Abstract
Obesity is a major public health issue resulting from an interaction between genetic and environmental factors. Genetic risk scores (GRSs) are useful to summarize the effects of many genetic variants on obesity risk. In this study, we aimed to assess the association of previously well-studied genetic variants with obesity and develop a genetic risk score to anticipate the risk of obesity development in the Iranian population. Among 968 participants, 599 (61.88%) were obese, and 369 (38.12%) were considered control samples. After genotyping, an initial screening of 16 variants associated with body mass index (BMI) was performed utilizing a general linear model (p < 0.25), and seven genetic variants were selected. The association of these variants with obesity was examined using a multivariate logistic regression model (p < 0.05), and finally, five variants were found to be significantly associated with obesity. Two gene score models (weighted and unweighted), including these five loci, were constructed. To compare the discriminative power of the models, the area under the curve was calculated using tenfold internal cross‐validation. Among the studied variants, ADRB3 rs4994, FTO rs9939609, ADRB2 rs1042714, IL6 rs1800795, and MTHFR rs1801133 polymorphisms were significantly associated with obesity in the Iranian population. Both of the constructed models were significantly associated with BMI (p < 0.05) and the area under the mean curve of the weighted GRS and unweighted GRS were 70.22% ± 0.05 and 70.19% ± 0.05, respectively. Both GRSs proved to predict obesity and could potentially be utilized as genetic tools to assess the obesity predisposition in the Iranian population. Also, among the studied variants, ADRB3 rs4994 and FTO rs9939609 polymorphisms have the highest impacts on the risk of obesity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Obesity is a complex health problem correlated with an increased risk of hypertension, type 2 diabetes, cardiovascular diseases, cancer, and poor quality of life (Mokdad Ford et al. 2003). In 2014, more than 40 million children and over 600 million adults were classified as obese worldwide. According to the World Health Organization (WHO), the number of obese adults reached 650 million in 2016 (Ren, Xu et al. 2019). It is estimated that over 30% of adults in the US are obese and, in most countries, the prevalence rate of this abnormality has increased over the last decades (Flegal Carroll et al. 2002; Wang, McPherson et al. 2011). Besides, studies have shown that more than 80% of Iranian adults are obese or overweight (Rahmani, Sayehmiri et al. 2015). Various environmental factors play important roles in developing obesity, such as overeating and lack of adequate physical activity (Carlos, Silva-Nunes et al. 2013). In addition to these factors, genetic risk factors could also contribute to the increase in the prevalence of obese adults. Genome-wide association studies (GWASs) have successfully identified the impact of various single nucleotide polymorphisms (SNPs) on obesity and being overweight (Rankinen, Zuberi et al. 2006; Lu and Loos 2013; Locke, Kahali et al. 2015). Among the investigated genetic variants, a common variant at the fat mass and obesity-associated (FTO) locus has powerful effect on body fat and mass (Loos and Yeo 2014). Also, several well-defined SNPs are contributing to obesity and metabolic syndrome, especially in Middle Eastern countries. For example, some studies have shown that methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms are related to obesity (Fu, Li et al. 2019), also polymorphisms in the vitamin D receptor (VDR) gene are involved in the pathogenesis of inflammation and obesity (Al-Daghri, Guerini et al. 2014). Moreover, interleukin 6 (IL6) variants on abdominal obesity have been largely studied (Berthier, Paradis et al. 2003). In addition, SNPs of antioxidant enzymes including glutathione peroxidase (GPX1), catalase (CAT), and superoxide dismutase 2 (SOD2) influence the endogenous free radicals’ clearance, which plays an important role in excess weight gain (Saravani, Miri et al. 2015; Hernández-Guerrero, Parra-Carriedo et al. 2018). Fatty acid-binding protein (FABP) is another critical gene that its polymorphisms cause an increased triglyceride transport in intestinal cells, which may result in metabolic disorders (Albala, Santos et al. 2004). The polymorphisms of β2 and β3 adrenergic receptors (ADRB2 and ADRB3, respectively) regulate the energy balance and are associated with obesity (Saliba, Reis et al. 2014). Many researchers have confirmed an association between transcription factor 7-like 2 (TCF7L2) variants and higher BMI and type 2 diabetes (T2D) risk (Cauchi, Choquet et al. 2008). The other potential risk locus is on the angiotensin-converting enzyme gene (ACE) and is linked to adiposity and metabolic outcomes (Bordoni, Marchegiani et al. 2017). Other potential candidates include a polymorphism in apolipoprotein C3 (APOC3) that has been associated with both dyslipidemia and insulin resistance (Miller, Rhyne et al. 2007), tumor necrosis factor-alpha (TNF- α) gene polymorphism which is associated with insulin resistance and obesity (Ghareeb, Abdelazem et al. 2021), the variant in caffeine metabolizing enzyme cytochrome P450 1A2 (CYP1A2) (Urry, Jetter et al. 2016) and finally, a polymorphism at the promoter region of the lactase gene (LCT) related to obesity and other anthropometric measurements (Corella, Arregui et al. 2011).
Since each individual allele could only have a limited effect on the risk of obesity, the impact of each variant should be considered in combination with other risk alleles. Thus, information from multiple risk alleles and a carefully constructed genetic risk score (GRS) is vital for obesity risk assessment. These translational genomic tools may be useful in tackling this public health issue (Belsky, Moffitt et al. 2013). Several studies in various ethnicities have suggested new genetic risk tools to predict obesity (Seral-Cortes, Sabroso-Lasa et al. 2021). However, a limited number of studies investigating the effect of these polymorphisms and providing an accurate GRS for Middle Eastern adults. The two main goals of this study were (1) to assess the association of sixteen genetic variants with obesity in the Iranian population and (2) to develop and implement a novel genetic model for this population to anticipate the risk of obesity development, more precisely.
Materials and methods
Subjects
This study was conducted on 968 Iranian subjects (male: 306 (31.6%), female: 662 (68.4%), aged 18–70) residents in Tehran. The proposed cut-off points of body mass index (BMI) for obesity focus on the factors which increase the risk of disease development, especially cardiovascular diseases. According to the WHO classification, a BMI of 25 kg/m2 is defined as a cut-off point for overweight, and a BMI of 30 kg/m2 is the cut-off point for obesity (Status 1995). However, Ali Babai et al. defined a new BMI cut-off for Iranians and indicated that Iranian men and women have a higher risk of developing metabolic syndromes with BMI of 25.7 and 27.05 kg/m2, respectively (Babai, Arasteh et al. 2016). Among 306 men, 239 persons (BMI > 25.7 kg/m2, 78.1%) were considered as obese cases and the remaining were considered as controls (BMI < = 25.7 kg/m2, 21.9%). Among 662 women, 360 participants were categorized as obese (BMI > 27.05 kg/m2, 54.4%), and the remaining as controls (BMI < = 27.05 kg/m2, 45.6%). All the procedures and the aims of the study were explained to the participants, and just those who signed the informed consent form were enrolled in the study. The ethics committee of Kawsar Human Genetic Research Center (2019) approved this research. All anthropometric parameters were measured by a trained person using standard techniques. Bodyweight and height measurements were done with light clothing and without shoes by Jawon BC 510 bio-electrical impedance analyzer (Jawon, South Korea). Waist circumference was measured at the point of waist narrowing. Hip circumference was determined as the maximum circumference around the hips. BMI was calculated by dividing weight in kilograms by height in meters squared. Moreover, the waist-to-height ratio and waist-to-hip ratio were calculated.
DNA isolation and genotyping
Participants’ saliva samples were collected using saliva collection kits (4N6 FLOQSwabs® genetics, Copan Diagnostics Inc.). Genomic DNA from the saliva samples was isolated using the salting-out method. We selected sixteen polymorphisms previously indicated to be associated with obesity, body weight regulation, or lipid metabolism, confirmed in the Middle Eastern population. These variants were within FTO, MTHFR, VDR, IL6, GPX1, FABP2, ADRB2, TCF7L2, ACE, APOC3, SOD2, LCT, TNF, CAT, CYP1A2, and ADRB3 genes. Detailed information about these polymorphisms is reported in Table 2. Genotyping for all the variants was conducted using the ARMS-PCR method. For each polymorphism, one common primer and two allele-specific primers were designed. Each PCR reaction contained 100 ng DNA, 15 µl KBC Alpha PCR Mix (Kawsar Biotech Co., KBC, Tehran, Iran), 1 U KBC Taq DNA polymerase, and 5 pM of each forward and reverse primers. PCR parameters were as follows: an initial denaturation step for 7 min at 95 °C, 1 min at 95 °C, 1 min at 62 °C, 1 min at 72 °C, and final extension for 15 min at 72 °C for 30 cycles.
Statistical analysis
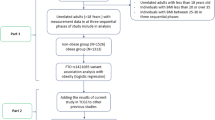
The deviation from Hardy–Weinberg equilibrium (HWE) was assessed using the X2 test. The test for linkage disequilibrium was performed using the ETDT program. Due to the difference in age and sex in case and control groups, the anthropometric characteristics of the studied individuals were analyzed by ANCOVA adjusted for sex and age. According to previous studies, genotypes of each variant were coded 0, 1, and 2 based on the number of risk alleles. For the first step, a generalized linear model (GLM) was used to establish a primary cut-off point (p < 0.25) as an initial filtration to refine the search to seven SNPs. This step was performed to exclude a set of SNPs that did not show a promising behavior in predicting BMI. Then for those seven SNPs that passed the initial filtration, a step-by-step logistic regression algorithm was used to select significant SNPs under the standard P value threshold (p < 0.05) and integrate them into a multivariate model. The goodness of fit of this model was analyzed by a calibration curve. The independent variables were age, sex, and the selected polymorphisms in both steps, and the dependent variables in the first and second steps were BMI and obesity status, respectively. The unweighted GRS was calculated by summing the number of risk alleles from the five significant SNPs. Also, to construct the weighted GRS, the odds ratio of each significant SNP was used as a measure of their effect sizes. The effect sizes were multiplied by the number of risk alleles at each locus and then the sum of these scores assigned to participants. As logistic regression was used at the final step of building the models, the output ranges of both models were between 0 and 1. A flow chart of this process is illustrated in Fig. 1. To estimate and compare the discriminative power of the constructed GRSs, the area under the receiver operating characteristic (ROC) curves was calculated (Dorfman and Alf Jr 1969). A tenfold cross-validation analysis was performed to validate the models internally. To perform this analysis, our dataset was divided into 10 groups, utilizing nine of them to create and train the models and the other one group to validate the models.
Analysis and model construction workflow. CV cross validation. (1) A step-by-step logistic regression was used to select significant SNPs (p < 0.05). At this step, obesity status was the dependent variable. (2) Final logistic regression model using the genetic risk scores. This model was utilized instead of threshold finding for the risk scores. (3) This logistic regression model was used to determine the effect size of each SNP which was used in the construction of the weighted model. At each step of the cross-validation, the effect sizes were calculated solely based on the training dataset
The effect sizes were calculated at each fold using the training data points for the weighted GRS model. Pearson’s correlation coefficient was calculated to evaluate the correlation between unweighted and weighted GRS models. Lastly, the associations between BMI and both unweighted and weighted GRSs were analyzed using a simple linear regression model to evaluate the reliability of these models. All statistical analyses were performed using SPSS software (V.28.0). P values less than 0.05 were considered statistically significant. Some plots were drawn using Python (V.3.9.5) with scikit-learn (V.1.0.2) and matplotlib (V.3.4.2) libraries.
Results
The characteristics of obese and control participants are presented in Table 1. According to these results, the differences between the age and sex of obese and control subjects were statistically significant. The obese group had significantly higher BMI, height, weight, waist, and hip circumference values. Initially, 16 SNPs previously indicated to be associated with obesity were selected. Genotype frequencies, HWE P values, and minor allele frequencies (MAF) of the genetic variants analyzed in this study are summarized in Table 2. The genotype frequencies of all the studied polymorphisms except FABP2 rs1799883 and LCT rs4988235 polymorphisms were in HWE.
Moreover, there was no linkage disequilibrium between the studied SNPs (data not shown). Seven SNPs were determined to be potentially associated with BMI under the p < 0.25 cut-off among these genetic variants. The results of the generalized liner model are shown in Table 3. Among the seven variants, FTO rs9939609, IL6 rs1800795, ADRB3 rs4994, ADRB2 rs1042714, and MTHFR rs1801133 polymorphisms were found to be statistically associated with the risk of obesity (p < 0.05). The multivariate model´s odds ratios (OR) and confidence intervals (CI) for these five polymorphisms are shown in Table 3. A forest plot of overall effect sizes is displayed in Fig. 2. The effect sizes that calculated at each fold are displayed in Supplementary Table 1.
Two GRSs (weighted and unweighted) including these five SNPs were developed. Both of the constructed models were significantly associated with BMI (p < 0.05). The area under the mean ROC curve in the weighted GRS was a little more than the unweighted GRS (Fig. 3). A more detailed analysis of the corresponding AUC, sensitivity, specificity, accuracy, and negative predictive value is shown in Table 4. The scatter plot of unweighted and weighted GRS models is shown in Supplementary Fig. 1. The Pearson’s correlation coefficient for these models was 0.9938 with 95% CI of (0.9930, 0.9946).
Discussion
This is the first study to investigate the cumulative effect of obesity-associated polymorphisms in a divergent Iranian population. Two GRSs (weighted and unweighted) consisting of five SNPs were developed. Previous studies published on the obesity-specific GRS mostly focused on the European population. In a recent cohort of 1069 European adolescents, Seral Cortes et al. constructed 2 weighted and unweighted GRSs using 21 BMI-related SNPs, which both were significantly associated with BMI (p < 0.001) (Seral-Cortes, Sabroso-Lasa et al. 2021). Belsky et al. developed a 32-locus GRS which was a predictor of obesity among white Americans (AUC = 0.57) (Belsky, Moffitt et al. 2013). For European ancestry, Song et al. constructed a GRS which was associated with BMI, and the associations were stronger in women (Song, Zheng et al. 2018). Besides, other GRS development studies in adults, adolescents, and preadolescents have been conducted in different European, Chinese, white American, black American, and African populations (Domingue, Belsky et al. 2014; Liang, Sun et al. 2016; Viljakainen, Dahlström et al. 2019), but none in the Iranian population of any age.
In this study, 16 genetic variants previously indicated to be associated with obesity were assessed in the Iranian population. Among the studied polymorphisms, ADRB3 rs4994, FTO rs9939609, ADRB2 rs1042714, IL6 rs1800795, and MTHFR rs1801133 polymorphisms were significantly associated with obesity. Among these SNPs, ADRB3 rs4994 is the locus that explains the largest association with BMI. The ADRB3 and ADRB2 genes are part of the adrenergic system, a major contributor to energy expenditure and body weight regulation. These receptors are expressed in adipose tissue and skeletal muscles, and stimulate lipolysis in fat cells (Rankinen, Zuberi et al. 2006). This explains why polymorphisms in these genes are among the most studied candidates for genetic obesity predisposition. The ADRB3 is one of the only 6 genes which showed reproducible association with obesity during 15 years of research (Loos and Yeo 2021). Xie and colleagues performed a meta-analysis on 12,497 case and controls of East Asia’s population, and indicated that the ADRB3 rs4994 polymorphism significantly increases the risk of childhood and adolescent overweight/obesity (Xie, Hua et al. 2020). Moreover, the molecular alteration at the codon 64 of the ADRB3 is related to weight gain, insulin resistance, and type 2 diabetes. A large meta-analysis consisting of 4864 T2D patients and 8779 controls demonstrated a significant association between rs4994 and T2D in Asians (Ryuk, Zhang et al. 2017). Another recent study proposes that rs4994 alters the body’s metabolic function into lower lipolytic activity and resistance to weight loss; hence an appropriate weight loss program for carries is necessary (González-Soltero, de Valderrama et al. 2021). The rs1042714 variant which occurs at codon 27 alters ADRB2 function. Although there are conflicting results about the relationship between rs1042714 and obesity, two meta-analysis studies involving this polymorphism and obesity have provided consistent results. First, Jalba, et al., were able to clarify their association in three genetic models (heterozygote, homozygote, and dominant) -in Asians, Pacific Islanders, and American Indians (Jalba, Rhoads et al. 2008). In the most recent meta-analysis which involved 17 studies, authors showed that rs1042714 (Gln27Glu) was significantly associated with increased obesity risk in the heterozygote and dominant model in all studied populations (Zhang, Wu et al. 2014). The findings from the current study also suggests an obvious role of the ADRB3 and ADRB2 alleles in the development of obesity (O.R. 1.878 and 1.464, respectively).
The well-known variant within the FTO gene had the second place for effect size among the five significant polymorphisms, which indicates its high impact in predicting obesity in our models. This genetic variant has been associated with the risk of obesity among distinct populations and ages, including European, African-American and Mexicans (Villalobos‐Comparán, Flores‐Dorantes et al. 2008; Liu, Zhu et al. 2010). Moreover, some studies indicate that this SNP can be considered a prognostic marker of obesity risk among Iranians (Koochakpour, Esfandiar et al. 2019; Mozafarizadeh, Mohammadi et al. 2019; Ostadsharif, Ebrahimi et al. 2019). The A Allele of rs9939609 has also been shown to associate with several metabolic syndrome components such as diabetes mellitus, coronary artery disease, and triglyceride and total cholesterol measurement in European and Asian populations. (Zhou, Liu et al. 2012), and thereby could be considered an important preventive marker. According to what Smemo et al. indicated, this gene is responsible for regulating IRX3 expression which is related to body composition (Smemo, Tena et al. 2014). This gene is highly expressed in the hypothalamus, adrenal and pituitary glands, and is involved in controlling food intake, which suggests a probable role in the hypothalamic–pituitary–adrenal axis (Todendi, Klinger et al. 2019). In a meta-analysis including 27 observational studies in different populations, Gholami et al., also concluded that IL6 rs1800795 increases the risk of obesity based on different genetic models (Gholami, Sharifi et al. 2019). IL6 (174G/C) gene polymorphism can affect obesity by playing an important role in IL6 expression (Ibrahim, Gabre et al. 2017). A higher concentration of IL6 is related to activation of adipose tissue inflammation, leading to lipid accumulation in adipocytes and obesity (Todendi, Klinger et al. 2015). It is well-established that this cytokine has a crucial role in physiological conditions such as immune function, metabolism regulation, and oncogenesis, thus the association between rs1800795 and hypertension, insulin resistance, type 2 diabetes mellitus and cancer progression is no surprise (Mauer, Denson et al. 2015). Barati et al. demonstrated an association between presence of IL6 variant and metabolic syndrome components in Iranian obese patients (Barati, Ghazizadeh et al. 2019). In line with these findings, our results confirmed that rs1800795 is a potential risk marker for obesity in our population (O.R. 1.368).
In addition, the fifth SNP included in our GRS is a functional variant located on the MTHFR gene, and substantially decreases the activity of the MTHFR enzyme (nearly 50%) and inhibits the efficient deoxynucleoside synthesis and intracellular methylation reactions (Cho, Amin et al. 2017). MTHFR rs1801133 polymorphism is responsible for the increased risk of obesity and also coronary artery diseases, but the exact mechanism of its influence on obesity is unclear. (Luo, Lu et al. 2018; Fu, Li et al. 2019). A meta-analysis by Meng et al. provided significant evidence for this MTHFR mutation role in T2DM susceptibility in Asians (Meng, Liu et al. 2019). Other studies have reported the same association in the Iranian population (Raza, Abbas et al. 2017; Poodineh, Saravani et al. 2019). MTHFR is responsible for maintaining methionine and homocysteine balance, through its role in the one-carbon cycle. Decreased activity of this enzyme leads to a reduction in the conversion of 5,10-methylenetetrahydrofolate (5,10-MeTHF) to 5-methyltetrahydrofolate (5-MTHF), resulting in an elevated homocysteine level. Hence, improving the homocysteine levels could be helpful in the prevention and treatment of obesity and other related diseases (Fu, Zhang et al. 2018).
Both unweighted and weighted GRSs were statistically associated with the risk of obesity (P < 0.05). The area under the mean ROC in the unweighted GRS was 70.19% and in the weighted GRS increased to 70.22%. Comparing the difference between AUCs for these two models, no significant difference in discriminatory ability to estimate obesity was observed between the two models. And the Pearson’s correlation coefficient for these models show that they are highly correlated (r2 = 0.988). ROC curve is proposed to evaluate the usefulness of genetic profiling, and the magnitude of the area under ROC (AUC) should be more than 0.8 to indicate an acceptable test for clinical screening of high-risk individuals (Janssens, Aulchenko et al. 2006). Although statistical power was low in the current study, it meets an acceptable discrimination power for a model (AUC equal to or more than 0.7). Our findings may significantly contribute to identifying people with susceptibility to developing greater BMI and obesity-related problems. Although the BMI is a highly heritable trait (between 40 and 70%) (Maes, Neale et al. 1997), almost all previous GRS models based only on genetic factors explained just a small fraction of BMI variance and a more holistic approach considering the complexity of the biological system is required (Eichler, Flint et al. 2010). Other studies suggest that risk scores incorporating healthy life style scores and childhood obesity have higher prediction ability. However, identification and replication of obesity-associated variants and GRS construction still provide further insights into weight regulation (Locke, Kahali et al. 2015). As a result, the current GRSs could be useful to minimizing T2D, cardiovascular events and other obesity-related concerns, especially in the Iranian population, which has been poorly studied before. However, our study has several potential limitations, including small sample size, a lack of the previous GWAS on the Iranian population, small number of investigated risk loci and not incorporating environmental factors before modeling. Even though the GRS models were validated by performing a tenfold cross-validation analysis, it is clear that the optimal validation process would be in an independent cohort. The odds ratios used to construct the weighted model were small which can reveal the influences of other genetic variants and also non-genetic factors such as depression status. This can be an explanation for the similarity of the results produced by our GRS models. Even though the results of our models were not significantly different, the weighted model performed better and is promising for future studies.
In conclusion, our findings suggest that ADRB3 rs4994, FTO rs9939609, ADRB2 rs1042714, IL6 rs1800795, and MTHFR rs1801133 SNPs are associated with the risk of obesity. Moreover, our results suggest that the developed GRSs in this study could be considered useful genetic tools for anticipating the risk of obesity development in the Iranian population. More studies with larger sample sizes and considering non-genetic factors are critical to constructing more accurate models. The alarming prevalence of obesity growing each year is a public health problem for many countries including Iran. This kind of study, which provides new insights into the biology and genetics of this abnormality, will assist us in developing and implementing novel therapies in the future.
References
Albala C, Santos JL et al (2004) Intestinal FABP2 A54T polymorphism: association with insulin resistance and obesity in women. Obes Res 12(2):340–345
Al-Daghri NM, Guerini FR et al (2014) Vitamin D receptor gene polymorphisms are associated with obesity and inflammosome activity. PLoS One 9(7):e102141
Babai MA, Arasteh P et al (2016) Defining a BMI cut-off point for the Iranian population: the Shiraz Heart Study. PLoS One 11(8):e0160639
Barati E, Ghazizadeh H et al (2019) Association of the IL6 gene polymorphism with component features of metabolic syndrome in obese subjects. Biochem Genet 57(5):695–708
Belsky DW, Moffitt TE et al (2013) Development and evaluation of a genetic risk score for obesity. Biodemography Soc Biol 59(1):85–100
Berthier M-T, Paradis A-M et al (2003) The interleukin 6–174G/C polymorphism is associated with indices of obesity in men. J Hum Genet 48(1):14–19
Bordoni L, Marchegiani F et al (2017) Obesity-related genetic polymorphisms and adiposity indices in a young Italian population. IUBMB Life 69(2):98–105
Carlos FF, Silva-Nunes J et al (2013) Association of FTO and PPARG polymorphisms with obesity in portuguese women. Diabetes, Metab Syndr Obes: Targets Therapy 6:241
Cauchi S, Choquet H et al (2008) Effects of TCF7L2 polymorphisms on obesity in European populations. Obesity 16(2):476–482
Cho K, Amin ZM et al. (2017) Methylenetetrahydrofolate reductase A1298C polymorphism and major depressive disorder. Cureus 9(10)
Corella D, Arregui M et al (2011) Association of the LCT-13910C> T polymorphism with obesity and its modulation by dairy products in a mediterranean population. Obesity 19(8):1707–1714
Domingue BW, Belsky DW et al (2014) Polygenic risk predicts obesity in both white and black young adults. PLoS One 9(7):e101596
Dorfman DD, Alf E Jr (1969) Maximum-likelihood estimation of parameters of signal-detection theory and determination of confidence intervals—rating-method data. J Math Psychol 6(3):487–496
Eichler EE, Flint J et al (2010) Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 11(6):446–450
Flegal KM, Carroll MD et al (2002) Prevalence and trends in obesity among US adults, 1999–2000. JAMA 288(14):1723–1727
Fu L, Zhang M et al (2018) Gene-gene interactions and associations of six hypertension related single nucleotide polymorphisms with obesity risk in a Chinese children population. Gene 679:320–327
Fu L, Li Y-n et al (2019) Plausible relationship between homocysteine and obesity risk via MTHFR gene: a meta-analysis of 38,317 individuals implementing Mendelian randomization. Diabetes, Metab Syndr Obes: Targets Therapy 12:1201
Ghareeb D, Abdelazem AS et al (2021) Association of TNF-α-308 G> A (rs1800629) polymorphism with susceptibility of metabolic syndrome. J Diabetes Metab Disord 20(1):209–215
Gholami M, Sharifi F et al (2019) Association of interleukin-6 polymorphisms with obesity: a systematic review and meta-analysis. Cytokine 123:154769
González-Soltero R, de Valderrama MJBF et al (2021) Can study of the ADRB3 gene help improve weight loss programs in obese individuals? Endocrinología Diabetes y Nutrición (english Ed) 68(1):66–73
Hernández-Guerrero C, Parra-Carriedo A et al (2018) Genetic polymorphisms of antioxidant enzymes CAT and SOD affect the outcome of clinical, biochemical, and anthropometric variables in people with obesity under a dietary intervention. Genes Nutr 13(1):1–10
Ibrahim O, Gabre A et al (2017) Influence of interleukin-6 (174G/C) gene polymorphism on obesity in Egyptian children. Open Access Maced J Med Sci 5(7):831–835
Jalba MS, Rhoads GG et al (2008) Association of codon 16 and codon 27 β2-adrenergic receptor gene polymorphisms with obesity: a meta-analysis. Obesity 16(9):2096–2106
Janssens ACJ, Aulchenko YS et al (2006) Predictive testing for complex diseases using multiple genes: fact or fiction? Genet Med 8(7):395–400
Koochakpour G, Esfandiar Z et al (2019) Evaluating the interaction of common FTO genetic variants, added sugar, and trans-fatty acid intakes in altering obesity phenotypes. Nutr Metab Cardiovasc Dis 29(5):474–480
Liang J, Sun Y et al (2016) Genetic predisposition to obesity is associated with insulin secretion in Chinese adults: the cardiometabolic risk in Chinese (CRC) study. J Diabetes Complicat 30(7):1229–1233
Liu G, Zhu H et al (2010) FTO variant rs9939609 is associated with body mass index and waist circumference, but not with energy intake or physical activity in European-and African-American youth. BMC Med Genet 11(1):1–12
Locke AE, Kahali B et al (2015) Genetic studies of body mass index yield new insights for obesity biology. Nature 518(7538):197–206
Loos RJ, Yeo GS (2021) The genetics of obesity: from discovery to biology. Nat Rev Genet 23(2):120–133
Loos RJ, Yeo GS (2014) The bigger picture of FTO—the first GWAS-identified obesity gene. Nat Rev Endocrinol 10(1):51–61
Lu Y, Loos RJ (2013) Obesity genomics: assessing the transferability of susceptibility loci across diverse populations. Genome Med 5(6):1–14
Luo Z, Lu Z et al (2018) Associations of the MTHFR rs1801133 polymorphism with coronary artery disease and lipid levels: a systematic review and updated meta-analysis. Lipids Health Dis 17(1):1–15
Maes HH, Neale MC et al (1997) Genetic and environmental factors in relative body weight and human adiposity. Behav Genet 27(4):325–351
Mauer J, Denson JL et al (2015) Versatile functions for IL-6 in metabolism and cancer. Trends Immunol 36(2):92–101
Meng Y, Liu X et al (2019) Association of MTHFR C677T polymorphism and type 2 diabetes mellitus (T2DM) susceptibility. Mol Genet Genom Med 7(12):e1020
Miller M, Rhyne J et al (2007) APOC3 promoter polymorphisms C-482T and T-455C are associated with the metabolic syndrome. Arch Med Res 38(4):444–451
Mokdad AH, Ford ES et al (2003) Prevalence of obesity, diabetes, and obesity-related health risk factors, 2001. JAMA 289(1):76–79
Mozafarizadeh M, Mohammadi M et al (2019) Evaluation of FTO rs9939609 and MC4R rs17782313 polymorphisms as prognostic biomarkers of obesity: a population-based cross-sectional study. Oman Med J 34(1):56
Ostadsharif M, Ebrahimi YB et al (2019) (2019) Gene Polymorphisms of human FTO and obesity in part of Iranian population. Recent Adv Biol Med 5:10205
Poodineh M, Saravani R et al (2019) Association of two methylenetetrahydrofolate reductase polymorphisms (rs1801133, rs1801131) with the risk of type 2 diabetes in South-East of Iran. Rep Biochem Mol Biol 8(2):178
Rahmani A, Sayehmiri K et al (2015) Investigation of the prevalence of obesity in Iran: a systematic review and meta-analysis study. Acta Med Iran 53(10):596–607
Rankinen T, Zuberi A et al (2006) The human obesity gene map: the 2005 update. Obesity 14(4):529–644
Raza ST, Abbas S et al (2017) Association between ACE (rs4646994), FABP2 (rs1799883), MTHFR (rs1801133), FTO (rs9939609) genes polymorphism and type 2 diabetes with dyslipidemia. Int J Mol Cell Med 6(2):121
Ren D, Xu JH et al (2019) Association study between LEPR, MC4R polymorphisms and overweight/obesity in Chinese Han adolescents. Gene 692:54–59
Ryuk JA, Zhang X et al (2017) Association of β3-adrenergic receptor rs4994 polymorphisms with the risk of type 2 diabetes: a systematic review and meta-analysis. Diabetes Res Clin Pract 129:86–96
Saliba LF, Reis RS et al (2014) Obesity-related gene ADRB2, ADRB3 and GHRL polymorphisms and the response to a weight loss diet intervention in adult women. Genet Mol Biol 37:15–22
Saravani S, Miri H et al (2015) Association of catalase (rs7943316) and glutathione peroxidase-1 (rs1050450) polymorphisms with the risk of type 2 diabetes (T2DM). Mol Genet Microbiol Virol 30(4):216–220
Seral-Cortes M, Sabroso-Lasa S et al (2021) Development of a genetic risk score to predict the risk of overweight and obesity in European adolescents from the HELENA study. Sci Rep 11(1):1–11
Smemo S, Tena JJ et al (2014) Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507(7492):371–375
Song M, Zheng Y et al (2018) Longitudinal analysis of genetic susceptibility and BMI throughout adult life. Diabetes 67(2):248–255
Status WP (1995) The use and interpretation of anthropometry world health organization. WHO Expert Committee, Geneva, Switzerland
Todendi PF, Klinger EI et al (2015) Association of IL-6 and CRP gene polymorphisms with obesity and metabolic disorders in children and adolescents. An Acad Bras Ciênc 87:915–924
Todendi PF, Klinger EI et al (2019) Genetic risk score based on fat mass and obesity-associated, transmembrane protein 18 and fibronectin type III domain containing 5 polymorphisms is associated with anthropometric characteristics in South Brazilian children and adolescents. Br J Nutr 121(1):93–99
Urry E, Jetter A et al (2016) Assessment of CYP1A2 enzyme activity in relation to type-2 diabetes and habitual caffeine intake. Nutr Metab 13(1):1–9
Viljakainen H, Dahlström E et al (2019) Genetic risk score predicts risk for overweight and obesity in finnish preadolescents. Clin Obes 9(6):e12342
Villalobos-Comparán M, Flores-Dorantes MT et al (2008) The FTO gene is associated with adulthood obesity in the Mexican population. Obesity 16(10):2296–2301
Wang YC, McPherson K et al (2011) Health and economic burden of the projected obesity trends in the USA and the UK. Lancet 378(9793):815–825
Xie C, Hua W et al (2020) The ADRB3 rs4994 polymorphism increases risk of childhood and adolescent overweight/obesity for East Asia’s population: an evidence-based meta-analysis. Adipocyte 9(1):77–86
Zhang H, Wu J et al (2014) Association of Gln27Glu and Arg16Gly polymorphisms in Beta2-adrenergic receptor gene with obesity susceptibility: a meta-analysis. PLoS One 9(6):e100489
Zhou D, Liu H et al (2012) Common variant (rs9939609) in the FTO gene is associated with metabolic syndrome. Mol Biol Rep 39(6):6555–6561
Acknowledgements
We thank all the research staff of Dr. Zeinali’s Medical Genetics Laboratory.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
ND was responsible for designing the study, data collection, interpreting results, model development and writing the report. SBZ and MHSA contributed to sample analysis, data interpretation, writing the report, and created the tables. AS contributed to data analyses, model constructing and writing the report. SZ provided supervision in all the steps and writing the report.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Shuhua Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Damavandi, N., Soleymaniniya, A., Bahrami Zadegan, S. et al. Development of a genetic risk score for obesity predisposition evaluation. Mol Genet Genomics 297, 1495–1503 (2022). https://doi.org/10.1007/s00438-022-01923-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-022-01923-0