Abstract
Root-knot nematodes (RKNs, genus Meloidogyne) affect a large number of crops causing severe yield losses worldwide, more specifically in tropical and sub-tropical regions. Several plant species display high resistance levels to Meloidogyne, but a general view of the plant immune molecular responses underlying resistance to RKNs is still lacking. Combining comparative genomics with differential gene expression analysis may allow the identification of widely conserved plant genes involved in RKN resistance. To identify genes that are evolutionary conserved across plant species, we used OrthoFinder to compared the predicted proteome of 22 plant species, including important crops, spanning 214 Myr of plant evolution. Overall, we identified 35,238 protein orthogroups, of which 6,132 were evolutionarily conserved and universal to all the 22 plant species (PLAnts Common Orthogroups—PLACO). To identify host genes responsive to RKN infection, we analyzed the RNA-seq transcriptome data from RKN-resistant genotypes of a peanut wild relative (Arachis stenosperma), coffee (Coffea arabica L.), soybean (Glycine max L.), and African rice (Oryza glaberrima Steud.) challenged by Meloidogyne spp. using EdgeR and DESeq tools, and we found 2,597 (O. glaberrima), 743 (C. arabica), 665 (A. stenosperma), and 653 (G. max) differentially expressed genes (DEGs) during the resistance response to the nematode. DEGs’ classification into the previously characterized 35,238 protein orthogroups allowed identifying 17 orthogroups containing at least one DEG of each resistant Arachis, coffee, soybean, and rice genotype analyzed. Orthogroups contain 364 DEGs related to signaling, secondary metabolite production, cell wall-related functions, peptide transport, transcription regulation, and plant defense, thus revealing evolutionarily conserved RKN-responsive genes. Interestingly, the 17 DEGs-containing orthogroups (belonging to the PLACO) were also universal to the 22 plant species studied, suggesting that these core genes may be involved in ancestrally conserved immune responses triggered by RKN infection. The comparative genomic approach that we used here represents a promising predictive tool for the identification of other core plant defense-related genes of broad interest that are involved in different plant–pathogen interactions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants must overcome a wide range of biotic stresses in their natural habitats using a sophisticated perception and immune response system. Under intensive agricultural production, biotic stress conditions can drastically affect plant growth and development, leading to a severe decrease in crop yield. From 2005 to 2015, the effects of climate change substantially affected the agriculture, causing losses of up to $100 billion dollars, in part because of the attack of plants by pests and pathogens favored by these climatic conditions (FAO report 2017).
Among the global threats to the sustainability of agrosystems (Singh et al. 2015), the root-knot nematodes (RKNs) are globally the most damaging plant-parasitic nematodes (Jones et al. 2013). RKNs are obligate endoparasites in the Meloidogyne genus, with approximately 100 species described (Decraemer and Hunt 2006) that can parasitize almost all cultivated plant species in tropical and temperate regions (Trudgill and Blok 2001). The four most common RKN species are Meloidogyne incognita, M. arenaria, M. javanica, and M. hapla, which cause the majority of the yield losses in crops (Agrios 2005). Another important RKN is M. graminicola, a damaging parasite of both upland and lowland Oryza spp. (De Waele and Elsen 2007). RKNs establish feeding sites in plant roots by inducing the formation of multinucleated giant cells inside galls (Bird 1961; Cabrera et al. 2015).
The responses to RKN infection depend on the plant species and genotype (Jones et al. 2013), with a few species showing high resistance levels, such as those mediated by R genes that lead to the hypersensitive response (HR) triggered in the early stages of the infection. However, the introgression of plant resistance by R-gene-based breeding may lead to yield losses due to trade-off effects (Mota et al. 2019). Therefore, identifying the molecular mechanisms by which the plant immune system responds to RKNs is pivotal for the development of stable resistance and a sustainable crop production (Nobori and Tsuda 2019).
Transcriptome analysis of plants submitted to biotic stresses has been widely used to identify responsive genes (Cui et al. 2016; Jain et al. 2016). However, this method is often species-specific and, with the increase in the number of complete plant genomes, has been rapidly been replaced by new approaches. One of these new approaches is comparative genomics, which allows the simultaneous analysis of several plants to identify putative evolutionarily conserved genes, including those involved in important functions such as defense mechanisms (Emms and Kelly 2015).
Different methods for inference of gene homology across taxa have been developed to identify, using sets of proteins, orthologs, and in-paralogous groups. Orthologs are defined as homologous genes originating from a speciation event, and in-paralogs are genes originating from a duplication event that occurred, after speciation and that are thus species-specific. Despite being derived from one common ancestral gene, orthologous genes might display functional divergence between species (Remm et al. 2001). Many orthology inference methods are based on the use of reciprocal best similarity relationships (e.g., by BLAST) to cluster proteins into groups of orthologs and in-paralogs. Currently, the genomes of at least 236 angiosperms have been completely sequenced (Chen et al. 2018). The availability of a variety of plant genomes from different orders and families constitutes an information-rich resource and allows a more accurate inference of orthologous relationships than was previously possible. In the present study, we compared the genomes of 9 monocot and 13 dicot plant species to identify core evolutionarily conserved genes. These species were selected based on the quality of their genome assembly, their economic relevance, and their responsiveness to different types of stress. To root our comparative genomics analysis, we included the genome of Amborella trichopoda, which is a closely related angiosperm outgroup (Albert et al. 2013).
The use of available transcriptome data from plants challenged with different types of stresses can subsequently identify specific stress-responsive orthologous groups and may promote biotechnological strategies for improving crop yield and suitability (Cohen and Leach 2019).
The present study aimed to identify RKN-responsive genes that are evolutionarily conserved across a wide range of plant species displaying distinct degrees of resistance. We used RNA-seq transcriptome data from different plant species challenged by RKNs to identify nematode-responsive genes and combined this information with homology inference to discover those that are evolutionarily conserved. The combination of strategies, including transcriptional and comparative genomics, will improve the prediction of gene and protein functions, and help to clarify general molecular mechanism of plant defense against root-knot nematodes.
The transcriptome data of four RKN-resistant plant species (Arachis stenosperma, Coffea arabica, Glycine max, and Oryza glaberrima) infected with three different Meloidogyne spp. (M. arenaria, M. incognita, and M. graminicola) (Albuquerque et al. 2010; Beneventi et al. 2013; Guimaraes et al. 2015; Petitot et al. 2017) were combined with the homology database generated in the present study, aiming to identify clusters of proteins commonly regulated during the response to RKN infection in the four pathosystems studied.
In addition, the generation of homologous groups (orthologous and in-paralogous) of proteins will produce a robust database of evolutionarily conserved plant genes, and a resource of general interest to be shared with the scientific community.
Methods
Comparative genomic analysis and orthogroup identification
For the comparative analysis, we chose 22 plant species based on their phylogenetic distribution and economic relevance. When a genome of sufficient quality was available, it was the preferred source with which to retrieve the corresponding predicted proteome (Suppl. Table 1). These predicted proteomes were retrieved from public and specialized databases (https://peanutbase.org and https://phytozome.jgi.doe.gov v11). For two plant species of interest (Medicago sativa and A. stenosperma), no genome was available and we had to retrieve the sets of proteins predicted from the transcriptomes. For M. sativa, the predicted proteome was retrieved from the transcriptome assembly published by Postnikova et al. (2015). For A. stenosperma, we assembled the transcriptome and predicted the corresponding proteins as explained in the corresponding method section. Overall, these species are representatives of Eudicotyledoneae and Monocotyledoneae, and include genotypes resistant to RKNs and/or tolerant to drought stress as well as plant models with well-characterized gene sets. We also used the Angiospermae species Amborella trichopoda as an outgroup species. To assess the completeness of the predicted proteomes, we used BUSCO v3 (Simão et al. 2015) in protein mode with the plant odb9 data set. To place the genome comparisons within a phylogenetic framework, we reconstructed a dated phylogeny of the 22 species based on Zanne et al. (2013), using TreeGraph v2 (Stöver and Müller 2010). The whole proteomes of the 22 selected species were compared and analyzed by OrthoFinder v1 (Emms and Kelly 2015) for the identification of orthogroups. The first step of OrthoFinder analysis consists of an all-against-all BLASTP step to find the best reciprocal hit for each protein. The blast results were processed by OrthoFinder using the default inflation values. The resulting file was used as input to Family-Companion software (Cottret et al. 2018), which generates statistical matrices of gene abundance per orthogroup, the presence and absence of orthogroups per species, and the percentages of proteins present in orthogroups. To determine whether the selected set of 22 species provided a good representation of the diversity present in angiosperms, we used the KinFin software v1.0 (Laetsch and Blaxter 2017), which produces a rarefaction curve as a function of the number of species present in the comparative analysis.
Functional annotation of plant proteomes
We used PfamScan v1.5 (Mistry et al. 2007) to predict conserved PFAM domains (version 30) in each of the 22 plant protein sets. The PFAM domains were then used to assign the corresponding Gene Ontology terms using the Pfam2Go association file (https://current.geneontology.org/ontology/external2go/pfam2go) with a custom script.
Transcriptome data analyses
To identify nematode-responsive genes, we used the transcriptome data of four RKN-resistant plant genotypes under Meloidogyne spp. infection previously generated by our groups: C. arabica (accession UFV 408-28) resistant to M. incognita, A. stenosperma (accession V10309) resistant to M. arenaria (Guimaraes et al. 2015), O. glaberrima (TOG5681) resistant to M. graminicola (Petitot et al. 2017), and G. max (cultivar PI 595,099) resistant to M. incognita.
RNA sample preparation and transcriptome sequencing
A. stenosperma inoculated with M. arenaria: Plants of A. stenosperma grown in a greenhouse were inoculated with M. arenaria infective juveniles (J2), and root samples were collected before inoculation (uninoculated control) and at three time-points (3, 6, and 9 days after inoculation; DAI) from two biological replicates according to Morgante et al. (2013). The total RNA isolation from samples, the cDNA synthesis, and the Illumina transcript sequencing were performed by Guimaraes et al. (2015).
O. glaberrima inoculated with M. graminicola: ten-day-old plantlets of O. glaberrima grown in a controlled growth chamber were inoculated with M. graminicola J2 juveniles. Root tip samples were collected before inoculation (uninoculated control) and at three time-points (2, 4, and 8 DAI) from two biological replicates, RNA was extracted, cDNA was synthesized, and the Illumina transcript sequences were produced by Petitot et al. (2017).
G. max inoculated with M. incognita: A total of 120 4-week-old plants of G. max grown in a greenhouse were challenged with 1,350 M. incognita J2 juveniles, and root samples were collected at zero (uninoculated control), 4, 8, and 12 DAI from three biological replicates. Total RNA was extracted from whole roots using a ReliaPrep™kit (Promega©, Wisconsin, USA) according to the manufacturer's instructions and the cDNA was produced using the SuperScript II enzyme and oligo (dT) 20 primer (Invitrogen, Carlsbad, USA). The TruSeq ™ SBS v5 protocol was used for the library construction (Illumina, San Diego, CA), and the 12 resulting libraries were sequenced on an Illumina HiSeq 4000 at the University of Illinois (USA).
C. arabica inoculated with M. incognita: A total of 30 6-week-old plants of C. arabica grown in a greenhouse were inoculated with 5000 M. incognita juveniles (J2) and root samples were collected at zero (uninoculated control) and 6 DAI from three biological replicates, as previously described (Albuquerque et al. 2010). Total RNA was extracted from the root samples using an RNeasy mini kit (Qiagen, Hilden, Germany), and the cDNA was produced using the SuperScript II enzyme and oligo (dT) 20 primer (Invitrogen, Carlsbad, USA). The TruSeq ™ SBS v5 protocol (Illumina, San Diego, CA) was used for library construction (Illumina, San Diego, CA), and the six resulting libraries were sequenced on an Illumina HiSeq 2500 at the University of Illinois (USA).
To enable reliable comparative and statistical analyses, all of the raw Illumina transcript raw sequences previously obtained for each of the plant–RKN interactions described above (12 libraries for each A. stenosperma, O. glaberrima, and G. max interaction and six libraries for C. arabica) were reanalyzed using the pipeline below:
Reads were trimmed using Trimmomatic v0.39 (Bolger et al. 2014) according to their quality, based on the Phred 33 score assessed by FastQC, using the following parameters: LEADING: 3, TRAILING: 3, SLIDINGWINDOW: 4:15, and MINLEN: 36. Trimmomatic was then used to eliminate Illumina adapters (TruSeq adapter primers).
For the four plant genotypes, the mapped reads were quantified into the corresponding predicted transcriptomes using Kallisto v0.46.1 (Bray et al. 2016) with the default settings to obtain transcript counts and abundances. For A. stenosperma, we used the genome-guided transcriptome assembly that we obtained as part of the current analysis. For G. max, transcripts predicted from the reference genome annotation were used (version Wm82.a2.v1; https://phytozome.jgi.doe.gov/). For O. glaberrima and C. arabica, the predicted transcriptomes from the closest available genomes were used (O. sativa and C. canephora, respectively; https://phytozome.jgi.doe.gov/).
The transcript counts obtained by Kallisto were used as inputs for two methods of differential expression detection: EdgeR v3.28.1 (Robinson et al. 2010) and DESeq2 v1.26.0 (Anders et al. 2013). Genes were considered differentially expressed genes (DEGs) when their relative gene expression levels had an adjusted p-value (false discovery rate, FDR) < 0.05 and at least a fourfold change (FC) value between inoculated and control samples (Log2FC > 2 or < − 2). Only the genes identified as DEGs in both DESeq and EdgeR statistical analyses were considered for further analyses.
Gene Ontology Enrichment in genes responsive to nematode infection
We used the hypergeometric test implemented in the FUNC package v.0.4.7 (Prüfer et al. 2007) to identify Gene Ontology terms that were significantly overrepresented in the group of genes responsive to nematode infection in comparison with the rest of the evolutionarily conserved genes. Two main analyses were conducted: a comparison between the differentially expressed genes from the four species and the whole set of genes belonging to orthogroups found in all the 22 species, and a second comparison against only the orthogroups shared among the four species. Only the terms with a family-wise error rate (FWER) < 0.05 for overrepresentation or underrepresentation were considered in the analysis.
MapMan functional analysis of differentially expressed genes
We conducted a MapMan analysis v3.6.0RC (Thimm et al. 2004) of the genes differentially expressed upon nematode infection for each of the four species studied (A. stenosperma, C. arabica, G. max, and O. glaberrima). The MapMan ontology was inferred using the online tool Mercator v3.6 (https://www.plabipd.de/portal/mercator-sequence-annotation). Mercator assigned MapMan BINs (functional categories/subcategories) to the transcript sequences of each transcript identified as a DEGs by the statistical analyses and incorporated BLAST and CDD searches as well as an InterProScan annotation for each species/time-point.
Validation by quantitative RT-PCR (qRT-PCR)
Differential gene expression upon nematode infection in each species was further validated by qRT-PCR using three technical replicates, a Platinum SYBR Green qPCR Super Mix-UDG w/ROX kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s recommendations and a StepOne Plus Real-Time PCR System (Applied Biosystem Foster City, CA, USA). The cDNA samples of A. stenosperma and O. glaberrima used were produced by Guimaraes et al. (2015) and Petitot et al. (2017), and the cDNA samples of G. max and C. arabica were produced as previous described. Only one time-point was used for each species [9 DAI (A. stenosperma), 6 DAI (C. arabica), 12 DAI (G. max), and 2 DAI (O. glaberrima)]. Specific primers were designed for a single nematode-responsive gene per orthogroup for each species using Primer3Plus (https://primer3plus.com/) software as previously described by Morgante et al. (2013). The specific primers and the reference gene sequences are listed in Suppl. Table 2. The cycle threshold (Cq) values were estimated with Real-Time PCR Miner (Zhao et al. 2005) and normalized using the specific reference genes for each species. The relative expression of the inoculated samples versus uninoculated control samples was analyzed with the REST 2009 software (Pfaffl et al. 2002).
Results
Identification of evolutionarily conserved plant genes
A dated phylogenetic tree was constructed based on Zanne et al. (2013) using 22 plant species representing the following botanical families: Fabidae, Malvidae, Asteridae, and Poaceae. These species were chosen as they are widely studied plant models (e.g., A. thaliana) with a wealth of genomic data available, or harbor resistance to different biotic and abiotic stresses, with sequenced genomes displaying high-quality parameters, such as high completeness and good gene annotation. Overall, the 13 species belonging to Eudicotyledoneae, the eight belonging to Monocotyledoneae, and the outgroup (A. trichopoda) spanned more than 214 Myr of plant evolution. Five genera had two or more representatives (Oryza spp., Solanum spp., Gossypium spp., Arachis spp., and Medicago spp.) due to their economic relevance (Fig. 1).
The predicted proteomes of the 22 plant species represented a total of 933,295 proteins (Suppl. Table 1), with the number of predicted proteins in each species varying from 25,574 (C. canephora) to 100,344 (T. aestivum) genes. Completeness scores assessed by BUSCO ranged from 70% (O. thomaeum) to 99% (A. thaliana). This high score for A. thaliana is expected as it is a plant model included in the BUSCO dataset, while less-studied and more distantly related species are expected to show a lower BUSCO score (Suppl. Table 1).
The comparative analysis of these 22 proteomes using OrthoFinder assigned the great majority of the proteins (773,931; 83%) to 35,238 orthogroups (Suppl. Table 1; Fig. 1). The percentage of proteins assigned to orthogroups varied according to the species, with the highest rates for O. glaberrima and G. raimondii (95% and 93%, respectively) and the lowest for A. stenosperma and Z. mays (57% and 59%, respectively) (Suppl. Table 1; Fig. 1).
Using KinFin, we showed that our selection of 22 plant species was representative of the diversity of proteins present in angiosperms. Indeed, the rarefaction curve showed that a plateau was reached at 20 species, indicating that the addition of more species to the homology inference would not provide any substantial novelty in the number and diversity of groups (Suppl. Fig. 1).
Of the 35,238 orthogroups, only 692 were entirely composed of species-specific in-paralogs (generated from a duplication event in one species and showing no orthologous in the other species). The rest of the orthogroups (98%) were thus shared by at least two different plant species and inherited from a common ancestor. Triticum aestivum harbored the largest number of species-specific in-paralogous groups (12.86%; 89 orthogroups), as it has undergone several events of duplication during its evolution (Pont et al. 2011).
We found 6,132 orthogroups conserved among all 22 plant species (PLAnt Common Orthogroups—PLACO) (Suppl. Fig. 2; Suppl. Table 3). These orthogroups represent a core of conserved genes and are likely involved in plant basal biochemical processes such as photosynthesis, respiration, and carbon metabolism.
The largest number of common orthogroups (PLACO) corresponded to the orthogroups shared among all 22 species (6,132), confirming the robustness of the data set for the identification of evolutionarily conserved core genes. The next most numerous orthogroups were those found in plant species belonging to the same genus, with Oryza spp. (5,275), Arachis spp. (1,543), Gossypium spp. (1,279), Medicago spp. (1,181), and Solanum spp. (989) showing the largest numbers of orthogroups (Suppl. Fig. 2).
Functional annotation using the PFAM conserved domains of the 933,295 proteins predicted in the 22 proteomes revealed 4,637 different domains in total, with 59% of proteins shared among all the species (2,753 domains) and 7% of those specific to M. truncatula, S. lycopersicum, and M. sativa showing the largest numbers of exclusive domains (Fig. 2).
Overall, this orthology analysis provides a resource with which to study core conserved genes and their corresponding functions as well as more species-specific gene families that may be related to traits specific to some taxonomic groups. In this analysis, we were interested in the genes that were universal to all the plant species included, because they could reveal central mechanisms of plant defense.
Identification of nematode-responsive genes conserved in RKN-resistant plant genotypes
To identify nematode-responsive genes conserved among several plant species known to be resistant to RKN infection, we used transcriptomes from the following plant species: C. arabica (Albuquerque et al. 2010), G. max (Beneventi et al. 2013), O. glaberrima (Petitot et al. 2017), and A. stenosperma (Proite et al. 2008). We identified 9323 orthogroups encompassing a total of 98,502 proteins that were common to at least the four RKN-resistant plant genotypes of interest (Fig. 3). In addition to these conserved orthologous genes, 8457 were in-paralogous groups specific to O. sativa, 2170 were specific to A. stenosperma, 1261 were specific to G. max, and 610 were specific to C. canephora (Fig. 3). The substantial difference in the number of in-paralogous groups among these species is probably due to duplication events that occurred after their differentiation. As expected, the two legume species (A. stenosperma and G. max), with the smallest evolutionary distance (Fig. 1), shared the largest number of common orthogroups (736).
Intersection of orthogroups among A. stenosperma (Aste), C. canephora (Ccan), G. max (Gmax), and O. sativa (Osat). The vertical bars represent the numbers of orthogroups. The horizontal bars represent the numbers of orthogroups per species. The black dots indicate the intersections among the four species
Identification of nematode-responsive genes in resistant plant cultivars
To identify genes differentially regulated upon nematode infection compared to uninfected controls, RNA-Seq data sets from the four RKN-resistant genotypes (A. stenosperma, coffee, soybean, and rice) were uniformly reanalyzed using the same pipeline. Only the genes identified as DEGs (FDR < 0.05 and Log2FC > 2 or < − 2) by two statistical software programs (EdgeR and DESeq) were used for further analyses. The overall analysis allowed the identification of 4,658 genes differentially expressed (DEGs) in response to RKN inoculation, with 2597 in O. glaberrima, 743 in C. arabica, 665 in A. stenosperma, and 653 in G. max unique genes distributed in all the time-points (Fig. 4a). The number of DEGs observed in O. glaberrima was four times larger than that observed in the other species, which might be due to the distinct infection mechanism of M. graminicola (Petitot et al. 2017). Most DEGs were upregulated upon nematode infection in A. stenosperma, G. max, and O. glaberrima, (486, 484 and 2,318 DEGs, respectively), with expression values varying from 5-to tenfold (Fig. 4b). In contrast, in C. arabica, most DEGs (598) were downregulated upon RKN infection (Fig. 4a, b). Since C. arabica and O. glaberrima do not have reference genomes available or sufficient transcriptome data with which to generate a comprehensive transcriptome database, the most closely related species from each genus, C. canephora, and O. sativa were used in the comparative analysis.
Differentially expressed genes (DEGs) in response to RKN inoculation in A. stenosperma, C. canephora, G. max, and O. sativa. a Bar plot representing the total numbers of DEGs in the four species. The red bars indicate upregulated genes, and the blue bars indicate downregulated genes. b Expression values of DEGs found in the four species studied. The expression values correspond to all the time-points for each plant species. Each species is represented by its own color, and the y-axis shows Log2FC values obtained from DESeq
Identification of evolutionarily conserved genes responsive to nematode infection
To identify evolutionarily conserved genes responsive to RKN infection, we combined the orthology analysis with differential expression analysis. The comparison of the 9323 orthogroups common to the four genotypes with the 4658 DEGs identified as RKN-responsive revealed 17 orthogroups containing at least one DEG from each of the four plant genotypes (Fig. 5). Interestingly, these 17 orthogroups were further conserved in the PLACO of all 22 plant species analyzed, suggesting core genes important for biotic stress response. Overall, these 17 orthogroups encompassed a total of 3661 proteins belonging to the four target species, with 364 proteins encoded by genes identified as DEGs in one unique time-point of each species [9 DAI (A. stenosperma), 6 DAI (C. arabica), 12 DAI (G. max), and 2 DAI (O. glaberrima)]. In addition to harboring the largest number of in-paralogous, O. sativa is the specie with the largest number of orthogroups in bilateral relations (only two species), with 95 orthogroups shared with C. canephora, 81 with A. stenosperma, and 80 with G. max (Fig. 5). On the other hand, the large number of in-paralogous groups of A. stenosperma might be due to the use of a comprehensive transcriptome in place of a reference genome. The expression profile analysis of the 364 RKN-responsive DEGs belonging to the 17 common orthogroups revealed that the majority (80%) were upregulated in A. stenosperma, G. max, and O. sativa. Conversely, in C. canephora, 73% of the DEGs were downregulated in response to RKN infection. Although expression values varied from 5- to 7.5-fold in G. max and O. sativa, respectively, most genes showed a two-to-threefold change in expression (Fig. 6).
Expression profile of 364 DEGs belonging to 17 common orthogroups in from four species in one unique time-point (A. stenosperma, C. canephora, G. max, and O. sativa). Different colors represent the four species. Each dot represents a different gene. Expression data obtained from DESeq (Log2FC) indicate upregulation or downregulation, as positive and negative values, respectively
Functional analysis of universally conserved nematode-responsive genes
Mercator analysis: The 364 RKN-responsive DEGs identified in the 17 orthogroups were grouped by Mercator into 12 functional categories (Fig. 7; Suppl. Table 4). Most of the DEGs (32%) were assigned to signaling, followed by cytochrome P450 (18%) and stress and secondary metabolism (11%), as well as different categories of cell wall-related functions (a total of 17%). Accordingly, the two orthogroups containing the largest numbers of genes were mainly assigned to PFAM domains characteristic of kinases and of cytochrome P450s. The signaling category encompasses plant proteins involved in the first responses to stress and pathogen perception, such as LRR-kinases and glycoprotein-like proteins, while assigned to the stress, cytochrome P450 and secondary metabolism categories are those related to common resistance responses to pathogens. In the transcription factor (TF) category, genes coding for the ethylene-response factor (ERF)/AP2 subfamily was induced in the four plant species. In addition, genes encoding heat shock transcription factors (HSFs) were also downregulated in the four plant species. Regarding hormone metabolism, only genes associated with the cytokinin metabolism were identified as regulated in all four species.
Identification of orthogroups containing universally conserved nematode-responsive genes
The numbers of DEGs identified in the 17 orthogroups conserved among the four plant genotypes were uneven among species, with G. max and O. sativa containing the largest numbers of genes (81 and 180, respectively) and A. stenosperma and C. canephora containing the fewest (44 and 59, respectively). Three orthogroups (OG0000001, OG0000003, and OG0000004) encompassed the majority (48.2%) of the 364 DEGs (Suppl. Table 5). However, each of these orthogroups contained thousands of genes, and each orthogroup was associated with multiple different PFAM domains. Nevertheless, these three orthogroups included important signaling proteins such as kinases and receptor-like proteins as well as proteins involved in secondary metabolite production, such as cytochrome P450. Thus, they could not be precisely functionally annotated and were not considered in further analysis. Among the remaining 14 orthogroups, OG0000019, OG0000035, and OG0000036 encode pectinesterases, α-expansins, and ERF/AP2 transcription factors, respectively, which correspond to widely known functions in biotic stress responses (An et al. 2008; Marowa et al. 2016; Xie et al. 2019). The orthogroup OG0000179 encodes members of the dirigent gene family that may be involved in defense responses to pathogens as well (Li et al. 2017). Interestingly, other orthogroups, such as OG0000309, which was highly enriched in G. max, include genes involved in reproduction and development, such as fasciclins (PF02469) (Johnson et al. 2003; Cagnola et al. 2018). The orthogroup OG0000056 contains members of the UDP-glucosyl transferase family known to regulate by glucosylation the activity of some compounds that play important roles in plant defense against pathogens, including salicylic acid (SA) and other phenylpropanoids (Chong et al. 2002). Two other groups, named OG0000057 and OG0000053, assigned to cupins and hydrophobic seed PFAM domains, respectively, were enriched in O. sativa compared to the other three species. These two orthogroups were previously described to play a role in stress tolerance (Dunwell et al. 2008; Finkina et al. 2016). Finally, orthogroup OG0000014 included a small set of genes encoding oligopeptide transporters putatively able to transport small peptides, including the tripeptide glutathione, as well as nitrate and amino acids (Pike et al. 2009).
Experimental validation of differential expression for the most interesting orthogroups
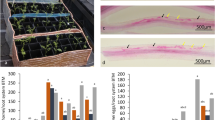
To validate the expression profiles of DEGs belonging to the orthogroups identified in the previous section, we designed specific primers according to Morgante et al. (2013). From all the DEGs in each orthogroups, only the genes with primers meting the parameters of Primer3 were kept. In total, 88 DEGs from the four species were tested by quantitative RT-PCR (qRT-PCR). Only the primers with specificity of amplification confirmed by the melt curve analysis were kept. The relative expression of the genes was assessed by the comparison of control and inoculated samples. For simplicity, only the earliest DAI showing an incompatible plant-nematode response was used for each genotype: 9 DAI (A. stenosperma), 6 DAI (C. arabica), 12 DAI (G. max), and 2 DAI (O. glaberrima). For each species, the uninoculated treatment was used as a control. The gene model of each species for one unique time-point is described in the Suppl. Table 6. The qRT-PCR analysis showed that, with one exception (OG0000053), all genes selected in the orthogroups showed significantly different gene expression upon nematode infection compared to the controls in at least two plant species (Fig. 8). In six orthogroups, the representative gene of three of the four species showed significantly differential expression upon nematode infection (OG0000014, OG0000019, OG0000036, OG0000050, OG0000056, and GO0000057). Finally, in one orthogroup (OG0000035), all the representative genes of the four plant species of interest showed significant differential gene expression following nematode infection. These genes were significantly upregulated in coffee, rice, and soybean, but downregulated in Arachis. The orthogroup OG0000309, assigned to a fasciclin, contained 20 G. max DEGs, all of which were upregulated. This was in a striking contrast to the other species, which showed only one fasciclin DEG per species (vs. 20 for G. max) and downregulation in both in silico RNA-seq and qRT-PCR analyses (Fig. 8).
Relative quantification of mRNA levels of DEG representatives of 13 orthogroups from A. stenosperma, C. arabica, G. max, and O. glaberrima inoculated with Meloidogyne spp. relative to the control (uninoculated). Values are the log2 of the expression levels, and significant (P < 0.05) differences between inoculated and control samples are marked with asterisks (*)
Discussion
Evolutionarily distant plant species have genes with similar functions
Despite the 22 species of monocots and dicots spanning more than 214 Myr of divergence and evolution from their last common ancestor, we identified 6132 orthogroups encompassing 3,939,809 genes universally conserved among the species. The evolutionarily conserved genes which we identified across the angiosperms probably correspond to core genes involved in essential common processes and functions. Although other studies have used this same strategy to detect orthogroups, this was the first time that a such a large number of plant species spanning > 200 Myr of evolution has been analyzed (Yoshida et al. 2017; Bel et al. 2018; Woldesemayat et al. 2018). The choice of species was based on their genome quality, phylogenetic diversity, economical relevance, and unique responses to different biotic and abiotic stresses.
Although orthology information across long evolutionary distances is informative and reveals core conserved genes, this information alone is not enough to directly transfer the functionality of a gene to an orthologous gene (Joshi and Xu 2007; Das et al. 2016; Liu et al. 2018; Yunes and Babbitt 2019).
Furthermore, duplications preceding or following separation of species can lead to functional divergence between paralogous gene copies, and this is important to take into account. In our study, out-paralogs that resulted from duplications occurring prior to separation of the species were placed in different orthogroups. However, in-paralogs that resulted from species-specific duplications after speciation were clustered together in the same orthogroup. As expected, in-paralogous groups were observed in all four species of interest, and some are probably involved in key functions, such as adaptive responses to different challenges, including defense responses to environmental conditions that each species faces, as observed by (Das et al. 2016).
Overall, the 35,238 orthogroups which we detected by comparing the 22 plant species constitute a rich and useful resource that can be further used for a variety of evolutionary or functional genomics projects. For instance, the 6123 universally conserved plant orthogroups (PLACO) that we identified could be used to infer the minimum ancestral plant gene set or study expansions and contractions of gene families across the plant tree of life. Similarly, orthogroups specific to certain phyla in the tree can point to functions specific to adaptations of the plants to some lifestyles, environments, or other peculiarities.
In this study, we combined the inference of orthologous groups and gene expression data from four RKN-resistant genotypes to identify evolutionarily conserved plant genes responsive to nematode infection. This could point to interesting candidate genes possibly involved in plant resistance to nematode infection. The previous studies using a similar approach identified commonly regulated genes in Sorghum, enabling the construction of a functional genomic database for the genus (Tian et al. 2016) and the identification of genes commonly regulated by drought, salt, cold, heat, and oxidative stresses (Woldesemayat et al. 2018).
In addition to orthology information, we also analyzed the conservation of functional protein domains based on PFAM annotation. Overall, we found that of the 4637 different PFAM domains identified in the 22 plant proteomes, 59% were conserved among all the species. This finding corroborates the previous works, stating that plants inherited most of their protein-coding genes from a common ancestor, and most likely have conserved most of the functions (Panchy et al. 2016; Qiao et al. 2019).
A number of DEGs for each species show the specialized responses
The differential gene expression analysis of four resistant plant genotypes (A. stenosperma, C. arabica, G. max, and O. glaberrima) infected with different Meloidogyne species (M. arenaria, M. incognita, and M. graminicola) revealed markedly different numbers of nematode-responsive genes among the species. These differences can be associated with a number of parameters, including the evolutionary distances between these species, the resistance levels, and mechanisms displayed by the genotypes used (HR, nematode penetration delay, and reduction in the numbers of galls and eggs) and the inoculum virulence. As expected, when phylogenetically closer species are studied together and apart from the rest (e.g., the legumes A. stenosperma, and G. max), the number of genes differentially expressed in common increases considerably. In addition, the use of different RKN species can activate slightly different and specialized responses, with different plant genotypes also showing different levels of responses (Jones et al. 2013).
The combination of comparative genomics and differential gene expression data from four resistant genotypes allowed the identification of 17 universally conserved orthogroups containing at least one nematode-responsive gene from each of the 4 plants of interest. These orthogroups harbor genes related to signaling, production of secondary metabolites, cell wall constitutive proteins or enzymes involved in cell wall modifications, peptide transport, transcription regulation, and plant defense, suggesting that, despite their evolutionary distance, these species have kept a set of commonly regulated genes to respond to nematode infection. The conservation of a network of common genes involved in plant defense responses in different species has also been demonstrated for multiple stresses by computational approaches comparing public databases (Breitling et al. 2005; Shaik and Ramakrishna 2014; Amrine et al. 2015).
Plant‐pathogenic nematodes secrete cell wall-degrading enzymes (CWDEs), representing a significant proportion of the encoded proteins of their genomes (Bird et al. 2009). Alterations to plant cell wall integrity by these CWDEs may lead to the regulation of a number of plant responses to reinforce the wall barrier, such as the production of pectinesterases (OG0000019), expansins (OG0000035), fasciclin-like arabinogalactan proteins (OG0000309), and dirigents (OG0000179). It is also known that alterations to plant cell wall trigger disease resistance responses, known as damage-associated molecular patterns (DAMP)-triggered immunity (Bacete 2018). Here, the most commonly regulated DEGs among the four species studied were involved in signaling and cell wall proteins, indicating that the first resistance response or barrier imposed by the plant on the pathogen is common to all these plant species, and does not trigger a plant species-specific response or a response specific to the nematode species.
One of the gene families associated with cell wall stress responses found in this study was the expansin gene family, which encodes cell wall-loosening proteins and that is known to respond to different stresses (Marowa et al. 2016). An Arachis spp. gene from the expansin-like b subfamily (AraEXLB8) is regulated upon infection by root-knot nematodes (Guimaraes et al. 2015). Soybean composite plants overexpressing the AraEXLB8 gene were less susceptible to RKNs than were plants without such overexpression (Guimaraes et al. 2017). Therefore, these genes may be promising candidates for improving nematode resistance in a wide range of crops. Another interesting orthogroup included the gene family encoding the fasciclin-like arabinogalactan proteins, with a large number of genes from G. max being upregulated upon nematode infection (20 genes), in contrast with the other three species. Although this gene family is often associated with ovary abortion in Arabidopsis, a study with Populus associated this gene family with salt response (Zang et al. 2015), suggesting its role in response to stress.
In addition to CWDEs, sedentary nematodes such as RKNs are able to secrete a series of proteins (called virulence effectors) inside root tissues to alter host metabolism, induce giant cell formation and repress plant immune responses (Rehman et al. 2016). Among the virulence effectors characterized so far are CLE-like peptides that mimic the plant CLAVATA3/ESR-related (CLE) protein family of Arabidopsis involved in the regulation of plant growth and development (Mitchum et al. 2012). Interestingly, the orthogroup OG0000014 included genes encoding oligopeptide transporters in the four host species studied. Plant oligopeptide transporters may display broad substrate specificity, such as the Arabidopsis OPT6 transporter, which is able to transport either reduced forms of glutathione or larger peptides from various origins, including nematode CLE-like peptides (Pike et al. 2009). The regulation of such transporter genes in nematode-resistant plants suggests that alteration of the transport of oligopeptides produced by the nematode might be one of the strategies used by the host to impair successful nematode infection.
Regarding plant transcription regulators, we found one orthogroup (OG:0000036) containing AP2/ERFs TFs regulated in common among the four species. AP2/ERFs are regulators of multiple ethylene-dependent plant pathways and can be associated with the ethylene-dependent and the jasmonic acid-dependent disease resistance pathways triggered by pathogen attack (Gu et al. 2017). Nevertheless, to date, only a few studies have associated AP2/ERF TFs with nematode infection resistance (Lee et al. 2018). Another TF family, the heat stress transcription factors (HSFs), was highlighted in the orthologous group OG0000050, which contained 5 rice DEGs and a unique DEG for Arachis, coffee, and soybean (Suppl. Table 3). HSFs play a crucial role in plant responses to several abiotic stresses, but some have also diversified to respond to biotic stresses. For instance, one of the five upregulated rice DEGs, LOC_Os09g28354 = CPuORF39, is the orthologue of the Arabidopsis key immune regulator TL1-binding transcription factor 1 (TBF1) (Hayden and Jorgensen 2007). TBF1 controls defense protein expression and acts as the growth-to-defense switch in the plant upon immune induction (Pajerowska-mukhtar et al. 2012; Xu et al. 2017).
Interestingly, no pathogen-related (PR) genes were commonly identified in this analysis, showing that, despite the large number of orthologous groups found in common, different nonorthologous PR genes may be involved in the different plant species in response to RKN infection (Snel et al. 2002).
Using comparative genomics, we identified core evolutionarily conserved genes and groups of orthologous and in-paralogous based on protein sequences from distantly related plant species. This constitutes an important resource for both evolutionary and functional genomics analyses. This method allowed the identification of more than 35,000 nonsingleton orthogroups, which clustered with high confidence, of which 6132 were universally conserved in all the plant genomes studied. The integration of the inference of orthologous groups and gene expression data from four RKN-resistant plant genotypes led to the identification of common genes involved in the response to nematode attack. This approach might be used for the identification of genes responsive to other stresses, constituting an important tool for crop improvement. In addition, this data set of core evolutionarily conserved genes in a large panel of plants of interest will constitute a valuable resource for future studies on the evolutionary determination of gene function.
References
Agrios, GN (2005). Plant diseases caused by nematodes. In: Introduction to plant pathology, fifth ed. Elsevier Academic Press, London pp. 826-872
Albert VA, Barbazuk WB, Der JP, et al (2013) The Amborella Genome and the Evolution of Flowering Plants. Science (80- ) 342
Albuquerque EVS, Carneiro RMDG, Costa PM et al (2010) Resistance to Meloidogyne incognita expresses a hypersensitive-like response in Coffea arabica. Eur J Plant Pathol 127:365–373
Amrine KCH, Blanco-ulate B, Cantu D (2015) Discovery of Core Biotic Stress Responsive Genes in Arabidopsis by Weighted Gene Co-Expression Network Analysis. PloS one, 10(3)
An SH, Sohn KH, Choi HW et al (2008) Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 228(1):61–78
Bacete L (2018) Plant cell wall-mediated immunity : cell wall changes trigger diseas resistanc responses. Plant J 614–636
Beneventi M, da Silva O, de Sa ME et al (2013) Transcription profile of soybean-root-knot nematode interaction reveals a key role of phythormones in the resistance reaction. BMC Genomics 14:322
Bird AF (1961) The ultrastructure and histochemistry of a nematode-induced giant cell. J Biophys Biochem Cytol 11:701–715
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics btu170
Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527
Breitling R, Armengaud P, Amtmann A (2005) Vector analysis as a fast and easy method to compare gene expression responses between different experimental backgrounds. BMC Bioinformatics 6(1):181
Cabrera J, Díaz-Manzano FE, Barcala M et al (2015) Phenotyping nematode feeding sites: three-dimensional reconstruction and volumetric measurements of giant cells induced by root-knot nematodes in Arabidopsis. New Phytol 206:868–880
Cagnola JI, Dumont de Chassart GJ, Ibarra SE et al (2018) Reduced expression of selected FASCICLIN-LIKE ARABINOGALACTAN PROTEIN genes associates with the abortion of kernels in field crops of Zea mays (maize) and of Arabidopsis seeds. Plant Cell Environ 41:661–674
Chen F, Dong W, Zhang J et al (2018) The Sequenced Angiosperm Genomes and Genome Databases. Front Plant Sci 9:1–14
Chong J, Baltz R, Schmitt C et al (2002) Downregulation of a Pathogen-Responsive Tobacco UDP-Glc : Phenylpropanoid Glucosyltransferase Reduces Scopoletin Glucoside Accumulation, Enhances Oxidative Stress, and Weakens Virus Resistance. Plant Cell 14:1093–1107
Cohen SP, Leach JE (2019) Abiotic and biotic stresses induce a core transcriptome response in rice. Sci Rep 9:6273
Cottret L, Rancurel C, Briand M, Carrere S (2018) Family-Companion: analyse, visualise, browse, query and share your homology clusters. bioRxiv 266742
Cui J, Luan Y, Jiang N et al (2016) Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J 89(3):577–589
Das M, Haberer G, Panda A et al (2016) Expression pattern similarities support the prediction of orthologs retaining common functions after gene duplication events. Plant Physiol 171:2343–2357
De Waele D, Elsen A (2007) Challenges in Tropical Plant Nematology
Decraemer W, Hunt D (2006) Structure and Classification. In: Plant Nematology. pp 3–32
Dunwell JM, Gibbings JG, Mahmood T et al (2008) Germin and Germin-like Proteins : Evolution, Structure, and Function. Critical Reviews in Plant Science 27(5):342–375
Emms DM, Kelly S (2015) OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16:1–14
Finkina EI, Melnikova DN, Bogdanov IV, Ovchinnikova TV (2016) Lipid Transfer Proteins As Components of the Plant Innate Immune System : Structure, Functions, and Applications. Acta Naturae 8:47–61
Gu C, Guo ZH, Hao PP, et al (2017) Multiple regulatory roles of AP2 / ERF transcription factor in angiosperm. Bot Stud 1–8
Guimaraes PM, Guimaraes LA, Morgante CV et al (2015) Root transcriptome analysis of wild peanut reveals candidate genes for nematode resistance. PLoS ONE 10:e0140937
Hayden CA, Jorgensen RA (2007) Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select factor-encoding genes. BMC Biol 30:1–30
Jain S, Chittem K, Brueggeman R et al (2016) Comparative transcriptome analysis of resistant and susceptible common bean genotypes in response to soybean cyst nematode infection. PLoS ONE 11:1–21
Johnson KL, Jones BJ, Bacic A, Schultz CJ (2003) The Fasciclin-Like Arabinogalactan Proteins of Arabidopsis. A Multigene Family of Putative Cell Adhesion Molecules Plant Physiology 1(133):1911–1925
Jones JT, Haegeman A, Danchin EGJ et al (2013) Top 10 plant-parasitic nematodes in molecular plant pathology. Mol Plant Pathol 14:946–961
Joshi T, Xu D (2007) Quantitative assessment of relationship between sequence similarity and function similarity. BMC Genomics 8(1):222
Laetsch DR, Blaxter M (2017) KinFin: Software for Taxon-Aware Analysis of Clustered Protein Sequences G3(7):3349–3357
Lee IH, Shim D, Jeong JC et al (2018) Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-resistant and susceptible sweetpotato cultivars. Planta 249:431–444
Li N, Zhao M, Liu T et al (2017) A Novel Soybean Dirigent Gene GmDIR22 Contributes to Promotion of Lignan Biosynthesis and Enhances Resistance to Phytophthora sojae. Front Plant Sci 8:1–12
Liu W, Liu J, Rajapakse JC (2018) Gene Ontology Enrichment Improves Performances of Functional Similarity of Genes. Sci Rep 1–12
Love M, Anders S, Huber W (2013) Differential analysis of RNA-Seq data at the gene level using the DESeq package. European Molecular Biology Laboratory (EMBL), Heidelberg, pp 1–32
Marowa P, Ding A, Kong Y (2016) Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep 35:1–17
Mistry J, Bateman A, Finn RD (2007) Predicting active site residue annotations in the Pfam database. BMC Bioinformatics 14:1–14
Mitchum MG, Wang X, Wang J, Davis EL (2012) Role of Nematode Peptides and Other Small Molecules in Plant Parasitism. Annu Rev Phytopathol 50:175–195
Morgante CV, Brasileiro ACM, Roberts PA et al (2013) A survey of genes involved in Arachis stenosperma resistance to Meloidogyne arenaria race 1. Funct Plant Biol 40:1298–1309
Mota APZ, Oliveira TN, Vinson CC et al (2019) Contrasting Effects of Wild Arachis Dehydrin Under Abiotic and Biotic Stresses. Front Plant Sci 10:1–16
Nobori T, Tsuda K (2019) The plant immune system in heterogeneous environments. Curr Opin Plant Biol 50:58–66
Pajerowska-mukhtar KM, Wang W, Tada Y et al (2012) The HSF-like transcription factor TBF1 is a major molecular switch for plant growth-to-defense transition. Curr Biol 22:103–112
Panchy N, Lehti-shiu M, Shiu S (2016) Evolution of Gene Duplication in Plants. Plant Physiol 171:2294–2316
Petitot AS, Kyndt T, Haidar R et al (2017) Transcriptomic and histological responses of African rice (Oryza glaberrima) to Meloidogyne graminicola provide new insights into root-knot nematode resistance in monocots. Ann Bot 119:885–899
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:36
Pike S, Patel A, Stacey G, Gassmann W (2009) Arabidopsis OPT6 is an Oligopeptide Transporter with Exceptionally Broad Substrate Specificity. Plant Cell Physiol 50:1923–1932
Pont C, Murat F, Confolent C, et al (2011) RNA-seq in grain unveils fate of neo- and paleopolyploidization events in bread wheat (Triticum aestivum L.). Genome Biol 12:R119.
Postnikova OA, Hult M, Shao J et al (2015) Transcriptome analysis of resistant and susceptible alfalfa cultivars infected with root-knot nematode Meloidogyne incognita. PLoS ONE 10:1–17
Proite K, Carneiro R, Falcao R et al (2008) Post-infection development and histopathology of Meloidogyne arenaria race 1 on Arachis spp. Plant Pathol 57:974–980
Qiao X, Li Q, Yin H, et al (2019) Gene duplication and evolution in recurring polyploidization – diploidization cycles in plants. Genome Biol 1–23
Rehman S, Gupta VK, Goyal AK (2016) Identification and functional analysis of secreted effectors from phytoparasitic nematodes. BMC Microbiol 1–18
Remm M, Storm CE V, Sonnhammer ELL (2001) Automatic Clustering of Orthologs and In-paralogs from Pairwise Species Comparisons. J Mol Biol 1041–1052.
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Shaik R, Ramakrishna W (2014) Machine learning approaches distinguish multiple stress conditions using stress-responsive genes and identify candidate genes for broad resistance in rice. Plant Physiol 164:481–495
Simão FA, Waterhouse RM, Ioannidis P et al (2015) BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212
Singh S, Singh B, Singh AP (2015) Nematodes: A Threat to Sustainability of Agriculture. Agric Clim Chang - Adapt Crop to Increased Uncertain (AGRI 2015) 29:215–216.
Snel B, Bork P, Huynen MA (2002) The identification of functional modules from the genomic association of genes. Proc Natl Acad Sci 99:5890–5895
Stöver BC, Müller KF (2010) TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7
Thimm O, Bläsing O, Gibon Y et al (2004) MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939
Tian T, You Q, Zhang L et al (2016) SorghumFDB: sorghum functional genomics database with multidimensional network analysis. Database (Oxford) 2016:1–16
Trudgill DL, Blok VC (2001) Apomictic, polyphagous root-knot nematodes: exceptionally successful and damaging biotrophic root pathogens. Annu Rev Phytopathol 39:53–77
Van BM, Diels T, Vancaester E et al (2018) PLAZA 4. 0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Res 46:1190–1196
Woldesemayat AA, Modise DM, Gemeildien J et al (2018) Cross-species multiple environmental stress responses: An integrated approach to identify candidate genes for multiple stress tolerance in sorghum (Sorghum bicolor (L.) Moench) and related model species. PLoS ONE 13:1–30
Xie Z, Nolan TM, Jiang H, Yin Y (2019) AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front Plant Sci 10:1–17
Xu G, Greene GH, Yoo H et al (2017) Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 545:487–490
Yoshida Y, Koutsovoulos G, Laetsch DR et al (2017) Comparative genomics of the tardigrades Hypsibius dujardini and Ramazzottius varieornatus. PLoS Biol 15(7):e2002266
Yunes JM, Babbitt PC (2019) Sequence analysis Effusion : prediction of protein function from sequence similarity networks. Bioinformatics 35:442–451
Zang L, Zheng T, Chu Y et al (2015) Genome-Wide Analysis of the Fasciclin-Like Arabinogalactan Protein Gene Family Reveals Differential Expression Patterns, Localization, and Salt Stress Response in Populus. Front Plant Sci 6:1–16
Zanne AE, Tank DC, Cornwell WK et al (2013) Three keys to the radiation of angiosperms into freezing environments. Nature 506:89–92
Zhao S, Fernald RD, Zhao S, Fernald DR (2005) Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol 12:1047–1064
Acknowledgements
This work was supported by grants/fellowships from EMBRAPA, UCB, CNPq-INCT, CAPES, and FAPDF. We are grateful to Dr. Regina Carneiro (Embrapa Cenargen, Brazil) for valuable contributions and for providing and characterizing Meloidogyne spp. We would like to thank the SPIBOC (INRA, Sophia-Antipolis) and CENARGEN (Embrapa, Brasília) Bioinformatics platform for technical help and advices.
Funding
APZM, BPMA, and FBMA received a PhD grant from the Brazilian Federal Agency for Support and Evaluation of Graduate Education (CAPES, Brazil), and APZM and FBMA also received CAPES mobility grants to France. DF was supported by a visiting scientist grant from the Science without Boarders program (project no. 400328/2012-7) from the Brazilian National Council for Scientific and Technological Development (CNPq, Brasil). ED was supported by a visiting scientist grant. Part of this research was funded by project 002/14 from the Agropolis Fondation (Montpellier, France), Embrapa and CAPES (Brazil) under the reference ID (1402-004) and under the program "Investissement d'avenir" ANR-10-LABX-001-01, Labex Agro, by the INCT PlantStress Biotech (project number 465480/2014-4); and the FAPDF-Distrito Federal Research Foundation (project number no.193001265/2017).
Author information
Authors and Affiliations
Contributions
ACBM, PMG, MFGS, EVSA, ASP, PG, and DF conceived the project. APZM, ACBM, PMG, EVSA, DF, PG, and MFGS designed the experiments. APZM and ED conceived, designed, and performed the bioinformatics pipelines, and analyzed the results. APZM, MAPS, ASP, BPM, DF, and EVSA executed the qPCR experiments. ACBM, PMG, MAPS, ASP, DF, EVSA, and MELS produced the plant samples for transcriptomics. APZM, EGJD, ACMB, PMG, EVSA, ASP, DF, and MFGS wrote and revised the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Communicated by Stefan Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mota, A.P.Z., Fernandez, D., Arraes, F.B.M. et al. Evolutionarily conserved plant genes responsive to root-knot nematodes identified by comparative genomics. Mol Genet Genomics 295, 1063–1078 (2020). https://doi.org/10.1007/s00438-020-01677-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-020-01677-7