.001).
In addition, AX-93833468 was located within the coding sequence of Glyma.15G015700, resulting in a nonsynonymous mutation. This SNP also divided the soybean natural population into two different haplotypes (Glyma.15G015700Hap I: AA; Glyma.15G015700Hap II: GG) (Fig. 2b). Glyma.15G015700Hap I and Glyma.15G015700Hap II included 168 accessions and 40 accessions from the soybean natural population, respectively (Supplementary Table 5). The mean DR of Glyma.15G015700Hap I and Glyma.15G015700Hap II in two environments (E1 and E2) was 0.30 and 0.55, respectively. The ANOVA revealed that the DR of Glyma.15G015700Hap I was significantly lower than that of Glyma.15G015700Hap II (P < 0.001). Interestingly, these results were highly consistent with the haplotype analysis results of the above four SNPs, all of which were highly linked. The results indicated that the polymorphisms of these four SNPs (AX-93833468, AX-93833469, AX-93833470 and AX-93833471) in Glyma.15G015700 and Glyma.15G015800, particularly AX-93833468, significantly affected the DRs of the soybean natural population.
In summary, Glyma.15G015700 and Glyma.15G015800 were identified as candidate genes related to SMV strain SC7. Glyma.15G015700 is 68% similar to the Arabidopsis gene AT3G16110 and encodes the protein disulfide isomerase like-3b (PDIL-3b), which belongs to the thioredoxin superfamily. Glyma.15G015800 is 65% similar to the Arabidopsis gene AT3G24600 and encodes a late embryogenesis abundant protein (LEA).
Expression patterns of Glyma.15G015700 and Glyma.15G015800
The experimental group was inoculated with SMV strain SC7 (SC7), while the control group was inoculated with PB (Mock). The expression levels of two candidate genes in the SC7-inoculated and mock-inoculated leaves are shown in Fig. 3.The expression levels of Glyma.15G015700 in Kefeng No.1 (4, 8, 12, 24 and 48 h) and Nannong 1138–2 (2, 4, 8, 12 and 24 h) were significantly increased than that at 0 h in the experimental group (SC7) (Fig. 3a).The expression levels of Glyma.15G015800 in Kefeng No.1 (4, 24 and 48 h) were significantly increased than that at 0 h in the experimental group (SC7), but the expression levels of Glyma.15G015800 in Nannong 1138–2 (2, 4, 8, 12, 24 and 48 h) were significantly decreased than that at 0 h in the experimental group (SC7) (Fig. 3b). In addition, in the control group (Mock), the expression levels of Glyma.15G015700 and Glyma.15G015800 in Kefeng No.1 and Nannong 1138–2 were also significantly increased or decreased, respectively (Fig. 3a, b).
After analyzing the above results, it was concluded that the expression levels of Glyma.15G015700 and Glyma.15G015800 were not only induced by the virus but also affected by the friction inoculation method and PB. Next, the expression levels of candidate genes were analyzed by determining the ratio of plants inoculated with SC7 to mock inoculated plants (Fig. 3c, d). The results indicated that the expression levels of Glyma.15G015700 in Kefeng No. 1 peaked at 4 h, with a significant increase compared to expression levels at 0 h. In contrast, the expression level of Glyma.15G015800 in Kefeng No. 1 was not significantly different at any time point compared to the expression level at 0 h.
The expression levels of Glyma.15G015700 and Glyma.15G015800 were relatively stable in the SMV-susceptible accession Nannong 1138-2, but changed significantly in the SMV-resistant accession Kefeng No. 1. These results suggested that the two candidate genes, particularly Glyma.15G015700, actively responded to the induction of SC7 to increase expression levels.
RNA-seq data from a public database (https://www.soybase.org/soyseq/) was used to analyze the expression patterns of candidate genes in different soybean tissues, which was helpful for understanding their potential functions. The results showed that Glyma.15G015700 and Glyma.15G015800 had the highest expression levels in roots, but had low expression levels in leaves, flowers and pods, with almost no expression in seeds and root nodules (Supplementary Fig. 2). Combined with the results of SC7-induced expression (Fig. 3), it showed that the expression levels of these two candidate genes, particularly Glyma.15G015700, will be significantly increased in the leaves of SMV-resistant accession after inoculation with SC7. These results indicated that Glyma.15G015700 may play a crucial role in SMV resistance.
Prediction of interacting proteins of Glyma.15G015700
The DNA coding sequence of Glyma.15G015700 is 1572 bp, encoding 523 amino acids, including the structural domain of protein disulfide isomerase like-3b (PDIL-3b), which is predicted to be located on the endoplasmic reticulum (http://linux1.softberry.com).
To further investigate the function of Glyma.15G015700, the interacting proteins were predicted, and the results showed that ten proteins may interact with Glyma.15G015700 (which is named Glyma15G01880 in Wm82.a1) (Fig. 4). Interestingly, there are six interacting proteins: GMBIP4 (expressed by Glyma05G36620, which is named Glyma.05G219400 in Wm82.a2), GMBIP2 (expressed by Glyma08G02960, which is named Glyma.08G025900 in Wm82.a2), GLYMA05G36600.1 (expressed by Glyma05G36600, which is named Glyma.05G219600 in Wm82.a2), GLYMA20G16070.1 (expressed by Glyma20G16070, which is named Glyma.20G065900 in Wm82.a2), GLYMA13G10700.1 (expressed by Glyma13G10700, which is named Glyma.13G003700 in Wm82.a2), and GLYMA08G02940.1 (LOC547838) (expressed by Glyma08G02940, which is named Glyma.08G025700 in Wm82.a2). These six proteins are all heat shock protein 70 (HSP70) family proteins, which have been shown to be associated with plant virus resistance (Satyanarayana et al. 2000; Jungkunz et al. 2011).
Predicted interacting proteins of Glyma.15G015700 (PDIL-3b). The colored nodes indicate query proteins and the first shell of interactors, the solid wathet blue lines indicate known interactions from curated databases, the solid pink lines indicate known interactions from experimentally determined data, the solid light green lines indicate gene neighborhoods, the solid navy-blue lines indicate gene co-occurrence, the solid black lines indicate co-expression, and the solid purple lines indicate protein homology
In conclusion, Glyma.15G015700 was identified as a candidate gene related to resistance to SMV strain SC7, and these results provided valuable information for further research on the function of SC7 resistance candidate genes.
The geographical distribution of different Glyma.15G015700 haplotypes
The geographical distribution of two haplotypes of Glyma.15G015700 in three major soybean production areas in China (NR, HR and SR) was investigated in 219 soybean accessions. Both Glyma.15G015700Hap I and Glyma.15G015700Hap II were present in all regions (Fig. 5a) (Supplementary Table 6). However, the proportion of the elite haplotype Glyma.15G015700Hap I with low DR in NR and HR was much greater than that in SR, especially in HR, indicating that Glyma.15G015700Hap I might be selected naturally or artificially in HR. As a result, some soybean accessions in HR have stronger resistance to SMV strain SC7, which is a prevalent strain in HR. This may led to the DR of soybean accessions in HR significantly lower than that in SR among the 219 soybean accessions (Fig. 5b).
The geographical distribution and DR variations of two haplotypes of Glyma.15G015700. a Distribution of two haplotypes in different regions of China. HR represents the Huang-Huai region of China, NR represents the northern region of China, and SR represents the southern region of China. b DR variations in different regions. n, the number of soybean accessions; ns, not significant; ∗∗Significant at P < 0.01
Discussion
SMV can seriously affect soybean yield and quality, and occurs in soybean production areas around the world (Song et al. 2016). Chemicals can only kill virus transmitted mediator-aphids and are less effective in controlling soybeans already infected with SMV (Usovsky et al. 2022). Therefore, the identification of SMV resistance genes and applying them to soybean molecular breeding will beneficial to improve the SMV resistance breeding in soybean. SMV resistance loci were mainly identified from SMV-resistant soybean varieties, and most of the resistance loci were located on chromosomes 2, 13 and 14 (Widyasari et al. 2020). Many potential resistance loci cannot be detected due to the narrow genetic background of SMV-resistant soybean varieties. In this study, a GWAS was performed on the DR-related trait of 219 soybean accessions with rich genetic background inoculated with SMV strain SC7 in two environments, aiming to identify new SMV resistance loci and resistance genes. Six newly significant SNPs were identified on chromosome 15 that could be repeatedly detected in two environments. One of these SNPs was located within the coding sequence of Glyma.15G015700 and caused a nonsynonymous mutation. In addition, Glyma.15G015700, as a candidate gene, can be strongly induced by SMV strain SC7.
Most of the identified SMV resistance genes are R genes containing leucine-rich repeats (LRRs). The SMV-resistant soybean varieties cultivated from R genes are usually resistant to only one or a few SMV strains, and the resistance is easily overcome by pathogens (Dodds and Rathjen 2010; Widyasari et al. 2020). Notably, Glyma.15G015700 does not belong to the R genes and encodes a protein disulfide isomerase (PDI). PDI and PDI-like (PDIL) proteins are members of the thioredoxin (TRX) superfamily (Chen et al. 2013). In plants, TRX can regulate the activity of many enzymes by regulating intracellular redox reactions and ultimately plays an important role in redox regulation, growth, development and disease resistance (Sun 2009). Rivas et al. (2014) reported that the resistant protein cf-9 could interact with CITRX (cf-9-interacting thioredoxin) in tomato, and virus-mediated CITRX silencing increased the content of reactive oxygen species in plants, thus improving disease resistance in tomato leaf mold. In addition, ZmTrxh could improve maize resistance to mosaic virus SCMV by inhibiting virus accumulation in the cytoplasm (Liu et al. 2017). According to the available studies, TRX can be used as a positive or negative regulatory protein in resistance to pathogenic bacteria. Furthermore, the proteins interacting with Glyma.15G015700 are heat shock proteins (HSPs). Studies have shown that HSPs can be involved in plant virus life through folding, translocation, localization, assembly or degradation of recruited proteins (Wang et al. 2018). HSP70 is a member of the HSP family and has been reported to be related to plant diseases, such as rice stripe virus (Satyanarayana et al. 2000) and turnip mosaic virus (Jungkunz et al. 2011).
In conclusion, a GWAS revealed that the TRX encoding gene Glyma.15G015700 was related to SMV resistance, and Glyma.15G015700 could be strongly induced by SMV strain SC7. Furthermore, the elite haplotype Glyma.15G015700Hap I with low DR was identified to be highly resistant to SC7 and might be selected naturally or artificially in the Huang-Huai region. Glyma.15G015700Hap I accounted for high percentage of the soybean population in the Huang-Huai region where SC7 is prevalent, which might enhance the resistance of soybean to SC7 in this region. These results indicated that Glyma.15G015700 may play an important role in regulating soybean resistance to SC7, but the related mechanism needs to be further explored. In further study, transgenic soybean plants with Glyma.15G015700-overexpressing lines and knockout mutants could be generated to further verify the function of Glyma.15G015700. Notably, Glyma.15G015700 is not an R gene. A variety of SMV strains could be utilized to verify whether Glyma.15G015700 has broad-spectrum resistance to SMV. These results could provide information and new gene resources for further SMV resistance breeding.
Data availability statement
All the data generated in the experiments are presented in the manuscript and its supplementary files.
References
Arkkelin D (2014) Using SPSS to understand research and data analysis. Valparaiso University, Indiana
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265. https://doi.org/10.1093/bioinformatics/bth457
Che ZJ, Zhang SY, Pu YX, Yang YM, Liu HL, Yang H, Wang L, Zhang YH, Liyu BH, Zhang HY, Wang H, Cheng H, Yu DY (2023) A novel soybean malectin-like receptor kinase-encoding gene, GmMLRK1, provides resistance to soybean mosaic virus. J Exp Bot 74(8):2692–2706. https://doi.org/10.1093/bioinformatics/bth457
Chen Z, Jiang Q, Zhu C (2013) Protein disulfide isomerise and PDI-like proteins in plant. Plant Physiol J 49(8):715–721. https://doi.org/10.13592/j.cnki.ppj.2013.08.014
Cheng RX, Mei RX, Yan R, Chen HY, Miao D, Cai LN, Fan JY, Li GR, Xu R, Lu WG, Gao Y, Ye WW, Su S, Han TF, Gai JY, Wang YC, Tao XR, Xu Y (2022) A new distinct geminivirus causes soybean stay-green disease. Mol Plant 15(6):927–930. https://doi.org/10.1016/j.molp.2022.03.011
Cho EK, Goodman RM (1979) Strains of soybean mosaic virus: classification based on virulence in resistant soybean cultivars. Phytopathology 69(5):467–470. https://doi.org/10.1094/PHYTO-69-467
Deng WK, Wang YB, Liu ZX, Cheng H, Xue Y (2014) HemI: a toolkit for illustrating heatmaps. PLoS ONE 9(11):e111988. https://doi.org/10.1371/journal.pone.0111988
Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11(8):539–548. https://doi.org/10.1038/nrg2812
Hayes AJ, Jeong SC, Gore MA, Yu YG, Buss GR, Tolin SA, Maroof MA (2004) Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics 166(1):493–503. https://doi.org/10.1534/genetics.166.1.493
Ishibashi K, Saruta M, Shimizu T, Shu M, Anai T, Komatsu K, Yamada N, Katayose Y, Ishikawa M, Ishimoto M, Kaga A (2019) Soybean antiviral immunity conferred by dsRNase targets the viral replication complex. Nat Commun 10(1):4033. https://doi.org/10.1038/s41467-019-12052-5
Jayaram C, Hill JH, Miller WA (1992) Complete nucleotide sequences of two soybean mosaic virus strains differentiated by response of soybean containing the Rsv resistance gene. J Gen Virol 73(8):2067–2077. https://doi.org/10.1099/0022-1317-73-8-2067
Jungkunz I, Link K, Vogel F, Voll LM, Sonnewald S, Sonnewald U (2011) AtHsp70-15-deficient Arabidopsis plants are characterized by reduced growth, a constitutive cytosolic protein response and enhanced resistance to TuMV. Plant J 66(6):983–995. https://doi.org/10.1111/j.1365-313X.2011.04558.x
Karthikeyan A, Li K, Jiang H, Ren R, Li C, Zhi HJ, Chen SY, Gai JY (2017) Inheritance, fine-mapping, and candidate gene analyses of resistance to soybean mosaic virus strain SC5 in soybean. Mol Genet Genomics 292(4):811–822. https://doi.org/10.1007/s00438-017-1310-8
Karthikeyan A, Li K, Li C, Yin JL, Li N, Yang YH, Song YP, Ren R, Zhi HJ, Gai JY (2018) Fine-mapping and identifying candidate genes conferring resistance to Soybean mosaic virus strain SC20 in soybean. Theor Appl Genet 131(2):461–476. https://doi.org/10.1007/s00122-017-3014-1
Kim H, Xing GN, Wang YF, Zhao TJ (2014) Constitution of resistance to common cutworm in terms of antibiosis and antixenosis in soybean RIL populations. Euphytica 196(1):137–154. https://doi.org/10.1007/s10681-013-1021-0
Li C, Karthikeyan A, Yuan Y, Yin JL, Ren R, Yang YQ, Zhi HJ (2017) Identification of candidate genes for resistance to soybean mosaic virus strain SC3 by using fine mapping and transcriptome analyses. Crop Pasture Sci 68(2):156–166. https://doi.org/10.1071/CP16353
Li K, Liu ZT, Li HC, Zhang K, Wang CK, Ren R, Lu WG, Zhi HJ (2013) Resistance to soybean mosaic virus and soybean cyst nematode of soybean cultivars from China national soybean uniform trials. Soybean Sci 32(5):670–675
Li K, Ren R, Karthikeyan A, Gao L, Yuan Y, Liu ZT, Zhong YK, Zhi HJ (2015) Genetic analysis and identification of two soybean mosaic virus resistance genes in soybean [Glycine max (L.) Merr]. Plant Breed 134(6):684–695. https://doi.org/10.1111/pbr.12315
Li K, Zhi HJ (2016) Advances in resistance to soybean mosaic virus disease in soybean. Soybean Sci 35(4):525–530. https://doi.org/10.11861/j.issn.1000-9841.2016.04.0525
Lipka AE, Tian F, Wang QS, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang ZW (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28(18):2397–2399. https://doi.org/10.1093/bioinformatics/bts444
Liu QQ, Liu HH, Gong YQ, Tao YF, Jiang L, Zuo WL, Yang Q, Ye JR, Lai JS, Wu JY, Lübberstedt T, Xu ML (2017) An atypical thioredoxin imparts early resistance tosugarcane mosaic virus in maize. Mol Plant 10(3):483–497. https://doi.org/10.1016/j.molp.2017.02.002
Ma Y, Li HC, Wang DG, Liu N, Zhi HJ (2010) Molecular mapping and marker assisted selection of soybean mosaic virus resistance gene RSC12 in soybean. Legum Genom Genet. https://doi.org/10.5376/lgg.2010.01.0008
Ma Y, Wang DG, Li HC, Zheng GJ, Yang YQ, Li HW, Zhi HJ (2011) Fine mapping of the RSC14Q locus for resistance to soybean mosaic virus in soybean. Euphytica 181(1):127–135. https://doi.org/10.1007/s10681-011-0457-3
Pu ZQ, Cao P, Xue BD (1983) Resistance in soybean varieties to six strains of soybean mosaic virus. J Nanjing Agric Univ 3:41–45
Rivas S, Rougon-Cardoso A, Smoker M, Schauser L, Yoshioka H, Jones JD (2014) CITRX thioredoxin interacts with the tomato Cf-9 resistance protein and negatively regulates defence. EMBO J 23(10):2156–2165. https://doi.org/10.1038/sj.emboj.7600224
Satyanarayana T, Gowda S, Mawassi M, Albiach-Martí MR, Ayllón MA, Robertson C, Garnsey SM, Dawson WO (2000) Closterovirus encoded HSP70 homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology 278(1):253–265. https://doi.org/10.1006/viro.2000.0638
Song YP, Li C, Zhao L, Karthikeyan A, Li N, Li K, Zhi HJ (2016) Disease spread of a popular soybean mosaic virus strain (SC7) in southern China and effects on two susceptible soybean cultivars. Philipp Agric Sci 99(4):355–364
Sun LJ (2009) Regulation and induction of plant defense by thioredoxin and bioelicitor. Dissertation, Nanjing Agricultural University
Sun W, Chou CP, Stacy AW, Ma H, Unger J, Gallaher P (2007) SAS and SPSS macros to calculate standardized Cronbach’s alpha using the upper bound of the phi coefficient for dichotomous items. Behav Res Methods 39(1):71–81. https://doi.org/10.3758/bf03192845
Team R (2010) R: a language and environment for statistical computing. Computing 14:12–21
Tran PT, Widyasari K, Seo JK, Kim KH (2018) Isolation and validation of a candidate Rsv3 gene from a soybean genotype that confers strain-specific resistance to soybean mosaic virus. Virology 513:153–159. https://doi.org/10.1016/j.virol.2017.10.014
Turner SD (2014) qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. Biorxiv. https://doi.org/10.21105/joss.00731
Usovsky M, Chen PY, Li DX, Wang AM, Shi AN, Zheng CM, Shakiba E, Lee DH, Vieira CC, Lee YC, Wu CJ, Cervantez I, Dong DK (2022) Decades of genetic research on soybean mosaic virus resistance in soybean. Viruses 14(6):1122. https://doi.org/10.3390/v14061122
Wang J, Chu SS, Zhang HR, Zhu Y, Cheng H, Yu DY (2016) Development and application of a novel genome-wide SNP array reveals domestication history in soybean. Sci Rep 6(1):20728. https://doi.org/10.1038/srep20728
Wang Y, Ren YF, Li DX, Chen Z (2018) The interaction of heat shock proteins and some components of plant viruses. Chin J Appl Environ Biol 24(2):415–424. https://doi.org/10.19675/j.cnki.1006-687x.2017.03013
Widyasari K, Alazem M, Kim K (2020) Soybean resistance to soybean mosaic virus. Plants 9(2):219. https://doi.org/10.3390/plants9020219
Yan HL, Wang H, Cheng H, Hu ZB, Chu SS, Zhang GZ, Yu DY (2015) Detection and fine-mapping of SC7 resistance genes via linkage and association analysis in soybean. J Integr Plant Biol 57(8):722–729. https://doi.org/10.1111/jipb.12323
Yang QH, Gai JY (2011) Identification, inheritance and gene mapping of resistance to a virulent soybean mosaic virus strain SC15 in soybean. Plant Breed 130(2):128–132. https://doi.org/10.1111/j.1439-0523.2010.01797.x
Yang YQ, Zhi HJ, Zhang MC (2014) Current status and prospect on resistance to soybean mosaic virus in Huang-Huai-Hai soybean region. Acta Agric Boreali-Sinica 29(S1):103–108. https://doi.org/10.7668/hbnxb.2014.S1.022
Yin JL, Wang LQ, Jin TT, Nie Y, Liu H, Qiu YL, Yang YH, Li BW, Zhang JJ, Wang DG, Li K, Xu K, Zhi HJ (2021) A cell wall-localized NLR confers resistance to soybean mosaic virus by recognizing viral-encoded cylindrical inclusion protein. Mol Plant 14(11):1881–1900. https://doi.org/10.1016/j.molp.2021.07.013
Zhang ZW, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu JM, Arnett DK, Ordovas JM, Buckler ES (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42(4):355–360. https://doi.org/10.1038/ng.546
Zhao L, Wang DG, Zhang HY, Shen YC, Yang YQ, Li K, Wang LQ, Yang YH, Zhi HJ (2016) Fine mapping of the RSC8 locus and expression analysis of candidate SMV resistance genes in soybean. Plant Breed 135(6):701–706. https://doi.org/10.1111/pbr.12428
Zheng CM, Chen PY, Gergerich R (2005) Characterization of resistance to soybean mosaic virus in diverse soybean germplasm. Crop Sci 45(6):2503–2509. https://doi.org/10.2135/cropsci2005.0114
Zheng GJ, Yang YQ, Ma Y, Yang XF, Chen SY, Ren R, Wang DG, Yang ZL, Zhi HJ (2014) Fine mapping and candidate gene analysis of resistance gene RSC3Q to soybean mosaic virus in Qihuang 1. J Integr Agric 13(12):2608–2615. https://doi.org/10.1016/S2095-3119(13)60738-8
Acknowledgements
This work was supported by the high-performance computing platform of Bioinformatics Center, Nanjing Agricultural University.
Funding
This work was supported by the National Natural Science Foundation of China (32090065, 32001507, 32260466), Jiangsu Province Agriculture Science and Technology Innovation [CX (22) 3088] and JBGS (2021) 060.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. YXP, HC and DYY conceived the research plan; YXP, RJY, DBJ, ZJC, RFY, CYY and HW performed the experiments; YXP analyzed the data and wrote the article with help from HC and DYY. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Bing Yang.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
438_2024_2151_MOESM2_ESM.xlsx
Supplementary file2 Expression profiles of two candidate genes in different soybean tissues. DAF, days after flowering. (XLSX 41 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pu, Y., Yan, R., Jia, D. et al. Identification of soybean mosaic virus strain SC7 resistance loci and candidate genes in soybean [Glycine max (L.) Merr.]. Mol Genet Genomics 299, 54 (2024). https://doi.org/10.1007/s00438-024-02151-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00438-024-02151-4