Abstract
Cisplatin is one of the most popular chemotherapeutic drugs in treating ovarian cancer. Resistance to cisplatin is a common clinical challenge that needs to be solved to increase its anti-tumor effects. The relation of miR-514 expression with prognosis in ovarian cancer patients was analyzed based on GSE73584 datasets. The regulation of miR-514 on proliferation and cisplatin chemosensitivity of ovarian cells was examined by MTT assay, colony-formation assay and soft-agar colony-formation assay. Dual luciferase assay was performed to detect the direct interaction of miR-514 with its downstream targets. Immunobloting and qRT-PCR were performed for target gene expression analysis. Low expression of miR-514 was related to poor prognosis in ovarian cancer patients. MiR-514 repressed proliferation and decreased cisplatin chemosensitivity in ovarian cancer cells by targeting ATP binding cassette subfamily. MiR-514 is of clinically significance in ovarian cancer by attenuating proliferation of ovarian cancer cells and decreasing chemoresistance of cisplatin by targeting ATP binding cassette subfamily.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cisplatin is one of the popular chemotherapeutic drugs in ovarian cancer (Rogers et al. 2018; Wang et al. 2017a; Xu et al. 2018). However, the long-term use of chemotherapeutic drugs, including Cisplatin will trigger chemoresistance. How to overcome the resistance is the key problem that need to be solved nowadays (Lee et al. 2005; Yang et al. 2006).
ATP binding cassette subfamily (ABC subfamily) encode ATP-binding proton pumps that consume ATP for substance transport to cells. ABC family members are generally highly expressed in cancer stem cells and are closely related to chemoresistance. Members of the ABC family can consume ATP to transport the chemotherapeutic drugs out of the cell, leaving the cell low or even free of medicine, thereby rendering the tumor cells resistant to chemotherapeutic drugs (Dean 2009; Glavinas et al. 2004; Lou and Dean 2007).
MicroRNAs (miRNAs) are small non-coding RNAs composed of 18–25 nucleotides regulating the expression of protein coding genes (Wang et al. 2017b). MicroRNA has been reported to participate in the development of multiple types of human cancers, including endometrial cancer, prostate cancer, papillary thyroid carcinoma, ovarian cancer and etc (Su et al. 2018; Todorovic et al. 2018; Wang et al. 2017b; Wu et al. 2018). For example, microRNA181c was reported to inhibit the growth and invasion of prostate cancer cells by targeting multiple ERK signaling pathway components (Su et al. 2018). MicroRNA-34a was found to inhibit cell proliferation and invasion by downregulating Notch1 in endometrial cancer (Todorovic et al. 2018).
We hypothesized that microRNAs that regulate ABC family members might play a role in the cisplatin chemoresistance of ovarian cancer. In this study, we aimed to investigate this hypothesis with miR-514 as an example by studying its effects on the chemotherapeutic drug resistance of ovarian cancer cells and its possible mechanism.
Materials and methods
Cell culture conditions
Two human ovary cancer cell lines SKOV3 and OVCA433 were purchased from American Type Culture Collection (Manassas, VA), and cultured in RPMI-1640 medium (Gibco-BRL, Gaithersburg, MD, USA) supplemented with 10% FBS, 100 µg/mL streptomycin and 100 U/mL penicillin (Jung et al. 2016). The cells were incubated in an incubator containing 5% CO2 and 95% air at 37 °C in a humidified atmosphere. Cisplatin was purchased from Sigma (St. Louis, MO, USA) and indicated concentrations and treatment period were applied.
RNA extraction and Real-Time PCR
Total RNA from cancer cell lines was isolated by TRIzol reagent according to the guidelines. Then cDNA was synthesized using Superscript III (Invitrogen, Carlsbad, CA) based on the instructions. For RT-PCR of miR-514, the reactions were performed utilizing TaqMan miRNA Assays (Applied Biosystems, Foster City, CA) according to the instructions with U6 spliceosomal RNA (U6 snRNA) as an input control and miScript PCR System (Qiagen, Valencia, CA, USA) was additionally used (Buechler et al. 2013). Primers for ATP binding cassette subfamily A member 1 (ABCA1) (Forward, ACCCACCCTATGAACAACATGA, Reverse, GAGTCGGGTAACGGAAACAGG), ATP binding cassette subfamily A member 10 (ABCA10) (Forward, GGAGACGACAAATCTATGCAGTG, Reverse, CCCAACAATGAGTTTCACGAGT), ATP binding cassette subfamily F member 2 (ABCF2) (Forward, CCTCTCACTTACCTTTCATGGTC, Reverse, ACTTCACGCTTCCCAATAGCA) and GAPDH (Forward, ACAACTTTGGTATCGTGGAAGG, Reverse, GCCATCACGCCACAGTTTC) were used together with SYBR Green PCR Master Mix (TaKaRa, Japan). The 2−ΔΔCT method was used to calculate the target gene expression levels with GAPDH used as the internal control.
3-(4,5-di methyl thiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay
Cells plated in 96-well plates at 10,000 per well were cultured for 24 h in the cell culture incubator. Then 10 µl of 5 mg/ml MTT was added to each well and then incubated in the dark for 4 h. Finally, the absorbance at 570 nm measured by a microplate reader was used to quantify the cell viability.
Colony-formation assay
Colony-formation assay was conducted according to the instruction as described with slight modifications (Franken et al. 2006). Briefly, cells with indicated treatments were plated to a 6-well plate at 100 cells/well. Ten days later, cell-forming colonies were fixed and stained with 6.0% glutaraldehyde supplemented with 0.5% crystal violet. Finally, the number of colonies was counted.
Soft-agar colony-formation assay
Soft-agar colony-formation assay was performed to identify the proliferation ability of ovary cancer cell lines as described (Horibata et al. 2015). Briefly, prepare the bottom layer with 0.6% agarose gel, and then prepare the cell-containing layer with 0.3% agarose gel by mixing the cells with the 0.6% agarose in a 1:1 ratio, finally put the feeding layer as 0.3% agarose gel on the top. Finally, count the number of colonies in each well using a light microscope.
Immunoblotting
10 µg total proteins from cell lysate were loaded in 8–12% SDS–polyacrylamide gels, and then were transferred to PVDF membranes. Membranes were blocked with 5% non-fat milk first, and then were incubated with primary antibody dilutions overnight at 4 °C, including anti-ABCA1 antibody (MAB10005, Millipore, Billerica, MA, USA, 1:1,000), anti-ABCA10 antibody (MA5-21013, Thermofisher Scientific, Waltham, MA, USA, 1:1000), anti-ABCF2 antibody (ab86178, Abcam, Cambridge, MA, USA, 1:2000), and anti-β-actin antibody (ab8226, Abcam, USA, 1:5,000). After 3× washes, membranes were incubated in corresponding secondary antibodies conjugated to HRP for 1 h. Then ECL reagents were used to develop the target bands.
Dual luciferase assay
Wild type and mutated sequences of ABCA1, ABCA10, ABCF2 3′-UTR mRNA were cloned into the downstream of the firefly luciferase gene in pMir-Report vector (Applied Biosystems, Waltham, MA, USA), respectively. SKOV3 cells were transfected by miR-514 and miR negative control, respectively, and were seeded into 24-well plates. Then cells were transfected with wild type (ABCA1, ABCA10, ABCF2) and mutated (ABCA1, ABCA10, ABCF2-mut) pMir-Report vector vectors using Lipofectamine LTX (Invitrogen, Carlsbad, CA, USA), respectively and co-transfecting pMir-Report Renilla vector as the transfection control. 24 h later, cells were lysed and firefly luciferase activity of each sample was measured, and normalized to Renilla luciferase activity by Dual-luciferase reporter assay kit (Promega, Madison, WI, USA) following the manufacturer’s protocol.
Gene knockdown by SiRNA
20 µM Stealth SiRNA of 3 genes, ABCA1 (HSS176437, Thermofisher Scientific, USA), ABCA10 (HSS115843, Thermofisher Scientific, USA) and ABCF2 (10061, Dharmacon) were separately transfected for target gene knockdown in OPTI-MEM media to 6-well cultured cells. After overnight, cells were trypsinized and plated to 96 well plates with or without the indicated drug for MTT assay.
Statistical analyses
All data shown in this study are presented as the mean ± SD (standard deviation). Student’s t tests were utilized to analyze the differences between two groups, and two-way ANOVA was used for analysis between three or more groups. P < 0.05 was considered as significant difference.
Results
Low expression of miR-514 is correlated with poor prognosis in ovarian cancer patients
By analyzing NCBI’s GEO datasets’ database among 2000 genes, we found that patients with miR-514 expression were obviously and significantly related with clinical prognosis in ovarian cancer. As shown, patients with low miR-514 expression had a poor prognosis both in overall survival (Fig. 1a) and in relapse-free survival (Fig. 1b). The expression of miR-514 in patients of G2 and G3 grade in ovarian tumors was also significantly lower than that in patients of G1 grade (Fig. 1c). Moreover, miR-514 expression in stage III patients was significantly lower than stage I by stage classification (Fig. 1d).
The relation of miR-514 expression with prognosis in ovarian cancer patients. a Kaplan–Meier plots of overall survival based on the miR-514 level in GSE73584 datasets. b Kaplan–Meier plots of distant relapse-free survival (DRFS) based on the miR-514 level in GSE73584 datasets. c The miR-514 levels in different grade of ovarian cancer patients in GSE73584 datasets. d The miR-514 levels in different stage of ovarian cancer patients in GSE73584 datasets. Data are shown as mean ± S.D. **P < 0.01; ***P < 0.001 (Student’s t test in figure c, d)
MiR-514 suppresses proliferation of ovarian cancer cells
The next step is to examine the effect of miR-514 on the proliferation of ovarian cancer cells. Transfection of miR-514 mimics into two ovarian cancer cell lines, SKOV3 and OVCA433, inhibited the proliferation of both cell lines (Fig. 2a, c). Transferring of miR-514 inhibitor into both ovarian cancer cells promotes the proliferation of both cell lines (Fig. 2b, d). Moreover, transfection of miR-514 mimics into SKOV3 cells inhibited cell proliferation by both clone formation assay (Fig. 2e, f) and soft agar assay (Fig. 2g, h).
Proliferation assay of ovarian cancer cells with transfection of miR-514. a The proliferation of SKOV3 cells transfected with miR-514 mimics was determined by MTT assay. b The proliferation of SKOV3 cells transfected with miR-514 inhibitor was determined by MTT assay. c The proliferation of OVCA433 cells transfected with miR-514 mimics was determined by MTT assay. d The proliferation of OVCA433 cells transfected with miR-514 inhibitor was determined by MTT assay. e The proliferation of SKOV3 cells transfected with miR-514 mimics was determined by colony-formation assay. f The statistical results of colony-formation assay. g The proliferation of SKOV3 cells transfected with miR-514 mimics was determined by soft-agar colony-formation assay. h The statistical results of soft-agar colony-formation assay. Data are shown as mean ± S.D. *P < 0.05; ***P < 0.001 (Student’s t test in figure f and h, and two-way ANOVA test in a–d panels)
MiR-514 decreases cisplatin chemosensitivity in ovarian cancer cells
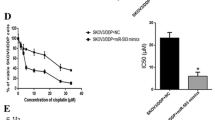
Next, we examined the effect of miR-514 on the resistance of ovarian cancer cells to chemotherapy with cisplatin. We treated SKOV3 cells with 4 µg/ml cisplatin for 6 months to establish a cisplatin-resistant SKOV3 cell line, named as SKOV3 CR (cisplatin resistance). Next, the expression of miR-514 in SKOV3 CR cells was detected by Real-Time PCR and lower expression of miR-514 was found compared to the untreated cells (Fig. 3a). The SKOV3 cells were transiently treated with 4 µg/ml of cisplatin for 48 h. The expression levels of miR-514 were detected and downregualtion of miR-514 was found (Fig. 3b). Next, MTT method was used to detect the effect of different concentrations of Cisplatin on cell growth. Figure 3c showed that SKOV3 CR cells transfected with miR-514 mimics can restore some degree of sensitivity in Ciplatin resistant cells. In Fig. 3d, miR-514 inhibitor was introduced into SKOV3 cells and was found to decrease the resistance to previously sensitive cells. In Fig. 3e, f plots, the effect of non-anchored growth of SKO cells, SKOV3 CR cells and SKOV3 CR cells transfected with miR-514 under the transient treatment of 4 µg/ml cisplatin was also examined by soft agar assay. Transfection of miR-514 was also found to restore some chemosensitivity in SKOV3 CR cells.
MiR-514 increases cisplatin sensitivity in cisplatin resistant ovarian cancer cells. a The expression levels of miR-514 in SKOV3 and cisplatin resistant SKOV3 CR cells were determined by Real-Time PCR. U6 spliceosomal RNA (U6 snRNA) was used as an input control. b The expression levels of miR-514 in SKOV3 cells treated with cisplatin for 48 h were determined by Real-Time PCR. U6 spliceosomal RNA (U6 snRNA) was used as an input control. c Cell viability of SKOV3, SKOV3 CR cells transfected with or without miR-514 mimics treated with different concentration of cisplatin was determined by MTT assay. d Cell viability of SKOV3 cells transfected with or without miR-514 inhibitor treated with different concentration of cisplatin was determined by MTT assay. e Cell viability of SKOV3, SKOV3 CR cells transfected with or without miR-514 mimics treated with 4ug/ml cisplatin was determined by soft-agar colony-formation assay. f The statistical results of soft-agar colony-formation assay. Data are shown as mean ± S.D. **P < 0.01; ***P < 0.001 (two-way ANOVA test in figure c, d, Student’s t test in other panels)
MiR-514 targets ATP binding cassette subfamily
Next, we utilized the targetcan prediction software to predict the target of miR-514. It was found that ten members of the ABC family are its potential targets, as shown in Fig. 4a, b in SKOV3 and OVCA433 cells. When the cells were transfected with miR-514 Mimics, only the mRNA expression levels of ABCA1, ABCA10 and ABCF2 were down-regulated. Figure 4c is a schematic diagram showing the binding site of miR-514 predicted by the targetscan and the 3′UTR of these three genes. In Fig. 4d, the 3′UTRs of these three genes were cloned into the psi-check2 vector for luciferase reporter assays. Also, the predicted binding sites on these 3′UTRs were mutated to reveal miR-514 inhibition. The expressing luciferase was inhibited by miR-514 in the vectors containing the 3′UTRs of these three genes, and the inhibition was disappeared when the predicted binding sites of these targets were mutated, indicating that these three genes are direct targets of miR-514. In Fig. 4e, immunoblotting was performed and showed that miR-514 also inhibited the protein expression of these three genes in SKOV3 cells.
ATP binding cassette subfamily are miR-514 targets. a The expression levels of ten members of ATP binding cassette subfamily were determined by Real-Time PCR in SKOV3 cells. b The expression levels of ten members of ATP binding cassette subfamily were determined by Real-Time PCR in OVCA433 cells. c The binding sites of miR-514 on ABCA1, ABCA10 and ABCF2 3′UTRs. d Luciferase reporter activities driven by wild-type or mutant miR-514 targets 3′UTRs were examined in SKOV3 cells transfected with miR-514 mimics or negative control (NC). e Immunoblotting analysis of ABCA1, ABCA10 and ABCF2 protein levels in SKOV3 cells transfected with miR-514 mimics. Data are shown as mean ± S.D. **P < 0.01; ***P < 0.001; ns, not significant (Student’s t test)
MiR-514 functional downstream effectors are ATP binding cassette subfamily
Next, we performed a series of combinatorial experiments to demonstrate whether miR-514 functions through these three targets ABCA1, ABCA10 and ABCF2. First, we examined the knockdown efficiency of siRNAs for these three genes (Fig. 5a). Transfection of all three siRNAs inhibited the proliferation of SKOV3 cells in Fig. 5b, which was consistent with the function of miR-514. Figure 5c showed that SKOV3 CR cells have some resistance to cisplatin compared to SKOV3 cells, however, all three siRNAs transfected into SKOV3 CR cells restored some sensitivity to the CR cells. In Fig. 5d, miR-514 inhibitor promoted proliferation of SKOV3 cells, but simultaneous transfection of three specific siRNAs of ABCA1, ABCA10 and ABCF2 showed that the promotion of proliferation disappeared. MiR-514 inhibitor in Fig. 5e increased the chemoresistance of SKOV3 to cisplatin. When the three siRNAs repressed their specific targets separately, the promoted resistance by miR-514 inhibitor is decreased. Taken together, these three genes are indeed downstream effector molecules that mediate function of miR-514.
ABCA1, ABCA10 and ABCF2 are the functional downstream effectors of miR-514. a The mRNA levels of ABCA1, ABCA10 and ABCF2 in SKOV3 cells transfected with their respective siRNAs. b Cell viability of SKOV3 cells transfected with ABCA1, ABCA10 and ABCF2 respective siRNAs was determined by MTT assay. c Cell viability of SKOV3, SKOV3 CR cells transfected with ABCA1, ABCA10 and ABCF2 respective siRNAs and treated with cisplatin was determined by MTT assay. d Cell viability of SKOV3 cells transfected with NC or miR-514 inhibitor and co-transfected with ABCA1, ABCA10 and ABCF2 respective siRNAs was determined by MTT assay. e Cell viability of SKOV3 cells transfected with NC or miR-514 inhibitor and co-transfected with ABCA1, ABCA10 and ABCF2 respective siRNAs and treated with or without cisplatin was determined by MTT assay. Data are shown as mean ± S.D. *P < 0.05; **P < 0.01; ***P < 0.001 (two-way ANOVA test in figure b, Student’s t test in other panels)
Discussion
In this study, we first analyzed the relationship between miR-514 expression and clinical prognosis of ovarian cancer. The reduced expression of miR-514 was found in advanced stages and is related to poor survival of ovarian cancer patients in GSE73584 datasets. miR-514 suppresses proliferation of ovarian cancer cells by MTT assay, colony-formation assay and soft-agar colony-formation assay. Next, we found that miR-514 increases cisplatin chemosensitivity in ovarian cancer cells by MTT assay and soft-agar colony-formation assay. Moreover, we found that miR-514 targets 3 ATP binding cassette subfamily members, including ABCA1, ABCA10 and ABCF2, which were confirmed by Real-Time PCR and luciferase assay. Finally, we confirmed that miR-514 exerts its regulation on cell viability through three above ATP binding cassette subfamily members by MTT assay and by utilizing miR-514 inhibitor, cisplatin treatment, and separate siRNA of ABCA1, ABCA10 and ABCF2. This study provides theoretical basis for a novel therapeutic strategy to minimize the chemoresistance in ovarian cancer treatment with Cisplatin.
Ovarian cancer is one of the most lethal types of gynecologic malignancy (Donninger et al. 2004; Proceedings of the Conference on Antilymphocyte Serum, Brook Lodge, Augusta, Michigan, May 2–3, 1969 1970). Ovarian cancer is the fifth leading cause of cancer deaths in women in the United States (Donninger et al. 2004; Jemal et al. 2003). Only 25% patients with advanced stage ovarian cancer survive to 5 years after initial diagnosis (Donninger et al. 2004; Friedlander 1998). The high case fatality rate is partially because that most of ovarian cancer patients were found with advanced stage disease, which is essentially incurable. Because of high malignancy, the chemoresistance is one of the most difficult questions to overcome for treating ovarian cancer.
Cisplatin has been wildly used in treating cancer, including lung cancer, ovarian cancer and many others (Kanaji et al. 2018; Saed et al. 2018; Staff 2018). As cisplatin is not effective for all patients and chemoresistance is a common problem, the chemoresistance of Cisplatin is a key problem to be studied with. Some microRNAs, for instance, miR-214 was found to reduce cisplatin resistance by targeting netrin-1 in bladder cancer cells (Liu et al. 2018). In addition, miR-133b was reported to reverse cisplatin resistance by targeting GSTP1 in cisplatin-resistant lung cancer cells (Lin et al. 2018). Reports on miR-514 were lacking. MiR-514 was only reported to play a role in tumor recurrence after nephrectomy and miR-514 might be a suitable adjunct marker for predicting tumor recurrence (Wotschofsky et al. 2013). In this study, we found miR-514 inhibition increases cisplatin chemosensitivity in ovarian cancer cells. As no findings suggesting relations on miR-514 and chemosensitivity was reported, this point is one of the novelty in this study.
ABCF2 were reported to contribute to cisplatin resistance in ovarian cancer cells (Bao et al. 2017). But ABCA1 and ABCA10 have not been reported to participate in this process, which is now reported in our study. We found that not only ABCF2, but also ABCA1 and ABCA10 all contribute to the chemoresistance of Cisplatin. MiR-361-5p was found to target ABCA1 in pulmonary artery smooth muscle cells (Zhang et al. 2018). In this study, for the first time, we found that miR-514 targets ABCA1, ABCA10, and ABCF2, through which miR-514 exerting its regulation on the chemosensitivity of Cisplatin in ovarian cancer cells. The findings in this study suggest that miR-514 may possibly be used as a marker for prognosis of ovarian cancer. Future work includes investigations on whether miR-514, ABCA1, ABCA10 and ABCF2 can be used as therapeutic targets for treating ovarian cancer.
Conclusions
Our findings in this study include that low expression of miR-514 has poor prognosis in ovarian cancer patients and that the reduced expression of miR-514 leads to increased expression of ABC family members, which in turn promotes tumor proliferation and reduce chemotherapeutic drug resistance.
References
Bao L, Wu J, Dodson M, Rojo de la Vega EM, Ning Y, Zhang Z, Yao M, Zhang DD, Xu C, Yi X (2017) ABCF2, an Nrf2 target gene, contributes to cisplatin resistance in ovarian cancer cells. Mol Carcinog 56:1543–1553. https://doi.org/10.1002/mc.22615
Buechler C, Eisinger K, Krautbauer S (2013) Diagnostic and prognostic potential of the macrophage specific receptor CD163 in inflammatory diseases. Inflamm Allergy Drug Targets 12:391–402
Dean M (2009) ABC transporters, drug resistance, and cancer stem cells. J Mammary Gland Biol Neoplasia 14:3–9. https://doi.org/10.1007/s10911-009-9109-9
Donninger H, Bonome T, Radonovich M, Pise-Masison CA, Brady J, Shih JH, Barrett JC, Birrer MJ (2004) Whole genome expression profiling of advance stage papillary serous ovarian cancer reveals activated pathways. Oncogene 23:8065–8077. https://doi.org/10.1038/sj.onc.1207959
Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C (2006) Clonogenic assay of cells in vitro. Nat Protoc 1:2315–2319. https://doi.org/10.1038/nprot.2006.339
Friedlander ML (1998) Prognostic factors in ovarian cancer. Semin Oncol 25:305–314
Glavinas H, Krajcsi P, Cserepes J, Sarkadi B (2004) The role of ABC transporters in drug resistance, metabolism and toxicity. Curr Drug Deliv 1:27–42
Horibata S, Vo TV, Subramanian V, Thompson PR, Coonrod SA (2015) Utilization of the soft agar colony formation assay to identify inhibitors of tumorigenicity in breast cancer cells. J Vis Exp. https://doi.org/10.3791/52727
Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ (2003) Cancer statistics, 2003. CA Cancer J Clin 53:5–26
Jung YS, Kim HJ, Seo SK, Choi YS, Nam EJ, Kim S, Kim SW, Han HD, Kim JW, Kim YT (2016) Anti-proliferative and apoptotic activities of mullerian inhibiting substance combined with calcitriol in ovarian cancer cell lines. Yonsei Med J 57:33–40. https://doi.org/10.3349/ymj.2016.57.1.33
Kanaji N, Kamitori K, Hossain A, Noguchi C, Katagi A, Kadowaki N, Tokuda M (2018) Additive antitumour effect of dallose in combination with cisplatin in non-small cell lung cancer cells. Oncol Rep. https://doi.org/10.3892/or.2018.6192
Lee S, Choi EJ, Jin C, Kim DH (2005) Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA amplification contributes to cisplatin resistance in an ovarian cancer cell line. Gynecol Oncol 97:26–34. https://doi.org/10.1016/j.ygyno.2004.11.051
Lin C, Xie L, Lu Y, Hu Z, Chang J (2018) miR-133b reverses cisplatin resistance by targeting GSTP1 in cisplatin-resistant lung cancer cells. Int J Mol Med. https://doi.org/10.3892/ijmm.2018.3382
Liu J, Bi J, Li Z, Li Z, Liu X, Kong C (2018) miR214 reduces cisplatin resistance by targeting netrin1 in bladder cancer cells. Int J Mol Med 41:1765–1773. https://doi.org/10.3892/ijmm.2018.3374
Lou H, Dean M (2007) Targeted therapy for cancer stem cells: the patched pathway and ABC transporters. Oncogene 26:1357–1360. https://doi.org/10.1038/sj.onc.1210200
Proceedings of the Conference on Antilymphocyte Serum, Brook Lodge, Augusta, Michigan, May 2–3, 1969 (1970) Fed Proc 29:93–229
Rogers LC, Davis RR, Said N, Hollis T, Daniel LW (2018) Blocking LPA-dependent signaling increases ovarian cancer cell death in response to chemotherapy. Redox Biol 15:380–386. https://doi.org/10.1016/j.redox.2018.01.002
Saed GM, Fletcher NM, Diamond MP, Morris RT, Gomez-Lopez N, Memaj I (2018) Novel expression of CD11b in epithelial ovarian cancer: potential therapeutic target. Gynecol Oncol. https://doi.org/10.1016/j.ygyno.2017.12.018
Staff P (2018) Correction: risk of thromboembolism in cisplatin versus carboplatin-treated patients with lung cancer. PLoS One 13:e0191454. https://doi.org/10.1371/journal.pone.0191454
Su Z, Zhang M, Xu M, Li X, Tan J, Xu Y, Pan X, Chen N, Chen X, Zhou Q (2018) MicroRNA181c inhibits prostate cancer cell growth and invasion by targeting multiple ERK signaling pathway components. Prostate. https://doi.org/10.1002/pros.23478
Todorovic L, Stanojevic B, Mandusic V, Petrovic N, Zivaljevic V, Paunovic I, Diklic A, Saenko V, Yamashita S (2018) Expression of VHL tumor suppressor mRNA and miR-92a in papillary thyroid carcinoma and their correlation with clinical and pathological parameters. Med Oncol 35:17. https://doi.org/10.1007/s12032-017-1066-3
Wang CW, Lee BH, Tai CJ (2017a) The inhibition of cordycepin on cancer stemness in TGF-beta induced chemo-resistant ovarian cancer cell. Oncotarget 8:111912–111921. https://doi.org/10.18632/oncotarget.22951
Wang Z, Wang W, Huang K, Wang Y, Li J, Yang X (2017b) MicroRNA-34a inhibits cells proliferation and invasion by downregulating Notch1 in endometrial cancer. Oncotarget 8:111258–111270. https://doi.org/10.18632/oncotarget.22770
Wotschofsky Z, Busch J, Jung M, Kempkensteffen C, Weikert S, Schaser KD, Melcher I, Kilic E, Miller K, Kristiansen G, Erbersdobler A, Jung K (2013) Diagnostic and prognostic potential of differentially expressed miRNAs between metastatic and non-metastatic renal cell carcinoma at the time of nephrectomy. Clin Chim Acta 416:5–10. https://doi.org/10.1016/j.cca.2012.11.010
Wu D, Lu P, Mi X, Miao J (2018) Downregulation of miR-503 contributes to the development of drug resistance in ovarian cancer by targeting PI3K p85. Arch Gynecol Obstet. https://doi.org/10.1007/s00404-018-4649-0
Xu Y, Miao C, Jin C, Qiu C, Li Y, Sun X, Gao M, Lu N, Kong B (2018) SUSD2 promotes cancer metastasis and confers cisplatin resistance in high grade serous ovarian cancer. Exp Cell Res. https://doi.org/10.1016/j.yexcr.2017.12.029
Yang X, Fraser M, Moll UM, Basak A, Tsang BK (2006) Akt-mediated cisplatin resistance in ovarian cancer: modulation of p53 action on caspase-dependent mitochondrial death pathway. Cancer Res 66:3126–3136. https://doi.org/10.1158/0008-5472.CAN-05-0425
Zhang X, Shao R, Gao W, Sun G, Liu Y, Fa X (2018) Inhibition of miR-361-5p suppressed pulmonary artery smooth muscle cell survival and migration by targeting ABCA1 and inhibiting the JAK2/STAT3 pathway. Exp Cell Res. https://doi.org/10.1016/j.yexcr.2018.01.015
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Not applicable.
Additional information
Communicated by S. Hohmann.
Rights and permissions
About this article
Cite this article
Xiao, S., Zhang, M., Liu, C. et al. MiR-514 attenuates proliferation and increases chemoresistance by targeting ATP binding cassette subfamily in ovarian cancer. Mol Genet Genomics 293, 1159–1167 (2018). https://doi.org/10.1007/s00438-018-1447-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-018-1447-0