Abstract
The gametophyte of moss exhibits a simple body plan, yet its growth is regulated by complex developmental phenomena similar to angiosperms. Because moss can be easily maintained under laboratory conditions, amenable for gene targeting and the availability of genome sequence, P. patens has become an attractive model system for studying evolutionary traits. Until date, there has been no Agrobacterium-mediated Tnt1 mutagenesis protocol for haploid protonemal filaments of moss. Hence, we attempted to use the intact tobacco Tnt1 retrotransposon as a mutagen for P. patens. Bioinformatic analysis of initiator methionyl-tRNA (Met-tRNAi), a critical host factor for Tnt1 transposition process, suggested that it can be explored as a mutagen for bryophytes. Using protonemal filaments and Agrobacterium-mediated transformation, 75 Tnt1 mutants have been generated and cryopreserved. SSAP analysis and TAIL-PCR revealed that Tnt1 is functional in P. patens and has a high-preference for gene and GC-rich regions. In addition, LTR::GUS lines exhibited a basal but tissue-specific inducible expression pattern. Forward genetic screen resulted in 5 novel phenotypes related to hormonal and gravity response, phyllid, and gamete development. SSAP analysis suggests that the Tnt1 insertion pattern is stable under normal growth conditions and the high-frequency phenotypic deviations are possibly due to the combination of haploid explant (protonema) and the choice of mutagen (Tnt1). We demonstrate that Agrobacterium-mediated Tnt1 insertional mutagenesis could generate stable P. patens mutant populations for future forward genetic studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Forward genetic screens have enabled biologists not only to understand phenotypes of interest at a molecular level, and they have facilitated the discovery of novel genes associated with the phenotype. The efficiency of any forward genetic screen is influenced by the choice of mutagen and the nature of the screen. Transfer DNA (T-DNA) insertions were preferred over traditional chemical mutagenesis tools, since known T-DNA borders aid in identifying the mutated locus in the genome. T-DNA insertions are random (Kim and Gelvin 2007) and mostly integrate into noncoding regions, because plant genomes contain repetitive sequences comprising up to 85% of the genome. A mutagenesis tool that preferably integrates into genic regions would reduce the required size of the mutant population to saturate plant genomes.
Retrotransposons are class I transposable elements which resemble a retrovirus in gene structure and mode of replication. Long terminal repeat (LTR)—retrotransposons contains genes essential for transposition activity and are flanked by direct repeats of LTR sequences (Finnegan 2012). These sequences act as both promoter and terminator for the synthesis of intermediate RNA transcripts and this integrates into a host genome leading to an increase in copy number (Finnegan 2012). Based on sequence complementarity, the host transfer RNA (tRNA) binds to the primer binding site (PBS) of retrotransposon and initiates reverse transcription of intermediate RNA transcripts (Feuerbach et al. 1997). Tnt1 retrotransposon was earlier isolated from tobacco (Grandbastien et al. 1989) and its transposition activity was studied extensively (Pouteau et al. 1991; Casacuberta and Grandbastien 1993; Vernhettes et al. 1997). Tnt1 has been shown to preferably transpose into gene-rich regions of Arabidopsis (Lucas et al. 1995), Medicago (d’Erfurth et al. 2003), soybean (Cui et al. 2013) and potato (Duangpan et al. 2013).
Moss (P. patens), a member of the bryophytes, has a simple body plan, yet its growth is regulated by many complex developmental phenomena like apical dominance (Fujita et al. 2008), phototropism (Jenkins and Cove 1983a), and gravitropism (Jenkins et al. 1986). In addition, mosses show the highest gametophytic tissue diversity (including spores, chloronema, caulonema, bud, rhizoid, gametophore axis, phyllids (blade and midrib), antheridium, and archegonium among all green plant lineages (Kofuji and Hasebe 2014). High regeneration capacity, desiccation tolerance, availability of well-annotated genome sequence, and the capacity for gene targeting techniques has established P. patens a unique model organism for understanding the evolution of developmental traits in land plants (Prigge and Bezanilla 2010). High homologous recombination frequency observed in P. patens allowed extensive reverse genetics studies on several orthologs of angiosperm genes (Schaefer and Zrÿd 1997). It has been shown that the gene regulatory networks controlling the development of haploid and diploid phases of plant lineages are distinct (Sakakibara et al. 2008, 2013; Caine et al. 2016; Chater et al. 2016). These studies indicate the challenges associated with candidate gene selection for studying gametophytic developmental processes in moss based on the knowledge available from the diploid phase of angiosperms. This suggests that there is a need for an efficient forward genetic screen to identify novel genes and to complement the reverse genetics approach. In this regard, the haploid dominant life phase of P. patens is an interesting feature as even a recessive allele will reveal the phenotype. Small colony size and simple growth conditions of P. patens provide an added advantage in carrying out a forward genetic screen. Cryostorage of mutant moss populations and maintenance of mutant lines with minimum effort have also been demonstrated (Schulte and Reski 2004).
Earlier, chemical mutagenesis on moss has resulted in many auxotrophic, phototropic, polarotropic, and various phytohormone resistant and sensitive lines (Ashton et al. 1979; Jenkins and Cove 1983b). Further studies on these mutant lines have implicated the role of hormones in moss and their functional conservation in non-vascular plants. Nishiyama et al. (2000) developed a shuttle mutagenesis tool for P. patens that specifically targets expressed regions of the genome, but complicated multiple insertions made identification of a causal insertion difficult (Hayashida et al. 2005). Using UV-C based mutagenesis and a positional cloning approach, the first successful characterization of a mutant in P. patens was recently reported (Stevenson et al. 2016).
When we began this work, there was no study regarding the usage of Tnt1 retrotransposon as a tool for mutagenesis of P. patens. Recently, Vives et al. (2016) have shown that a two-component system derived from Tnt1 retrotransposon when introduced in to moss protoplast, is capable of transposition into the P. patens genome. Authors demonstrated that modified Tnt1 eliminates further transposition activity in moss.
However, our approach is significantly different, since we used an intact retrotransposon carried by Agrobacterium to mutagenize haploid protonemal filaments and relied on a cryostorage method to reduce the occasional possibility of transposition. Haploid protonemal filaments, high gene preference of Tnt1, and the cryostorage of mutant lines have offered unique advantages to moss forward genetic studies. Another advantage of using protonemal explants instead of protoplasts is that it eliminates the risk of diploid formation.
In this study, using Agrobacterium-mediated transformation of protonemal filaments, we developed a Tnt1 insertional mutant population and validated its transposition activity in P. patens under various physiological conditions. We have also analyzed the transposition preference of Tnt1 insertions to gene and GC rich regions. In addition, LTR::GUS lines were developed to evaluate the transcriptional regulation of tobacco Tnt1 in moss. Our forward genetic screen identified several novel mutants related to phyllid development, hormonal and gravity response and phenotypes associated with gamete development. Using SSAP-PCR, we have demonstrated the stability of Tnt1 insertions and viability of our Tnt1 mutagenesis protocol, respectively. Our results suggest that Agrobacterium tumefaciens-mediated Tnt1 insertional mutagenesis could also generate moss mutants for forward genetic studies.
Materials and methods
Moss culture and maintenance
Physcomitrella patens ecotype Gransden was grown in vitro as described by Cove et al. (2009). Homogenized protonemal tissue was grown on cellophane-overlaid BCDAT agar medium. Post homogenization, tissues were incubated for 4–5 days in tissue culture incubators at 16:8 h light–dark cycle at 24 °C for all experiments.
Met-tRNAi sequence analysis
To analyze if P. patens has necessary host factors for successful Tnt1 transposition, all the available green plant genomes (55 species) from Phytozome v12.0 (https://phytozome.jgi.doe.gov/pz/portal.html) (Goodstein et al. 2012) were downloaded. Klebsormidium nitens draft genome was downloaded from the project website (Hori et al. 2014). Transfer RNA genes were predicted from the genomic sequences using tRNAscan-SE version 1.3.1 with default parameters (Pavesi et al. 1994). Predicted methionyl-tRNA (Met-tRNA) (initiator and elongator) sequences were further tested for the stability of the tRNA acceptor arm by checking the 7 bp complementarity between the 5ʹ (5ʹ ATCAGAG 3ʹ) and the 3ʹ end (3ʹ A-TAGTCTC 5ʹ). Verified Met-tRNA 3ʹ end sequences were analyzed for the degree of complementarity with the 9 bp of Tnt1 PBS (Primer Binding Site) (5ʹ TGG-TATCAGAGC 3ʹ). The remaining first 3 bp (5ʹ TGG-TATCAGAGC 3ʹ) complementarity was not assessed, since the 3ʹ end CCA is not coded in the tRNA gene, but is subsequently added by an enzymatic reaction. Representative organisms were arranged on a cladogram using PhyloT online tool (http://phylot.biobyte.de/) and visualized in iTOL (http://www.itol.embl.de). PhyloT tool generates trees based on NCBI taxonomy database.
Cloning and moss transformation
Using Agrobacterium tumefaciens-mediated transformation, pCAMBIA-1391Xc-Tnt1 (vector Entrez accession number: AF234311.1 and Tnt1-94 element Entrez accession number: X13777) vector was used to transform into P. patens protonema, as described in PHYSCObase (http://www.moss.nibb.ac.jp/protocol.html). Moss tissue surviving hygromycin selection (20 mg/L) was further subjected to PCR confirmation using Tnt1 (LTR_F and LTR_R) and hygromycin phosphotransferase II (HPTII) gene-specific primers (Hyg_F and Hyg_R). Confirmed Tnt1 mutant lines were propagated independently on BCDAT medium. To generate LTR::GUS lines, the LTR promoter of Tnt1 was amplified from pCAMBIA-1391Xc-Tnt1 using primers XbaI-LTR-F and XmaI-LTR-R as forward and reverse primers, respectively. A 610 bp amplified product was digested with XbaI and XmaI restriction enzymes and ligated into the binary vector pBI101 upstream to the GUS gene. LTR::GUS-pBI101 construct was introduced into wild-type P. patens protonemal tissue using Agrobacterium-mediated transformation. Mutants were cryopreserved for long-term storage as described by Schulte and Reski (2004) and revived when required. All primer details were provided in Table S1.
SSAP PCR
To further analyze the number of T-DNA and Tnt1 insertions, we utilized SSAP (Sequence-Specific Amplified Polymorphism). Altogether, five different SSAP analyses were carried out with mutant lines. In the beginning, three mutant lines (1, 3, and 6) were subjected to SSAP for Tnt1 insertion events. To determine the stability of Tnt1 insertions in transformed tissue, we tested (i) individual protonemal colonies established from randomly selected protonemal filaments of Tnt1 lines (1 and 3) maintained independently over 10 homogenization cycles; (ii) tissue regenerated from single gametophore leaves for 10 cycles; and (iii) individual cryopreserved mutant lines (after 12 months cryostorage) by SSAP. To determine the number of T-DNA insertions, altogether, 13 lines were analysed by SSAP. Five hundred nanograms (500 ng) of CTAB-extracted genomic DNA (Doyle 1990) from the above samples was digested with Csp6I. This was ligated with a Csp6I adapter prepared by annealing the Csp6I adapter1 primer and the 5ʹ phosphorylated Csp6I adapter2 primer (Syed and Flavell 2006). The adapter-ligated DNA was pre-amplified with adapter specific primer C00, and finally, SSAP PCR was performed with gamma [32P]dATP end labeled OL13 primer (Tnt1-specific) or SSAP RB primer (T-DNA specific) and C00 primer (Waugh et al. 1997; Tam et al. 2005). PCR products were resolved in a 3.5% UREA-PAGE gel along with a 50 bp ladder (B7025; New England Biolabs) and detected by autoradiography. All radiographic images were interpreted as explained in Fig. 2a.
TAIL-PCR
To determine the Tnt1 insertion loci in the transgenic lines, Thermal Asymmetric Interlaced PCR (TAIL-PCR) was performed using three nested primers (LTR3, LTR4, and LTR7) designed from the known LTR sequence and used in consecutive reactions together with one of the arbitrary degenerate primers (AD1–AD6). Genomic DNA (50 ng) isolated from each Tnt1 mutant line was used in primary PCR reaction using HiMedia Taq polymerase as described by the manufacturer’s protocol. A fourfold diluted primary PCR product (1 µl) was used as a template in consecutive (secondary and tertiary) PCR reactions. The tertiary TAIL-PCR products were cloned into pGEM-T Easy (Promega, MA, USA) and sequenced. The sites of Tnt1 insertion were identified by BLASTN analysis of Physcomitrella genome (http://www.cosmoss.org) (Lang et al. 2005).
GC content analysis
To assess if Tnt1 has specificity for GC-rich regions, altogether, 72 Tnt1 retrotransposon insertion sites in moss genome were taken for GC content analysis. To increase the robustness of our analysis, we have used 26 insertion data from the published report of Vives et al. (2016) in addition to 37 insertions reported in the present study and the remaining 9 insertions were not shown here. The nature of the insertion (gene or intergene) and the GC content of insertion sites over a span of 10, 20, 30, and 50 bp were calculated (Table S2), since GC content distribution would vary depending on the fragment size. In an in silico experiment, 10, 20, 30, and 50 bp were randomly chosen (n = 100,000) from the P. patens genome and their GC content was calculated. Furthermore, the nature of the randomly chosen site (gene or intergene) was determined using P. patens genome annotation version 3.3. In this study, coding regions along with 1000 bp up and downstream were considered as genic regions and remaining genome as intergenic. Student’s t test was performed between the simulated random insertions and experimentally determined Tnt1 insertions based on the mean of local GC content of genic, intergenic, and combined insertions. All the simulations and calculations were performed using NumPy 1.11.2—a Python scientific computing package (Walt et al. 2011). The data were plotted using Matplotlib 2.0—a Python 2D plotting library (Hunter 2007).
Tnt1 retrotransposon expression analysis
To assess if Tnt1 retrotransposon is inducible upon CuCl2 treatment, 7-day-old cultures of WT (strain Gransden) and transgenic lines (LTR::GUS line 1 and line 4) of P. patens were transferred for 24 h to liquid BCDAT medium containing 0, 0.5, and 2.0 µM CuCl2. For GUS assay, tissue was transferred to a GUS-staining buffer and incubated at 37 °C overnight (Jefferson et al. 1987). Images were obtained using a Leica S8 APO Stereomicroscope (Leica, Wetzlar, Germany). For microtome sectioning, alcohol was serially replaced with xylene and then paraffin wax. Thin (10 µm) sections were taken using a Leica RM2265 microtome. Sections were counterstained with Safranin–O to increase the visibility of background tissue and imaged using a Zeiss ApoTome microscope (Zeiss, Oberkochen, Germany).
For qRT-PCR analysis, total RNA was extracted from treated protonemal tissue using RNAiso Plus (Takara Bio USA Inc., CA, USA). Crude RNA samples were incubated with Ambion® DNase I (Thermo Fisher Scientific, MA, USA) to remove residual genomic DNA. Two micrograms of the DNA-free RNA samples were reverse-transcribed using oligo dT primers and M-MLV reverse transcriptase (Invitrogen, CA, USA). Specific PCR primers were designed to detect endogenous β-actin (Act F, Act R) and GUS gene (Gus F, Gus R) transcripts. cDNA was diluted to 1:10 concentration only during β-actin transcript amplification and relative quantification of transcripts were performed using Bio-Rad CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, CA, USA). Cycler conditions were as follows 95 °C for 10 s; 40 cycles of 95 °C for 5 s and 60 °C for 30 s and an additional step for melting curve analysis at 95 °C for 10 s. SYBR green used for detection of transcripts was SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) from Takara (Takara Bio USA Inc., USA). Each plate was run with samples including no template control. Relative target gene expression levels were carried out using β-actin as a reference gene.
Screening of Tnt1 insertion lines
Moss tissue was homogenized and grown for 5 days to get homogeneous protonemal filaments and was subjected to hormone assay. Protonemal explants were inoculated on BCDAT medium with and without BAP (1 µM) or NAA (1 µM) and grown for 2 weeks at 24 °C, under continuous light before analyzing the phenotype. For gravitropism experiments, 5-day-old protonemal tissue was pre-cultured on BCDAT agar plates supplemented with glucose (0.5%) and incubated at 24 °C in a tissue culture incubator under continuous light conditions for 10 days. Post-incubation, plates were arranged vertically in darkness for 2 weeks for scoring the phenotype (Cove and Quatrano 2006). Wild-type was inoculated next to mutant lines in Petri dishes for all the assays.
Results
When we initiated this work, there was no report available regarding Tnt1 transposition activity in moss. Recently, Vives et al. (2016) have shown that Tnt1 is functional in P. patens. However, in the present report, we have explored an intact tobacco Tnt1 retrotransposon for mutagenesis in P. patens using haploid protonemal explants unlike protoplasts used in the previous study. We also characterized Tnt1 transcription activity, transposition preference, and stability in detail. The possibility of employing Tnt1 as a mutagen for all green plant lineages by comparing host initiator methionyl-tRNA (Met-tRNAi) sequences was also assessed. Agrobacterium-mediated transformation of haploid protonemal filaments was the method of choice to introduce tobacco Tnt1 in P. patens and the forward genetic screen resulted in several novel developmental mutants.
Tobacco Tnt1 is functional in P. patens
Because tobacco Tnt1 retrotransposon is a useful mutagenesis tool in many heterologous angiosperm model organisms such as Arabidopsis, Medicago, etc, we aimed to use it to mutagenize the bryophyte model plant P. patens. Therefore, we analyzed if P. patens has the necessary host factors critical for successful transposition. In our analysis, Met-tRNAi from all heterologous model organisms, in which Tnt1 has been shown to transpose successfully, was found to have absolute complementarity with primer binding site (PBS) of Tnt1 (Fig. 1; Table S3). Despite the evolutionary distance between bryophytes and angiosperms, the 3ʹ end of Met-tRNAi is found to be conserved in P. patens, Sphagnum fallax, and Marchantia polymorpha, but not in algal species (Fig. 1; Table S3).
P. patens has the necessary host factor for Tnt1 transposition activity. Sequence alignment of 3ʹ end Met-tRNAi gene from representative organisms of Viridiplantae and their degree of sequence complementarity with 9 bp from the 3ʹ end of Tnt1 primer binding site (PBS) 5ʹ TGGTATCAGAGC 3ʹ. Drosophila melanogaster was used as an outgroup
Detection of T-DNA integration and Tnt1 transposition activity in mutant lines of moss generated by Agrobacterium-mediated transformation. a Schematic diagram of pCAMBIA-1391Xc-Tnt1 insertion cassette and a transposed Tnt1 element (LB–left border; RB–right border). For SSAP analysis, mutant moss genome was digested by the Csp6I enzyme and Tnt1 LTR-specific primer (OL13) or SSAP-RB primer was used. OL13 and Csp6I adapter primers will amplify 410 bp and 388 bp bands from T-DNA linked Tnt1 and only 410 bp band from a transposed Tnt1. b SSAP analysis of wild-type (W) and Tnt1 lines (1, 3, 4, 6, 7, 10, 11, 15, 17, 18, 19, 8, and 22) displays the polymorphism that is specific to the T-DNA right border. Wild type (W) did not show any SSAP bands, while Tnt1 line 22 showed maximum 5 bands and line 8 did not show any bands. Arrowheads denote reproducible, unique T-DNA insertions and arrow marks 200 bp band. c SSAP analysis of wild-type (W) and Tnt1 lines 1, 3, and 8. Nicotiana benthamiana (T), water (H), and 50 bp ladder (L) were used as positive control, negative control, and DNA ladder, respectively. Arrowheads denote unique Tnt1 insertions and arrow marks 500 bp band
Using Agrobacterium-mediated transformation, T-DNA harbouring an intact Tnt1-94 retroelement and a hygromycin selection cassette (HPTII) from the pCAMBIA-1391Xc binary vector was introduced into P. patens protonemal filaments. These filaments were subjected to medium containing hygromycin (20 mg/L) for 2 weeks followed by antibiotic-free relaxation incubation for further 2 weeks. Out of 84 lines subjected to a second selection on hygromycin medium (20 mg/L), 75 stable transgenic lines were generated harbouring the Tnt1 retrotransposon. These lines were confirmed for the presence of Tnt1 retrotransposon fragments (LTR) as well as the hygromycin (Hyg) phosphotransferase II (HPTII) gene (Fig. S1a). All these mutant lines were cryopreserved and revived when required for further study (Fig. S1b-e). Using SSAP technique, we analyzed 13 lines for the polymorphism along the T-DNA right border and determined the average number of T-DNA insertions of the moss mutant population as 1.25 per mutant line. Except for line 8, all the other lines showed minimum one insertion. Exceptionally, line 22 showed bands corresponding to five different T-DNA insertions (Fig. 2b). We employed the sequence-specific amplified polymorphism (SSAP) technique, which is more robust and extensively used to detect Tnt1 retrotransposon insertions (Waugh et al. 1997; Courtial et al. 2001; Tam et al. 2005; Vives et al. 2016). Lines 1 and 3 showed the characteristic 388 bp and 426 bp bands amplified from proper T-DNA integration (Fig. 2c). Line 8 represents a rare incomplete T-DNA integration found in our mutant screen, where only the 388 bp band was observed (Fig. 2c). Apart from the low-intensity nonspecific bands, two insertions for line 1, and five insertions for line 3 were observed indicating that tobacco Tnt1 has transposed in the moss genome from the initial T-DNA insertion. Line 8, which had an incomplete T-DNA integration, did not show any specific Tnt1 insertion (Fig. 2c).
Tnt1 preferentially transposes into genes and GC-rich regions in P. patens genome
To identify the transposed loci in moss genome, Thermal Asymmetric Interlaced PCR (TAIL-PCR) analysis was performed on stable T-DNA insertion lines containing a Tnt1 retrotransposon. Out of 37 insertions detected, 26 were found to be in the genic regions [including promoter (1), exon (11), intron (12)], which were dispersed among 17 chromosomes of P. patens (Fig. 3; Table S4). Though the moss genome has only 22.74% genic region, we found 70.2% of Tnt1 insertions in gene-rich regions indicating preferential transposition. The total number of insertions per individual line varied from 1 to 9. In Tnt1 line 23, all the 9 insertions were found to be dispersed over 8 chromosomes indicating a lack of hot spots (Fig. 3; Table S4).
Tnt1 retrotransposon insertions in P. patens genome determined by TAIL-PCR analysis. Chromosomes are labeled Pp01 to Pp27 and the insertion coordinates are marked on the chromosomes. Each insertion is labeled as line number: insertion number (23:1 is the 23rd line: first insertion). Red triangle—promoter; blue star—exon; pink square—intron; black circle—intergenic. (Color figure online)
To understand if there is a GC content bias in Tnt1 transposition events, we compared the local GC content of Tnt1 insertion loci with the simulated random insertions. In all fragment sizes (10, 20, 30, and 50 bp) that we analyzed, mean GC content was significantly different between combined random insertions versus combined Tnt1 insertions (Fig. 4; Table S5) indicating the GC bias. In all simulations, mean GC content of genic regions of random insertions and Tnt1 insertions was also significantly different. To distinguish GC preference from gene preference, intergenic insertions were further compared. We found a significant difference of mean GC content of intergenic regions between random insertions and Tnt1 insertions (Fig. 4; Table S5) indicating the preference for GC-rich regions. From our analysis, a critical GC value (GC*) of (44–57) % was determined as the cutoff (Fig. S2). If Tnt1 transposition site is chosen only based on local GC content, then any insertion below GC* would more likely be intergenic (I) than genic (G) and vice versa. Tnt1 insertion data indeed showed a considerable increase in intergenic insertions, whose local GC content value was below GC* (Table S6). However, in both categories (above and below GC*), the number of genic insertions was higher than intergenic insertions. This suggests that high GC content is not the sole criterion for choosing a Tnt1 insertion site, but may act parallel to other factors resulting in a preference for genic regions.
Tnt1 transposition sites are biased towards high local GC content regions of P. patens genome. Grey and white bars represent mean local GC content of simulated random insertions and experimentally determined Tnt1 insertions. Student’s t test was performed to check for the statistically significant difference between the means of random insertions and Tnt1 insertions in different categories combined (genic + intergenic regions), genic regions and intergenic regions. In all 10, 20, 30, and 50 bp, sampling fragment sizes, local GC content of combined, genic, and intergenic regions were significantly different between simulated insertions and Tnt1 insertions. All statistical data including sample size are given in Table S5. Error bars are SE. *Denotes p value < 0.001
Tnt1 promoter expression is tissue-specific and inducible
To check the expression pattern of Tnt1, we cloned the entire LTR sequence (610 bp) upstream of GUS reporter gene and generated five LTR::GUS lines of P. patens. All transgenic lines showed similar expression pattern under normal growth conditions, we rarely detected any GUS activity in protonemal filaments. Interestingly, gametophores and buds showed expression at apical regions and lateral branches (Fig. 5a; ii–iv). This basal tissue-specific expression pattern of LTR promoter was further confirmed by histological studies and expression was found to be localized to gametophore lateral branch apex only (Fig. 5b). To test if the LTR promoter is inducible, the entire gametophore was treated with 0.5 and 2 µM CuCl2. The intensity of GUS expression increased in the gametophore apex and lateral branches upon CuCl2 treatment (Fig. 5a; iii–viii) suggesting the inducibility of Tnt1 promoter. Furthermore, we have analyzed the abundance of GUS transcripts in uninduced and CuCl2 induced gametophores of two independent LTR::GUS lines by qRT- PCR. LTR::GUS lines 1 and 4 showed 2.7- and 1.4-fold increase in GUS transcript accumulation, respectively, upon 0.5 µM CuCl2 induction while both lines showed 2.1 and 0.9 fold change upon 2 µM CuCl2 induction.
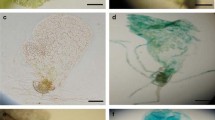
Tissue-specific inducible expression pattern of tobacco Tnt1 in P. patens. a LTR::GUS shows basal expression in bud, gametophore and branch apical cells. Compared to wild-type (i), LTR promoter showing GUS expression pattern in bud apical cells (arrowhead) (ii), and apex and branching points of gametophore (iii and vi). LTR::GUS line 1 and LTR::GUS line 4 basal LTR promoter expression (iii and vi) and tissue-specific induction using 0.5 µM CuCl2 (iv and vii) and 2.0 µM CuCl2 (v and viii) (scale bar 1 mm). b Compared to wild-type (i) histological sections showing basal GUS expression (blue) in LTR::GUS line 1 gametophore stem tangential section (ii) and cross-section at the base of gametophore (iii and iv) (arrowheads denote the GUS expression in lateral branches). All the sections were counterstained with Safranin-O (scale bar 50 µm). c Relative transcript abundance of GUS transcripts in LTR::GUS lines (P. patens) upon CuCl2 induction. Fifteen-day-old gametophore colonies of each line (LTR::GUS line 1 and LTR::GUS line 4) were incubated for 4 days in 0.5 and 2.0 µM CuCl2 solution prepared in BCDAT liquid media. Data are representative of independently performed three biological replicates. Error bars represent standard deviation. Statistical analysis was done for individual lines using t test (paired two sample for means). The difference is statistically significant at p < 0.05 indicated by an asterisk (*) for LTR::GUS line 1 and (a) for LTR::GUS line 4
Isolation of mutants impaired in gametophyte development
Altogether, 75 mutants were developed and were subjected to various screens to isolate mutations in the broad gametophyte developmental pathways. Our analyses revealed a short-leaf mutant (Tnt1 line 5), which produced shorter leaves compared to wild-type (Fig. 6a). When all these lines were induced for gametogenesis and sporophyte formation, Tnt1 line 27 formed multiple organs at the gametophore apex (Fig. 6c), whereas a single sporophyte was developed in wild-type (Fig. 6b). These organs appeared to be either unfertilized archegonia that continued to grow mimicking immature sporophyte or they are just poorly developed sporophytes. Various phytohormone assays were performed for phenotypic characterization. We observed that Tnt1 line 9 showed partial recovery of normal gametophore development even in the presence of 1.0 µM BAP (Fig. 6d–g). Tnt1 line 26 showed excessive rhizoid formation and defective gametophore development in BCD minimal media (Fig. 6h, i). In the presence of either exogenous 1 µM NAA (Fig. 6j, k) or 0.5 mM ammonium tartrate (Fig. 6l, m), both wild-type and Tnt1 line 26 were indistinguishable. Gravitropism analysis revealed that the Tnt1 line 56 protonemal filaments were insensitive to gravity (Fig. 6n, o). Overall, we could isolate 5 novel phenotypes related to gametophyte development out of 75 mutants identified in this study.
Novel phenotypes isolated from the Tnt1 insertional mutant population. (a) Tnt1 line 5 (right) exhibits short-leaf phenotype compared to wild-type (left). Wild-type (b) gametophore apex bearing mature single sporophyte and Tnt1 line 26 mutant (c) gametophore producing multiple immature sporophytes were shown. (d–g) Tnt1 line 9 shows cytokinin resistance phenotype. Wild-type (d) and Tnt1 line 9 (e) grown without exogenous BAP. Wild-type (f) showing stunted bud development and Tnt1 line 9 mutant (g) showing partial recovery of gametophores when grown in the presence of 1.0 µM BAP. (h–m) Tnt1 line 26 shows conditional rhizoid overproduction phenotype. Wild-type (h, j, and i) and Tnt1 line 26 (i, k, and m) growth in BCD media (h, i), BCD supplemented with 1.0 µM NAA (j, k) or 0.5 mM ammonium tartrate (l, m). (n, o) Tnt1 line 56 has failed to align to the gravity vector. Wild-type (n) caulonemal filaments grow upwards, whereas Tnt1 line 56 mutant (o) caulonemal filaments grow randomly. Arrow indicates the gravity vector (scale bar 1 mm)
Viability of Tnt1 as a mutagenesis tool for P. patens
To demonstrate the viability of Tnt1 as a mutagenesis tool, we checked the stability of Tnt1 insertions through SSAP-PCR. We analyzed the Tnt1 transposition activity using SSAP-PCR at the protonemal growth stage, gametophore apex stage, and with cryopreserved tissues. The number of insertions for Tnt1 did not change for both lines 1 and 3 when we assessed if the protonemal subculturing process is causing any Tnt1 induction (Fig. 7a, d). Again, the number of Tnt1 insertions in line 1 and line 3 remained unchanged when we assessed if the tissue-specific activity of LTR promoter in gametophore apex correlates to high mutation load (Fig. 7c, e). Similarly, there were no changes in insertional patterns when we revived and tested the 12-month-old cryopreserved tissue of moss for Tnt1 lines 1 and 3 (Fig. 7b, d). In our analysis, we found that Tnt1 line 5 had three Tnt1 and one T-DNA insertions. One of them was found to be inserted in the coding region of a cell wall Extensin gene, and the second one was an uncharacterized protein, while the third Tnt1 insertion as well as the single T-DNA insertion were found in intergenic regions. The detailed investigation of Tnt1 line 5 and these potential candidate genes is in progress.
SSAP analysis to assess the stability of Tnt1 insertional mutants. SSAP analysis of wild-type (W) and Tnt1 lines 1 (a–c) and 3 (d, e). Routinely maintained lines (p) or untreated lines (u) of Tnt1 lines 1 and 3 were subjected to SSAP analysis along with four biological replicates (i–iv) of clonally propagated lines that are derived from a protonemal plate (a, d). Similarly, tissues revived from a cryo-stock (b, d) or gametophore apex derived tissues (c, e) were analyzed for mutant stability. In all the experiments Nicotiana benthamiana (T), water (H) and 50 bp ladder (L) were used as positive control, negative control and DNA ladder respectively. Arrowheads denote unique Tnt1 insertions and arrow marks 500 bp band
Discussion
The tobacco Tnt1 retrotransposon is functional in several heterologous angiosperm species (Lucas et al. 1995; d’Erfurth et al. 2003; Cui et al. 2013; Duangpan et al. 2013). During the Tnt1 transposition process, the 3ʹ end of host Met-tRNAi binds to primer binding site (PBS) of Tnt1 and acts as a primer for initial cDNA synthesis (Feuerbach et al. 1997). Considering the evolutionary distance between tobacco and moss, we wanted to determine if P. patens has the necessary host factor for Tnt1 transposition. We noted that the Met-tRNAi 3ʹ end sequence from all heterologous model organisms, wherein Tnt1 transposes successfully, exhibited absolute complementarity with PBS, suggesting that Met-tRNAi is a critical host factor. Furthermore, in our analysis, we observed that all embryophytes (land plants) including P. patens and representative organisms such as Marchantia polymorpha and Selaginella moellendorffii shared this conservation of the 3ʹ end Met-tRNAi sequence (Fig. 1; Table S3). Though basal algal species had multiple mismatches that clearly distinguish algae and embryophytes, only a single mutation was observed in Klebsoridium nitens (a charophycean algae), which is consistent with the fact that charophycean algae are sister group to land plants. Our Met-tRNAi analysis suggests that Tnt1 is a potential candidate for mutagenesis studies in bryophytes.
Tnt1 retrotransposon is delivered through an Agrobacterium-mediated T-DNA insertion protocol. Both T-DNA and Tnt1 insertions are capable of contributing to mutant phenotype. By targeting the T-DNA right-border flanking regions using SSAP analysis, we determined the average number of T-DNA integration to be 1.25 per mutant line as most of the lines had only a single T-DNA insertion (Fig. 2b). However, we also found abnormal T-DNA insertions in the line number 22 (five T-DNA insertions) and line 8 (no T-DNA insertion).
To check the number of Tnt1 insertions, we used SSAP-PCR, indicating maximum five independent Tnt1 transposition events per line (Fig. 2c). Again, line 8 showed an anomalous result with only the 5′-LTR detectable, and no Tnt1 transposition events (Fig. 2c). This indicates the possibility of a rare incomplete T-DNA integration event comprising the left T-DNA border, HPTII gene, and 5ʹ LTR but with neither the 3ʹ LTR nor right T-DNA border remaining in the moss genome. Tnt1 has been demonstrated to preferentially transpose into gene-rich regions in the angiosperm model organisms tested (Lucas et al. 1995; d’Erfurth et al. 2003). Consistent with this and the recent report of Vives et al. (2016), we noted that approximately 70% Tnt1 insertions were in genic regions of P. patens genome (Fig. 3). In addition, Tnt1 insertions were found to have a preference for regions of high local GC content independent of gene preference (Fig. 4). Though high GC preference does not qualify as a sole criterion for choosing transposition site (Table S6), it could contribute to increasing the chance of transposition into gene-rich regions acting in conjunction with other factors.
The 5ʹ- LTR sequence acts as a promoter that drives the expression of Tnt1 to initiate the first step of transposition process (Grandbastien et al. 1989). Using a GUS reporter gene cloned downstream of LTR promoter and the first 25 amino acid coding sequence of Tnt1 (Pouteau et al. 1991), it was shown that the Tnt1 promoter is induced by plant defense signaling and abiotic stress such as CuCl2 in sporophytic tissues (Pouteau et al. 1994; Mhiri et al. 1997). This response is mainly mediated by the BII sequence repeats present in the U3 region of the LTR promoter (Vernhettes et al. 1997). Hence, we used entire LTR sequence from Tnt1-94 retroelement to characterize its promoter activity. Though all the properties of Tnt1 transposition in P. patens were reminiscent of studies on angiosperms, LTR promoter activity was distinct. Surprisingly, in gametophyte of P. patens, we observed a basal low expression of LTR::GUS in gametophore apical cell and lateral branch apical cells (Fig. 5a; iii–vi). This tissue-specific expression pattern of LTR mimics the previously reported PpLFY1 and PpLFY2 expression pattern (Tanahashi et al. 2005) in gametophores of P. patens. As previously reported in tobacco (Mhiri et al. 1997), the LTR::GUS promoter is also inducible in P. patens by abiotic factors such as CuCl2. We noticed that the increased GUS transcript accumulation at 0.5 µM and the inhibitory effect at high concentration of CuCl2 (2 µM) is consistent with the previous studies in tobacco (Vernhettes et al. 1997) (Fig. 5c). Though Vives et al. (2016) reported the expression of Tnt1 in protonemal filaments, we rarely observed GUS expression in protonemal filaments in our study. This could be either because of the expression level is less than the detectable limit or protonemal expression is regulated by cis-regulatory elements present downstream of LTR promoter.
In this investigation, we could isolate 5 novel mutants out of 75 lines screened that showed deviation from the wild-type in different gametophyte developmental pathways (Fig. 6). Though four out of five mutants exhibited novel phenotypes, agravitropic mutants like that of our Tnt1 line 56, were isolated from P. patens (Jenkins et al. 1986) and Ceratodon purpureus (Cove and Quatrano 2006) earlier.
Earlier, Vives et al. (2016) have modified the retrotransposon and developed a two-component system in a mini-Tnt1 retroelement that can transpose only when the proteins required for Tnt1 transposition were supplemented by a separate vector. As we have used an intact Tnt1 retrotransposon for mutagenesis, high-frequency phenotypic deviation observed in the mutant population prompted us to study the stability and the number of mutagenic transpositions. We assessed the stability of Tnt1 insertional mutants and found that the Tnt1 insertional pattern does not change over normal subculturing of protonema, gametophore apex-derived tissues, where LTR promoter is active, and finally tissues stored via cryopreservation. We hypothesize that when tobacco Tnt1 was introduced into P. patens cells, LTR promoter might be active and able to complete transposition events. However, when the host machinery could detect it as an active retrotransposon, Tnt1 transposition might be a rare event due to the tight gene regulation through a siRNA-mediated pathway like other endogenous moss LTR retrotransposons (Coruh et al. 2015). Even though retrotransposon insertions are rare events, continuous regular subculture of even wild-type lines can result in genetic and physiological instability (Kartha and Engelmann 1994). Hence, we took advantage of the cryopreservation method to avoid undesirable and rare transposition events, since cryoconditions would likely arrest all metabolic activities.
In this analysis, we have tested both T-DNA and Tnt1 insertions in the moss mutant population and showed that Tnt1 insertions have preference to gene as well as GC rich regions and stable under all the physiological conditions tested. Even though T-DNA insertions have been traditionally used for mutagenesis and more stable, they are known to integrate randomly. The low T-DNA insertion number (1.25 insertions per mutant line) compared to Tnt1 (4 insertions per mutant line) observed in this study suggests that Tnt1 as a mutagen could possibly yield mutants with interesting phenotypes over T-DNA insertions. However, T-DNA-based delivery of Tnt1 can be performed on protonemal explants to avoid diploid formation through undesirable protoplast fusion. Tnt1 line 5 showed 2 out of 3 Tnt1 insertions in genic regions and a T-DNA insertion of intergenic nature. This highlights the preferential integration of Tnt1 to genic regions and random T-DNA insertions.
We demonstrate an alternative approach for P. patens mutagenesis using an intact tobacco Tnt1 retrotransposon for forward genetic studies. This has an added advantage since, we used haploid protonemal filaments for Agrobacterium-mediated transformation. Unlike the protoplast transformation used in the previous report, this approach should maintain haploidy throughout the mutagenesis process. While all major characteristics of Tnt1 retrotransposition appeared to be quite similar between angiosperm and moss, the basal tissue-specific expression pattern of the Tnt1 promoter (LTR) detected in moss was a striking observation. Despite the fact that the moss mutants exhibited on average 4 Tnt1 insertions and 1.25 T-DNA insertions per line, we could eventually isolate 5 interesting phenotypes from a total population of 75 individuals. In addition, through SSAP analysis, we showed that the high-frequency phenotypes obtained in this study could be because of the high gene preference of Tnt1 transposition and random T-DNA insertions in combination with the haploid nature of protonemal filaments of moss, but does not seem to be caused by highly proliferative Tnt1 insertions. Together, these data suggest that our intact Tnt1 insertional mutagenesis tool is a viable approach to perform forward genetics screens in moss.
Data availability
The data supporting the results of this article are provided as additional files. All the data pertaining to the present study has been included in the tables/figures of the manuscript.
Abbreviations
- GC* :
-
Critical GC value
- Met-tRNAi :
-
Initiator methionyl-tRNA
- PBS:
-
Primer-binding site
- P. patens :
-
Physcomitrella patens
- LTR:
-
Long-terminal repeats
- T-DNA:
-
Transfer DNA
- TAIL-PCR:
-
Thermal asymmetric interlaced PCR
- tRNA:
-
Transfer RNA
References
Ashton NW, Grimsley NH, Cove DJ (1979) Analysis of gametophytic development in the moss, Physcomitrella patens, using auxin and cytokinin resistant mutants. Planta 144:427–435
Caine RS, Chater CC, Kamisugi Y et al (2016) An ancestral stomatal patterning module revealed in the non-vascular land plant Physcomitrella patens. Development 143:3306–3314
Casacuberta JM, Grandbastien M-A (1993) Characterisation of LTR sequences involved in the protoplast specific expression of the tobacco Tnt1 retrotransposon. Nucl Acids Res 21:2087–2093
Chater CC, Caine RS, Tomek M et al (2016) Origin and function of stomata in the moss Physcomitrella patens. Nat Plants 2:16179
Coruh C, Cho SH, Shahid S et al (2015) Comprehensive annotation of Physcomitrella patens small RNA loci reveals that the heterochromatic short interfering RNA pathway is largely conserved in land plants. Plant Cell 27:2148–2162
Courtial B, Feuerbach F, Eberhard S et al (2001) Tnt1 transposition events are induced by in vitro transformation of Arabidopsis thaliana, and transposed copies integrate into genes. Mol Genet Genom 265:32–42
Cove DJ, Quatrano RS (2006) Agravitropic mutants of the moss Ceratodon purpureus do not complement mutants having a reversed gravitropic response. Plant Cell Environ 29:1379–1387
Cove DJ, Perroud P-F, Charron AJ et al (2009) Culturing the moss Physcomitrella patens. Cold Spring Harb Protoc 2009:pdb.prot5136. https://doi.org/10.1101/pdb.prot5136
Cui Y, Barampuram S, Stacey MG et al (2013) Tnt1 retrotransposon mutagenesis: a tool for soybean functional genomics. Plant Physiol 161:36–47. https://doi.org/10.1104/pp.112.205369
d’Erfurth I, Cosson V, Eschstruth A et al (2003) Efficient transposition of the Tnt1 tobacco retrotransposon in the model legume Medicago truncatula. Plant J 34:95–106
Doyle JJ (1990) Isolation of plant DNA from fresh tissue. Focus (Madison) 12:13–15
Duangpan S, Zhang W, Wu Y et al (2013) Insertional mutagenesis using Tnt1 retrotransposon in potato. Plant Physiol 163:21–29. https://doi.org/10.1104/pp.113.221903
Feuerbach F, Drouaud J, Lucas H (1997) Retrovirus-like end processing of the tobacco Tnt1 retrotransposon linear intermediates of replication. J Virol 71:4005–4015
Finnegan DJ (2012) Retrotransposons. Curr Biol 22:R432–R437. https://doi.org/10.1016/j.cub.2012.04.025
Fujita T, Sakaguchi H, Hiwatashi Y et al (2008) Convergent evolution of shoots in land plants: lack of auxin polar transport in moss shoots. Evol Dev 10:176–186
Goodstein DM, Shu S, Howson R et al (2012) Phytozome: a comparative platform for green plant genomics. Nucl Acids Res 40:D1178–D1186
Grandbastien M-A, Spielmann A, Caboche M (1989) Tnt1, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature 337:376–380
Hayashida A, Takechi K, Sugiyama M et al (2005) Isolation of mutant lines with decreased numbers of chloroplasts per cell from a tagged mutant library of the moss Physcomitrella patens. Plant Biol 7:300–306
Hori K, Maruyama F, Fujisawa T et al (2014) Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun 5
Hunter JD (2007) Matplotlib: a 2D graphics environment. Comput Sci Eng 9:90–95
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901
Jenkins GI, Cove DJ (1983a) Phototropism and polarotropism of primary chloronemata of the moss Physcomitrella patens: responses of mutant strains. Planta 159:432–438. https://doi.org/10.1007/BF00392079
Jenkins GI, Cove DJ (1983b) Phototropism and polarotropism of primary chloronemata of the moss Physcomitrella patens: responses of the wild-type. Planta 158:357–364
Jenkins GI, Courtice GRM, Cove DJ (1986) Gravitropic responses of wild-type and mutant strains of the moss Physcomitrella patens. Plant Cell Environ 9:637–644
Kartha KK, Engelmann F (1994) Cryopreservation and germplasm storage. In: Vasil IK, Thorpe TA (eds) Plant cell and tissue culture. Springer, Dordrecht, pp 195–230
Kim Sang-Ic, Gelvin stanton B (2007) Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J 51:779–791. https://doi.org/10.1111/j.1365-313X.2007.03183.x doi
Kofuji R, Hasebe M (2014) Eight types of stem cells in the life cycle of the moss Physcomitrella patens. Curr Opin Plant Biol 17:13–21
Lang D, Eisinger J, Reski R, Rensing SA (2005) Representation and high-quality annotation of the Physcomitrella patens transcriptome demonstrates a high proportion of proteins involved in metabolism in mosses. Plant Biol 7:238–250
Lucas H, Feuerbach F, Kunert K et al (1995) RNA-mediated transposition of the tobacco retrotransposon Tnt1 in Arabidopsis thaliana. EMBO J 14:2364
Mhiri C, Morel J-B, Vernhettes S et al (1997) The promoter of the tobacco Tnt1 retrotransposon is induced by wounding and by abiotic stress. Plant Mol Biol 33:257–266
Nishiyama T, Hiwatashi Y, Sakakibara K et al (2000) Tagged mutagenesis and gene-trap in the moss, Physcomitrella patens by shuttle mutagenesis. DNA Res 7:9–17
Pavesi A, Conterio F, Bolchi A et al (1994) Identification of new eukaryotic tRNA genes in genomic DNA databases by a multistep weight matrix anaylsis of transcriptional control regions. Nucl Acids Res 22:1247–1256
Pouteau S, Huttner E, Grandbastien MA, Caboche M (1991) Specific expression of the tobacco Tnt1 retrotransposon in protoplasts. EMBO J 10:1911
Pouteau S, Grandbastien M-A, Boccara M (1994) Microbial elicitors of plant defence responses activate transcription of a retrotransposon. plant J 5:535–542
Prigge MJ, Bezanilla M (2010) Evolutionary crossroads in developmental biology: Physcomitrella patens. Development 137:3535–3543
Sakakibara K, Nishiyama T, Deguchi H, Hasebe M (2008) Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evol Dev 10:555–566
Sakakibara K, Ando S, Yip HK et al (2013) KNOX2 genes regulate the haploid-to-diploid morphological transition in land plants. Science 339:1067–1070
Sambrook J, Fritsch EF, Maniatis T et al (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Schaefer DG, Zrÿd J-P (1997) Efficient gene targeting in the moss Physcomitrella patens. Plant J 11:1195–1206. https://doi.org/10.1046/j.1365-313X.1997.11061195.x
Schulte J, Reski R (2004) High throughput cryopreservation of 140 000 Physcomitrella patens mutants. Plant Biol 6:119–127
Stevenson SR, Kamisugi Y, Trinh CH et al (2016) Genetic analysis of Physcomitrella patens identifies ABSCISIC ACID NON-RESPONSIVE (ANR), a regulator of ABA responses unique to basal land plants and required for desiccation tolerance. Plant Cell 28:1310–1327
Syed NH, Flavell AJ (2006) Sequence-specific amplification polymorphisms (SSAPs): a multi-locus approach for analyzing transposon insertions. Nat Protoc 1:2746
Tam SM, Mhiri C, Vogelaar A et al (2005) Comparative analyses of genetic diversities within tomato and pepper collections detected by retrotransposon-based SSAP, AFLP and SSR. Theor Appl Genet 110:819–831
Tanahashi T, Sumikawa N, Kato M, Hasebe M (2005) Diversification of gene function: homologs of the floral regulator FLO/LFY control the first zygotic cell division in the moss Physcomitrella patens. Development 132:1727–1736
Vernhettes S, Grandbastien M-A, Casacuberta JM (1997) In vivo characterization of transcriptional regulatory sequences involved in the defence-associated expression of the tobacco retrotransposon Tnt1. Plant Mol Biol 35:673–679
Vives C, Charlot F, Mhiri C et al (2016) Highly efficient gene tagging in the bryophyte Physcomitrella patens using the tobacco (Nicotiana tabacum) Tnt1 retrotransposon. New Phytol 212:759–769
Walt S van der, Colbert SC, Varoquaux G (2011) The NumPy array: a structure for efficient numerical computation. Comput Sci Eng 13:22–30
Waugh R, McLean K, Flavell AJ et al (1997) Genetic distribution of Bare–1-like retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP). Mol Gen Genet MGG 253:687–694
Acknowledgements
We sincerely thank Prof. Pascal Ratet (CNRS, France) for providing us tobacco Tnt1 retrotransposon (pCAMBIA-1391Xc-Tnt1) construct. BM acknowledges fellowship support from CSIR, New Delhi. All authors thank IISER Pune and DST Govt. of India for core funding support for this investigation. We are thankful to Prof. Meenu Kapoor for her timely help in moss establishment at IISER Pune. Thanks to Mr. Nitish Lahigude for his support in moss maintenance and Ms. Kavya Mohan for her technical help in cryopreservation of moss mutants. We also thank Prof. David Hannapel, Iowa State University for critical reading of our manuscript.
Funding
The present study was supported by a grant (Grant No. EMR/2016/004852) from Department of Science and Technology (DST), Government of India to AKB. Core funding and infrastructure was provided by Indian Institute of Science Education and Research (IISER) Pune, India.
Author information
Authors and Affiliations
Contributions
BM and AKB have conceived and designed the experiments. BM, VR, SJ, and AB have carried out all experiments. BM and AKB have written the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by S. Hohmann.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mohanasundaram, B., Rajmane, V.B., Jogdand, S.V. et al. Agrobacterium-mediated Tnt1 mutagenesis of moss protonemal filaments and generation of stable mutants with impaired gametophyte. Mol Genet Genomics 294, 583–596 (2019). https://doi.org/10.1007/s00438-019-01532-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-019-01532-4