Abstract
Parasitic infection is one of the many challenges facing livestock production globally. Cysticercosis tenuicollis is a common parasitic disease in domestic and wild ruminants (intermediate host) caused by the larval stage of Taenia hydatigena that primarily infects dogs (definitive host). Although genetic studies on this parasite exist, only a few describe the genetic variation of this parasite in Mongolia. Our aim was thus, to identify the mitochondrial differences in ovine isolates of Cysticercus tenuicollis entering China from Mongolia and comparison with existing Chinese isolates from sheep and goats based on the recently described PCR–RFLP method and mitochondrial genes of NADH dehydrogenase subunit 4 (nad4) and the NADH dehydrogenase subunit 5 (nad5). Sixty-nine isolates were collected during routine veterinary meat inspections from sheep that originated from Mongolia, at the modern slaughterhouses in Erenhot City, Inner Mongolia. Additional 114 cysticerci were also retrieved from sheep and goats from northern (Inner Mongolia Autonomous Region, Ningxia Hui Autonomous Region, and Gansu Province), western (Tibet Autonomous Region), and southern (Jiangxi Province and Guangxi Province) China. The PCR–RFLP approach of the nad5 showed nine mitochondrial subclusters A1, A2, A3, A5, A8, A9, A10, A11, and B of T. hydatigena isolates from sheep and goats from Mongolia and China. Meanwhile, haplogroup A1 RFLP profile was more widespread than other variants. These data supplements existing information on the molecular epidemiology of T. hydatigena in China and Mongolia and demonstrate the occurrence of similar genetic population structures in both countries.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Taenia belongs to the class Cestoda, the order Cyclophyllidea, and the family Taeniidae. Members of this genus infect carnivores and humans while their larval stages occur in a wide variety of intermediate hosts causing cysticercosis and coenurosis (Nguyen et al. 2016). Carnivores (primarily dogs) are the final hosts for T. hydatigena; meanwhile, wild and domestic ruminants act as intermediate hosts (Omar et al. 2016). The infection caused by the larval stages is of veterinary and medical importance with significant economic losses in undeveloped countries (Scala et al. 2015; Shamsaddini et al. 2017).
Taenia hydatigena infection in wild carnivores has been reported in 17 provinces of Mongolia. For the parasites to complete the life cycle, ecological regions, the existence of wild carnivores, and the behavior of local herders all play important roles. The number of livestock in Mongolia has reached 66.5 million consisting primarily of goats and sheep, followed by horses, cattle, and camels. Most herders in rural areas of Mongolia own animals, which are kept in common open pasturages (Ulziijargal et al. 2020). Slaughter of livestock for sale or personal consumption without veterinary inspection is common in Mongolia, especially in rural regions, and this action can promote the transmission of Taenia species (Davaasuren et al. 2014). The prevalence of T. hydatigena in Mongolia according to copro-DNA analysis of the cox1 and 12S rRNA nuclear genes reaches about 61.3% in dogs, 19.9% in wolves, 13.8% in red foxes, 4.8% in corsac foxes, and 7.5% in snow leopards (Ulziijargal et al. 2020). Cysticercus tenuicollis has also been reported in 30% of goats in the forest-steppe zone (Sharkhuu 2001). Cysticercus tenuicollis have also been detected in Mongolian cattle, yaks, sheep, roe deer, and ibex (Sharhuu and Sharkhuu 2004; Tazieva et al. 1981; Temuujin et al. 2022).
In more than 20 provinces of western and north-western areas of China, where livestock husbandry is relatively developed, Cysticercus tenuicollis is mainly an enzootic disease. The rates of C. tenuicollis infection in sheep differ by province (Li et al. 2013; Zhang et al. 2018). The prevalence of C. tenuicollis was as high as 46.94%, 62.77%, and 43.93% in goats, sheep, and pigs, respectively, in the Tibet Autonomous Region of China (Luo et al. 2017; Xia et al. 2014).
Molecular characterization and description of the genetic population structure of C. tenuicollis in sheep, camels, goats, pigs, deer, and wild ungulates (wild boar and wolves) are available in several countries (Abbas et al. 2021; Alvi et al. 2020; Boufana et al. 2015; Cengiz et al. 2019; Filip et al. 2019; Luo et al. 2017; Omar et al. 2016). In a previous report, analyses of the complete mitochondrial demonstrated two major ancestors of T. hydatigena, namely, A and B. Moreover, a significant degree of genetic variation was also reported among T. hydatigena found in Asian and African countries (China, Nigeria, Pakistan, and Sudan) (Ohiolei et al. 2021a, 2021b).
Although the distribution and prevalence of T. hydatigena have been widely investigated in final and intermediate hosts in Mongolia, the detailed genetic structure/variation requires detailed study since it shares a large land border with China with frequent animal movement. For China, while information is relatively available, additional and comparative studies with isolates from other countries will provide robust data for understanding the parasite transmission pattern and genetic population structure. Phylogenetic network analysis and the recent PCR–RFLP assay were thus applied to assess the mitochondrial genetic population structure of T. hydatigena metacestodes originating from Mongolia and China.
Methods
Study area and sample collection
Mongolia is a country in East and Central Asia, bordering the People’s Republic of China and the Russian Federation with a population of 2.7 million as of 2011. About 40% of the rural population are nomadic or semi-nomadic herdsmen; thus, the livestock sector consists of 90% of the total agricultural production. More than 70 million open-range pastoral livestock are present in the country. The temperature is 25 to 30 °C in the summer and can reach − 45 to − 50 °C in the winter (Tumur et al. 2020; Ulziijargal et al. 2020).
Cysticerci of T. hydatigena were collected from sheep entering China from Mongolia in the months of November and December 2020 during inspection at the modern slaughterhouses in Erenhot City, Inner Mongolia. An additional 114 cysticerci or cysts were also retrieved from sheep and goats from northern (Inner Mongolia Autonomous Region, Ningxia Autonomous Region, and Gansu Province), western (Tibet Autonomous Region), and southern (Jiangxi Province and Guangxi Province) China. The infected visceral organs were separated from the carcass to record the size and number of Cysticercus tenuicollis cysts. The sizes were measured by a ruler. All samples were processed and preserved in 70% alcohol and stored at − 70 °C before DNA extraction (Sarvi et al. 2020).
DNA extraction, amplification, and sequencing
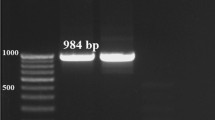
Cysticercus tenuicollis cysts (usually scoleces) were rinsed with phosphate buffer saline three times to remove the ethanol (Bhattacharya et al. 2007). Genomic DNA extraction was performed from each cysticercus or cyst using a genomic DNA isolation kit that was supplied by TIANamp Genomic DNA Kit (Tiangen, Beijing, China) following the manufacturer’s instructions. The DNA concentration of each extraction was assessed using a spectrophotometer (Thermo Scientific Inc. Nano Quant) and kept at − 20 °C until PCR amplification. The nad4 and nad5 genes were amplified using the following respective primer pairs, Thn4F (forward): 5′-GTCTATTACTCGTTTTCTAAACAG-3′ and Thn4R (reverse): 5′-AATCTCAACCAGACACATAAGTG-3′ and Thn5F (forward): 5′-TAGGATTAATTTATGACTAGAGTCTCT-3′ and Thn5R (reverse): 5′-CTTCTCTCAATCTACCACTAGAAGAGG-3′ (Ohiolei et al. 2021b). In a PCR reaction cocktail containing 12.5 μl premix Ex Taq™ version 2.0 (Takara Bio, Kusatsu, Japan), 10 pmol of each primer, 0.5 μl of genomic DNA extract (concentration (c) ≥ 10 ng), and DNase-free water up to the final volume of 25 μl for nad4. For nad5 amplification, PrimeSTAR GXL master mix was used instead of premix Ex Taq™ version 2.0 (Takara Bio, Kusatsu, Japan). For the PCR reaction, 5 μl of 5 × PrimeSTAR GXL Buffer (Takara Bio, Kusatsu, Japan), 10 pmol of each primer, 2 μl of dNTP mixture (2.5 mM each), 0.5 μl PrimeSTAR GXL DNA Polymerase (1.25 U/μl), 0.5 μl of gDNA (c. ≥ 10 ng), and DNase-free water up to the final volume of 25 μl for nad4. The PCR conditions were as previously described (Ohiolei et al. 2019). Then, 5 μl of the PCR amplicons was visualized by electrophoresis in 1.5% (w/v) Tris 0.09 M, borate 0.09 M, and EDTA 0.02 M agarose gels in 1 × TAE buffer (40 mM Tris–acetate, 2 mM EDTA, pH 8.5), stained with Gel Red™ and viewed under a UV transilluminator. A 2000-bp stair was used as a DNA size marker in each gel for estimating the size of the amplicons. The PCR products were then sequenced (Beijing Tsingke Biotechnology Co., Ltd., Beijing, China).
RFLP-PCR assay for differentiation of haplogroups
The described RFLP-PCR assay for T. hydatigena targeting 1444 bp of the NADH dehydrogenase subunit 5 gene (nad5) (Ohiolei et al. 2021b) was employed. Briefly, the final PCR products amplified with the forward 5′-AGGAGTTACATTGTGTGTGACTGTG-3′ and reverse 5′-CCACTTATAAAATTGATTCCTG-3′ primers were digested with the following restriction endonuclease enzymes: Rsa1 (Thermo Fisher Scientific, Vilnius, Lithuania) and Acc1 (Thermo Fisher Scientific, Vilnius, Lithuania). Restriction reactions were performed in 25 µl using a concentration, 1 µl Rsa1 (10 U/µl), 1 µl Acc1 (10 U/µl), 2 µl Tango buffer, 10 µl PCR product, and 11 µl nuclease-free water. The reaction mixture was incubated at 37 °C for 16 h and the fragment patterns were examined after fractionation by 3% (w/v) Tris 0.09 M, borate 0.09 M, and EDTA 0.02 M agarose gels in 1 × TAE buffer, stained with Gel Red™ under a UV transilluminator.
Molecular analysis
All generated nucleotide sequences were manually checked for misread nucleotide bases and aligned with reference sequence in Unipro UGENE v1.29.0 software (Okonechnikov et al. 2012). nBLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to confirm the identity of each sequence while the population indices, including the number of haplotypes (h), nucleotide diversity (π), and haplotype diversity (Hd) were estimated in DnaSP v6 (Rozas et al. 2017). Pairwise fixation index or Fst was calculated using the Arlequin 3.5.2.2 software package (Excoffier et al. 2005). Median-joining network was constructed with sequences of the nad4, nad5, and their concatenated genes (nad4-nad5) using PopART (http://popart.otago.ac.nz) (Harigai et al. 2020). The general time reversible model and a gamma-shaped distribution of rates across sites with a proportion of invariable sites (GTR + G + I) were used as the best model of isolate evolution as determined by JModelTest (Posada 2008). A maximum likelihood (ML) phylogeny was used to construct phylogenetic tree with sequences of concatenated genes (nad4-nad5) in the program MEGA-X (Kumar et al. 2018). The Interactive Tree of Life (https://itol.embl.de) was used to display the phylogenetic tree.
Result and discussion
Cyst characteristics and sequence variation
The majority of the cysts were from sheep (172 out of 183 isolates) with only 11 from goats. The proportion of C. tenuicollis in slaughtered small ruminants was 46% in the omentum, 36.6% in the mesentery, 16.8% in the liver, and 0.6% in the lungs (Table 1). The high proportion of cysts collected from omentum followed by mesentery is a common observation (Alvi et al. 2020; Radfar et al. 2005). Cysts were round with a white thin wall containing a scolex (bladder worms) which appeared as a white dot. The sizes range from 1 to 11 cm in the omentum and 1 to 2 cm in the liver and lung which is similar to previous cyst sizes reported in Al-Diwaniyah Province Abattoirs in Iraq by Al-Hamzawi and Al-Mayali (2020).
PCR amplification of 114 isolates from China was 100% (114 from 114 isolates) and 85.1% (97 out of 114 isolates) successful for the complete nad4 (1254 bp) and nad5 (1569 bp) mitochondrial genes, respectively. Examination of the entire nad4, nad5, and concatenated nad4-nad5 (2823 bp) genes sequences showed 182, 223, and 385 segregating sites or polymorphic sites, respectively.
PCR amplification of the sixty-nine isolates from Mongolia demonstrated 100% (69 from 69) and 85.5% (59 out of 69) successful amplification of nad4 (1254 bp) and nad5 (1569 bp) genes, respectively. Examination of the complete nad4 (1254 bp), nad5 (1569 bp), and concatenated nad4-nad5 (2823 bp) gene sequences indicated 161, 168, and 318 polymorphic sites, respectively, for each gene (Table 2).
The nBLAST search for the resulting sequences accurately identified all isolates as T. hydatigena with 98.7 to 100% similarity to GenBank deposited sequences.
Population and diversity indices
The highest number of mutations, haplotypes, and nucleotide diversity were observed in sheep from China compared to Mongolia (Table 2).
The overall number of mutations in Chinese sheep and goats was 191 (177 and 69 for sheep and goats, respectively), and in Mongolian sheep was 165 for nad4. Evaluation of nad5 gene showed 233 mutations in Chinese sheep and goats (223 and 31 for sheep and goats, respectively), and 173 in Mongolian sheep for nad5. A previous study on sheep (Ohiolei et al. 2021a) from China demonstrated lower mutations (93). Compared to another study from Nigeria, fewer mutations were observed in sheep (1) and fairly higher in goats (33) (Ohiolei et al. 2019). For the spliced genes (nad4-nad5), the number of mutations observed was 400 in Chinese sheep and goats (375 and 94 for sheep and goats, respectively), and 326 in Mongolian sheep. Fewer mutations from Pakistan based on cox1 mitochondrial gene (30 and 26 for goats and sheep, respectively) (Alvi et al. 2020) and from Sudan based on cox1 (735 bp) and nad1 (764 bp) from sheep (2 and 4, respectively) were reported (Muku et al. 2020). The disparity in sample sizes may be responsible for the number of mutations observed in all these studies.
The overall parsimony informative sites in Chinese sheep and goats were 106 (97 and 44 for sheep and goats, respectively), and 81 in Mongolian sheep for nad4. The nad5 gene gave 118 parsimony informative sites in Chinese sheep and goats (116 and 7 for sheep and goats, respectively), and 58 in Mongolian sheep. The parsimony of informative sites from a previous investigation in northern China provinces and autonomous regions (Qinghai, Gansu, Inner Mongolia, Xinjiang, and Beijing) was lower (52) (Ohiolei et al. 2021a). Compared to the current study, moreover, the present study comprised isolates from the southern part of the country. Similarly, lower parsimony informative sites were reported from Pakistan based on cox1 (387 bp) mitochondrial gene in goats (16) and sheep (15) (Alvi et al. 2020) and from Sudan using cox1 (0) and nad1 (1) in sheep (Muku et al. 2020). The sample size differences and the size of the mitochondrial genes used may have contributed to this variation. The spliced nad4-nad5 gave a total of 217 parsimony informative sites for sheep (206) and goats (48) from China, and 128 for sheep from Mongolia.
The number of haplotypes in Chinese sheep and goats was 87 and 11, respectively, and 62 in Mongolian sheep based on nad4. For nad5, it was 69 and 10 in Chinese sheep and goats, respectively, and 53 in Mongolian sheep. Previous reports showed fewer haplotypes in Chinese and Nigerian sheep and goats (Ohiolei et al. 2019, 2021a). On a general note, the current indices were genetically incomparable to existing data from other countries due to the application and analysis of mitochondrial genes other than the nad4/nad5 genes. For instance, from Pakistan analysis of the cox1 gene of isolates found in goats and sheep gave 28 and 23 haplotypes, respectively (Alvi et al. 2020); Sudan, 2 and 3 for cox1 and nad1, respectively, in sheep (Muku et al. 2020); Sardinia (21 and 16) using nad1 and cox1 mitochondrial gene, in sheep, goat, and wild boar were reported (Boufana et al. 2015); and Southern Italy (21) using cox1 (386 bp) gene in wild boar (Sgroi et al. 2020).
Overall, haplotype diversity (Hd) in Chinese sheep and goats was 0.995 and 1.000, respectively, and was 0.994 in Mongolian sheep based on nad4. For nad5, 0.987 and 1.000 in Chinese sheep and goats, respectively, and 0.991 in Mongolian sheep. Lower diversities are also found in sheep in other regions of China (Ohiolei et al. 2021a), and likewise in Nigerian sheep and goats (Ohiolei et al. 2019). The lack of nad4/nad5 data from other countries with existing genetic data did not allow proper comparison. Nonetheless, results from Pakistan based on cox1 mitochondrial gene in goats and sheep were 0.893 and 0.908, respectively (Alvi et al. 2020) and Sardinia (0.988 and 0.947) for nad1 and cox1 mitochondrial gene in sheep, goat, and wild boar (Boufana et al. 2015). In Sudan, much lower Hd has been demonstrated for cox1 (Hd = 0.222) and nad1 (Hd = 0.345) in sheep (Muku et al. 2020).
Low nucleotide diversity (π) was observed in Chinese sheep and goats 0.013 (0.013 and 0.017 for sheep and goats, respectively), and in Mongolian sheep 0.011 using the nad4 gene and even lower for nad5 gene with 0.009 and 0.004 for Chinese sheep and goats, respectively and 0.007 for Mongolian sheep. Mitochondrial genes other than those analyzed in this study have also demonstrated lower π in Europe (π = 0.012 and π = 0.007) (Boufana et al. 2015) and Africa (π = 0.001) (Muku et al. 2020).
The overall nad4 Tajima’s D was negative for all Chinese isolates − 1.733 (− 1.709 and − 0.226 for sheep and goats, respectively), as well as in Mongolian sheep − 1.956. According to nad5, higher negative values were observed in Chinese sheep and goats (− 2.199 and − 1.483, respectively) with an overall value of − 2.257 and − 2.464 in Mongolian sheep (Table 2). Elsewhere in China, reports show higher Tajima’s D − 1.583 in sheep (Ohiolei et al. 2021a). In contrast, a Nigeria study showed a positive Tajima’s D in sheep (1.166) and a similar negative value in goats (− 1.646) (Ohiolei et al. 2019) and negative values in sheep (− 1.657) and goats (− 1.323) from Pakistan based on cox1 (Alvi et al. 2020), Sardinia (− 0.748 and − 1.486) using nad1 and cox1 mitochondrial gene, respectively (Boufana et al. 2015), and Sudan in sheep using cox1 (− 1.362) and nad1 (− 1.321) (Muku et al. 2020).
The overall Fu’s Fs were negative with very high values across all studied populations (Table 2) supporting the existence of rare haplotypes as expected from a recent population expansion. This feature is common in virtually all T. hydatigena populations in Asia and Europe (Alvi et al. 2020; Boufana et al. 2015; Ohiolei et al. 2021a) except in certain geography in Africa where very low values were reported from a rather small population (Muku et al. 2020; Ohiolei et al. 2019).
Average number of differences (K) was 17.14, 14.84, and 30.77 for nad4 (1254 bp), nad5 (1569 bp), and concatenated nad4-nad5 (2823 bp) mitochondrial genes, respectively, in China, and 14.96, 11.24, and 24.14 for nad4, nad5, and nad4-nad5, respectively, for the population from Mongolia.
Pairwise fixation index or Fst between China and Mongolia was also estimated to determine the genetic differences between populations from both countries and was 0.000 and 0.003 for nad4, respectively and 0.000 and 0.007 for China and Mongolia, respectively, for nad5. For the combined genes, it was 0.000 and 0.005, respectively (Table 2).
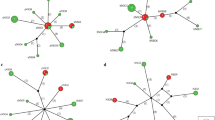
Haplotype networks
The haplotype network of nuclear nad4 and nad5 genes and their concatenation of C. tenuicollis showed haplotypes arranged in a star-like configuration with three extending groups (A1, A2, and B) (Fig. 1a, b, and c). For nad4, the network formed a star-like configuration of 153 haplotypes of C. tenuicollis of Mongolian and Chinese origin, of which seven haplotypes H6, H69, H5, H8, H46, H61, and H57 were present in both countries. For nad5, of the 125 haplotypes, four haplotypes H8, H15, H51, and H47 appeared in both countries. For the spliced nad4-nad5 gene network, only one haplotype H76 of the 144 was common to both countries, and interestingly, similar findings were reported previously (Sgroi et al. 2020). The majority of sequences from 93 out of 114 Chinese isolates were as haplogroup A variants, while 15 and 6 were as haplogroups B and A2 for the nad4 gene. However, out of 69 isolates from Mongolia 60, 5, and 4 belonged to haplogroups A, A2, and B, respectively. The most isolates, 85 out of 97, were as haplogroup A variants, 8 as haplogroup B variants, and 4 as haplogroup A2 for the nad5 gene from China, whereas from Mongolia 55, 1, and 3 from 59 isolates were as haplogroups A, A2, and B, respectively. On the other hand, for concatenated nad4-nad5, 85, 8, and 4 out of 97 Chinese isolates were grouped in to haplogroups A, B, and A2 variants whereas for Mongolia, 55, 1, and 3 out of 59 isolates were as haplogroups A, A2, and B, respectively (Fig. 1a, b, and c).
Restriction fragment length polymorphism
We digested a portion of the nad5 gene as recently described (Ohiolei et al. 2021a, b). The in silico RFLP fragment generated for 156 sequences showed nine RFLP profiles, namely, A1, A2, A3, A5, A8, A9, A10, A11, and B. A comparison of the in silico RFLP fragments of the Chinese and Mongolian isolates revealed five common haplogroups (A1, A2, A3, A5, and B). Besides four additional profiles (A8, A9, A10, A11), others were previously encountered in either one or all of the following countries: China, Nigeria, Pakistan, and Sudan (Ohiolei et al. 2021b). The fragment sizes and profiles of the haplogroups are showed in Table 3. Haplogroup A1 was more widespread than other haplogroups as previously described (Ohiolei et al. 2021b). Figure 2a, b depict PCR–RFLP profiles of all variants.
Phylogenetic analyses
In total, 156 spliced (nad4-nad5) nucleotide sequences with other homologous sequences from GenBank were used for the phylogenetic analysis. All obtained sequences diverged significantly from other taeniid species, T. solium (AB086256), T. asiatica (AF445798), T. saginata (AY684274), T. multiceps (GQ228818), T. pisiformis (GU569096), T. ovis (NC_021138), and E. granulosus (AB786664), which is used as an outgroup. The majority of nucleotide sequences of Cysticercus tenuicollis concatenated nad4-nad5 genes within phylogenetic tree, except for these isolates (haplotypes H136, H83, H125, H76, H104, H45, H131, H8, H69, H41, H40, H93, H38, H82, H120, H23, H127, H36, H98, H16, H97, H96, H139, H39, H14, H129, H143, H116, H9, H101, H92, H90, H79, H87, H21, H123, H91, H81, H51, H112, H124, H77, H109, H33, H56, and H32), clustered together and were divided in two subclades (Fig. 3). This study showed H119, H72, H109, H133, H2, H33, H100, H28, H56, H32, H117, and H50 as haplogroup B, and H11, H70, H102, H29, H112, H124, and H77 from Mongolia and China as haplogroup A2 of T. hydatigena, as represented in Fig. 3, and others as haplogroups A including A1, A3, A5, A8, A9, A10, and A11.
Conclusion
In conclusion, analysis of the mitochondrial loci demonstrate similar genetic composition of C. tenuicollis in sheep and goats in both countries compared to other geographical locations suggesting that animal movement and migration between the border countries contribute to the existing genetic population structure. These data provide supplementary information on the genetic epidemiology of T. hydatigena in China and Mongolia. Nonetheless, the lack of nad4 and nad5 genetic information from other enzootic regions limited a robust comparative outlook in the affected regions.
Data availability
All data supporting the conclusions of this article are included in this article and its additional files. Representative nucleotide sequences of nad4 and nad5 genes from the current study have been deposited in the GenBank database under the following accession numbers ON379092–ON379244 and ON379245–ON379369, respectively.
References
Abbas I, El-alfy ES, Janecek-erfurth E, Strube C (2021) Molecular characterization of Cysticercus tenuicollis isolates from sheep in the Nile Delta, Egypt and a review on Taenia hydatigena infections worldwide. Parasitology 148:913–933. https://doi.org/10.1017/S0031182021000536
Al-hamzawi EG, Al-mayali HMH (2020) Morphological characterizations of Cysticercus tenuicollis of Taenia hydatigenia isolated from sheep and goats slaughered in Al-Diwaniyah Province Abattoirs, Iraq. Eur Asian J Biosci 14:2261–2266
Alvi MA, Ohiolei JA, Saqib M, Li L, Muhammad N, Tayyab MH, Qamar W, Alvi AA, Wu YD, Li XR (2020) Preliminary information on the prevalence and molecular description of Taenia hydatigena isolates in Pakistan based on mitochondrial cox1 gene. Infect Genet Evol 85:104481. https://doi.org/10.1016/j.meegid.2020.104481
Bhattacharya D, Bera A, Bera B, Maity A, Das S (2007) Genotypic characterisation of Indian cattle, buffalo and sheep isolates of Echinococcus granulosus. Vet Parasitol 143:371–374. https://doi.org/10.1016/j.vetpar.2006.09.007
Boufana B, Scala A, Lahmar S, Pointing S, Craig PS, Dessì G, Zidda A, Pipia AP, Varcasia A (2015) A preliminary investigation into the genetic variation and population structure of Taenia hydatigena from Sardinia, Italy. Vet Parasitol 214:67–74. https://doi.org/10.1016/j.vetpar.2015.08.003
Cengiz G, YucelTenekeci G, Bilgen N (2019) Molecular and morphological characterization of Cysticercus tenuicollis in red deer (Cervus elaphus) from Turkey. Acta Parasitol 64:652–657. https://doi.org/10.2478/s11686-019-00085-1
Davaasuren A, Dorjsuren T, Yanagida T, Sako Y, Nakaya K, Davaajav A, Agvaandaram G, Enkhbat T, Gonchigoo B, Dulmaa N (2014) Recent situation of taeniasis in Mongolia (2002–2012). Korean J Parasitol 52:211. https://doi.org/10.3347/kjp.2014.52.2.211
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50. https://doi.org/10.1177/117693430500100003
Filip KJ, Pyziel AM, Jeżewski W, Myczka AW, Demiaszkiewicz AW, Laskowski Z (2019) First molecular identification of Taenia hydatigena in wild ungulates in Poland. EcoHealth 16:161–170. https://doi.org/10.1007/s10393-019-01392-9
Harigai W, Saito A, Suzuki H, Yamamoto M (2020) Genetic diversity of Ligidium isopods in Hokkaido and Niigata, northern Japan, based on mitochondrial DNA analysis. Zool Sci 37:417–428. https://doi.org/10.2108/zs200017
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547. https://doi.org/10.1093/molbev/msy096
Li WH, Jia WZ, Qu ZG, Xie ZZ, Luo JX, Yin H, Sun XL, Blaga R, Fu BQ (2013) Molecular characterization of Taenia multiceps isolates from Gansu Province, China by sequencing of mitochondrial cytochrome C oxidase subunit 1. Korean J Parasitol 51:197–201. https://doi.org/10.3347/kjp.2013.51.2.197
Luo H, Zhang H, Li K, Rehman MU, Mehmood K, Lan Y, Huang S, Li J (2017) Epidemiological survey and phylogenetic characterization of Cysticercus tenuicollis isolated from Tibetan pigs in Tibet. China Biomed Res Int 2017:6. https://doi.org/10.1155/2017/7857253
Muku RJ, Yan HB, Ohiolei JA, Saaid AA, Ahmed S, Jia WZ, Fu BQ (2020) Molecular identification of Taenia hydatigena from sheep in Khartoum. Sudan Korean J Parasitol 58:93. https://doi.org/10.3347/Kjp.2020.58.1.93
Nguyen MTT, Gabriel S, Abatih EN, Dorny P (2016) A systematic review on the global occurrence of Taenia hydatigena in pigs and cattle. Vet Parasitol 226:97–103. https://doi.org/10.1016/j.vetpar.2016.06.034
Ohiolei JA, Luka J, Zhu GQ, Yan HB, Li L, Magaji AA, Alvi MA, Wu YT, Li JQ, Fu BQ, Jia WZ (2019) First molecular description, phylogeny and genetic variation of Taenia hydatigena from Nigerian sheep and goats based on three mitochondrial genes. Parasit Vectors 12:1–11. https://doi.org/10.1186/s13071-019-3780-5
Ohiolei JA, Yan HB, Li L, Li WH, Wu YD, Alvi MA, Zhang NZ, Fu BQ, Wang XL, Jia WZ (2021a) A new molecular nomenclature for Taenia hydatigena: mitochondrial DNA sequences reveal sufficient diversity suggesting the assignment of major haplotype divisions. Parasitology 148:311–326. https://doi.org/10.1017/S003118202000205X
Ohiolei JA, Yan HB, Li L, Alvi MA, Muku RJ, Wu YD, Zhang NZ, Li WH, Guo AM, Wang XL, Fu BQ, Jia WZ (2021b) PCR-RFLP assay confirms the existence of different mitochondrial lineages of Taenia hydatigena including a possible geographically restricted group. Transbound Emerg Dis. https://doi.org/10.1111/tbed.141551
Okonechnikov K, Golosova O, Fursov M, U Team (2012) Unipro UGENE: a unified bioinformatics toolkit. Bioinform 28:1166–1167. https://doi.org/10.1093/bioinformatics/bts091
Omar MAE, Elmajdoub LO, Al-aboody MS, Elsify AM, Elkhtam AO, Hussien AA (2016) Molecular characterization of Cysticercus tenuicollis of slaughtered livestock in Upper Egypt governorates. Asian Pac J Trop Biomed 6:706–708. https://doi.org/10.1016/j.apjtb.2016.06.009
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. https://doi.org/10.1093/molbev/msn083
Radfar MH, Tajalli S, Jalalzadeh M (2005) Prevalence and morphological characterization of Cysticercus tenuicollis (Taenia hydatigena cysticerci) from sheep and goats in Iran. Vet Arh 75:469–476
Rozas J, Ferrer-mata A, Sánchez-delbarrio JC, Guirao-rico S, Librado P, Ramos-onsins SE, Sánchez-gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302. https://doi.org/10.1093/molbev/msx248
Sarvi S, Behrestaghi LE, Alizadeh A, Hosseini SA, Gohardieh S, Bastani R, Charati JY, Daryani A, Amouei A, Spotin A (2020) Morphometric, genetic diversity and phylogenetic analysis of Taenia hydatigena (Pallas, 1766) larval stage in Iranian livestock. Parasitology 147:231–239. https://doi.org/10.1017/S0031182019001434
Scala A, Pipia AP, Dore F, Sanna G, Tamponi C, Marrosu R, Bandino E, Carmona C, Boufana B, Varcasia A (2015) Epidemiological updates and economic losses due to Taenia hydatigena in sheep from Sardinia, Italy. Parasitol Res 114:3137–3143. https://doi.org/10.1007/s00436-015-45-32-x
Sgroi G, Varcasia A, D’alessio N, Varuzza P, Buono F, Amoroso MG, Boufana B, Otranto D, Fioretti A, Veneziano V (2020) Taenia hydatigena cysticercosis in wild boar (Sus scrofa) from southern Italy: an epidemiological and molecular survey. Parasitology 147:1636–1642. https://doi.org/10.1017/S00311820001559
Shamsaddini S, Mohammadi MA, Mirbadie SR, Rostami S, Dehghani M, Sadeghi B, Harandi MF (2017) A conventional PCR for differentiating common taeniid species of dogs based on in silico microsatellite analysis. Rev Inst Med Trop São Paulo 2017:59. https://doi.org/10.1590/S1678-9946201759066
Sharhuu G, Sharkhuu T (2004) The helminth fauna of wild and domestic ruminants in Mongolia—a review. Eur J Wildl Res 50:150–156. https://doi.org/10.1007/s10344-004-0050-3
Sharkhuu T (2001) Helminths of goats in Mongolia. Vet Parasitol 101:161–169. https://doi.org/10.1016/S0304-4017(01)00508-8
Tazieva ZH, Isabaev MI, Sharhuu G (1981) Legočnye nematody kopytnyh Mongolii. Tezisy dokladov nauč. konf. vsesoűzn. obŝestva gel’mintol. Akad Nauk SSSR 33:74–76
Temuujin J, Ulziijargal G, Yeruult C, Amarbayasgalan Z, Mungunzaya T, Bayarsaikhan U, Khulan J, Wandra T, Sato MO, Gantstetseg C (2022) Distribution and prevalence of Taenia hydatigena, Taenia multiceps, and Mesocestoides spp. in Mongolian sheepdogs. Vet Parasitol Reg Stud Rep 28:2. https://doi.org/10.1016/j.vprsr.2021.100680
Tumur E, Heijman W, Gurjav EA, Agipar B, Heerink N (2020) Options for increasing Mongolia’s livestock sector exports-a revealed comparative advantage analysis. Mong J Agric Sci 30:38–50. https://doi.org/10.5564/mjas.v30i2.1490
Ulziijargal G, Yeruult C, Khulan J, Gantsetseg C, Wandra T, Yamasaki H, Narankhajid M (2020) Molecular identification of Taenia hydatigena and Mesocestoides species based on copro-DNA analysis of wild carnivores in Mongolia. Int J Parasitol Parasites Wildl 11:72–82. https://doi.org/10.1016/j.ijppaw.2019.12.004
Xia C, Liu J, Yuan Z, Feng J, Song T (2014) Investigation of infectious prevalence of major metacestodes in livestock in Tibet. Chinese Vet Sci Zhongguo Shouyi Kexue 44:1205–1209
Zhang Y, Zhao W, Di yang YT, Zhang W, Liu A (2018) Genetic characterization of three mitochondrial gene sequences of goat/sheep-derived Coenurus cerebralis and Cysticercus tenuicollis isolates in Inner Mongolia. China Parasite 25:2. https://doi.org/10.1051/parasite/2018002
Acknowledgements
The authors would like to acknowledge the Graduate School of Chinese Academy of Agricultural Sciences for providing the scholarship. We would like also thank to Lanzhou Veterinary Research Institute for all the facilitations. Finally, our thanks goes to those who help during sample collection and processing.
Funding
This work was funded by Central Public-Interest Scientific Institution Basal Research Fund (Y2022GH13; 1610312020016), National Key Research and Development Program of China (2021YFE0191600), and Cultivation of Achievements of State Key Laboratory of Veterinary Etiological Biology (SKLVEB2020CGPY01).
Author information
Authors and Affiliations
Contributions
SA Qurishi and WZ Jia did the conceptualization; SA Qurishi, HB Yan, L Li, and JA Ohiolei designed the research work, data analysis; SA Qurishi, LS Zhang, HaDa, HM Qiao, BaoHua, BX Bai, WJ Tian, and JM Xu did the sample collection and processing; SA Qurishi, JA Ohiolei, MA Alvi, and NA Shumuye review the manuscript; HB Yan, L Li, BQ Fu, and WZ Jia supervised the overall activities of the research. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
All animals were handled in strict accordance with good animal practice according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China, and the study was approved by the Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (No. LVRIAEC2012-007).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Handling Editor: Una Ryan
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Qurishi, S.A., Yan, HB., Li, L. et al. Comparison of mitochondrial genetic variation of Taenia hydatigena cysticerci from China and Mongolia. Parasitol Res 121, 3455–3466 (2022). https://doi.org/10.1007/s00436-022-07669-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-022-07669-3