Abstract
The presence of Sarcocystis cysts in the muscle tissue of domestic dogs (Canis familiaris), which normally serve as definitive hosts, is unusual and infrequent. Here, S. caninum sarcocysts were identified for the first time in two of 37 dogs (2.7%) from China. Examination using light microscopy found that the S. caninum sarcocysts were up to 1520 μm long and up to 147 μm wide and contained numerous 1.5–3.3 μm wedge-like villar protrusions (vp). Transmission electron microscopy revealed that the sarcocysts had pleomorphic vp that closely resembled those of “type 9c.” Five loci, 18S rDNA, 28S rDNA, mitochondrial cox1, ITS1 and ropB, were sequenced and characterized in S. caninum sarcocysts. The sequences of the five loci shared similarities of 99.9–100%, 99.0–100%, 99.4–100%, 99.6–100%, and 99.7–100%, respectively, with those of S. arctica. Phylogenetic analysis based on the sequences of 28S rDNA and mitochondrial cox1 indicated that S. caninum and S. arctica are closely related to Sarcocystis species that use a raptorial bird as their definitive host.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sarcocystis spp. are cyst-forming intracellular protozoan parasites with an obligate two-host life cycle that alternates between various predators, which serve as definitive hosts, and prey animals, which serve as intermediate hosts. The intermediate hosts become infected with Sarcocystis species by ingesting sporocysts and/or oocysts that have been excreted by a definitive host, while definitive hosts become infected by ingesting sarcocysts present in the muscle tissue of an intermediate host species (Dubey et al. 2016).
The domestic dog (Canis familiaris) commonly serves as a definitive host of numerous species of Sarcocystis. However, it is considered only incidental for dogs to serve as intermediate hosts of Sarcocystis (Dubey et al. 2016). Recently, two species, S. caninum and S. svanai, have been proposed to be responsible for the sarcocysts found in dogs by Dubey et al. (2015) based on investigation of the sarcocyst wall ultrastructure, limited molecular characterization, and clinical findings. To date, Sarcocystis cysts have been detected in only nine dogs worldwide (Dubey et al. 2016; Hagner et al. 2018); however, the prevalence of Sarcocystis spp. in domestic dogs in China is unknown.
Currently, sequence analysis has proved to be an essential tool to delineate and identify Sarcocystis species found in the same or different hosts. Different genetic markers have shown varying levels of intra- or interspecific sequence diversity (Gjerde 2013). However, there are only two partial 18S rDNA sequences, one ITS1 sequence, and two ropB sequences from Sarcocystis spp. that were obtained from muscle samples from dogs that have been deposited in GenBank.
Therefore, the aims of the present study were to (i) investigate the prevalence of Sarcocystis in domestic dogs in China based on morphological characteristics, and (ii) characterize the parasite via the analysis of five loci (18S rDNA, 28S rDNA, mitochondrial cox1, ITS1, and ropB). This is the first investigation of Sarcocystis in muscle tissues from dogs in China.
Materials and methods
Examination of sarcocysts from domestic dogs
The present study was approved by the Animal Ethics Committee of Yunnan University (permission number AEC2015021). Skeletal muscle samples from 37 dogs (100 g each) were purchased from a peddlers’ market in Yulin City, Guangxi Zhuang Autonomous Region, southwestern China, in November 2017. In this area, the consumption of dog meat is a long-standing culinary tradition for the local residents. All of the samples were from adult dogs, and the histories of these dogs were unknown. In our laboratory, 40 0.5-mm pieces of muscle from each animal were squeezed between two glass slides and examined for sarcocysts using a stereomicroscope. Individual sarcocysts were then isolated from muscular fibers using dissection needles and processed for examination using light microscopy (LM), transmission electron microscopy (TEM), and DNA analysis.
For TEM, six sarcocysts (three from dog no. 14 and three from dog no. 21) were fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) at 4 °C, postfixed in 1% osmium tetroxide in the same buffer, then dehydrated using a series of graded alcohols and embedded in a mixture of epon-araldite. Ultrathin sections were stained with uranyl acetate and lead citrate and examined using a JEM 100-CX transmission electron microscope (JEOL Ltd., Tokyo, Japan) at 80 kV.
Isolation, polymerase chain reaction (PCR) amplification, cloning, and sequence analysis of DNA
Genetic DNA was extracted from four individual sarcocysts (cysts #1 and #2 from dog no. 14 and cysts #3 and #4 from dog no. 21) using the TIANamp Genomic DNA Kit (Tiangen Biotech Ltd., Beijing, China) according to the manufacturer’s instructions. Sarcocystis species were characterized at five loci located in 18S rDNA, 28S rDNA, mitochondrial cox1, ITS1, and rpoB. The sequences at these loci were amplified using the primer pairs shown in Table 1. The PCR amplifications were performed as previously described (Kirillova et al. 2018). The PCR products were purified, cloned, sequenced, and analyzed using the methods described in a previous report (Hu et al. 2017).
Results
LM and TEM examination of S. caninum sarcocysts
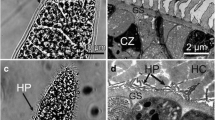
Only sarcocysts resembling those of S. caninum were found in two of 37 (2.7%) dogs. LM examination revealed that sarcocysts appeared to be slender and thin-walled and had numerous 1.5–3.3 μm wedge-like villar protrusions (vp) (Fig. 1a). Mature sarcocysts were 546–1520 × 93–147 μm (n = 20) in size, were septated, and contained bradyzoites that were 7.1–11.3 × 1.8–3.7 μm in size.
Morphological characteristics of Sarcocystis caninum isolated from skeletal muscle in domestic dogs. a LM micrograph of a sarcocyst (unstained). Note the short wedge-like villar protrusions (vp) (arrow). b TEM micrograph of a sarcocyst. Note the pleomorphic vp, thick ground substance (gs), septa (arrowhead), and host cell (hc). c TEM micrograph of a sarcocyst. Note the pleomorphic, anastomosed (arrow) vp, microfilaments (arrowhead), and wavy parasitophorous vacuolar membrane (pvm) lined by an electron-dense layer (edl)
Six sarcocysts from the both dogs in which S. caninum was found were examined using TEM, all of which appeared to have walls that were ultrastructurally similar and that closely resembled “type 9c.” The sarcocyst walls contained wavy parasitophorous vacuolar membranes (pvm) that contained pleomorphic vp and were lined with an approximately 70-nm-thick electron-dense layer. The vp were found to be up to 1.3 μm long and up to 1.0 μm wide and contained scattered microfilaments and numerous fine granules. Depending on the plane of the section, some vp were narrowed at their base and laterally expanded, and appeared to be anastomosed. A layer of ground substance measuring 0.7–1.1 μm in thickness was located immediately beneath the sarcocyst wall (Fig. 1b, c).
Molecular analysis
PCR amplification and sequencing were successful at all five loci in four S. caninum cysts (#1–4). The nucleotide sequences at each of the five loci that were obtained from the four individual sarcocysts were completely identical; therefore, only one sequence for each of the five loci was deposited in GenBank. The 18S rDNA (1803 bp long), 28S rDNA (3285 bp), mitochondrial cox1 (1085 bp), ITS1 (953 bp), and rpoB (807 bp) sequences obtained from the S. caninum cysts were deposited in GenBank under the accession numbers MH469238, MH469239, MH469240, MH469241, and MH469242, respectively.
The 18S rDNA sequence obtained from the S. caninum cyst was 100% identical to that of S. caninum (KM362427) found in a domestic dog from the USA, and 99.9–100% identical with those of S. arctica (KX022100–KX022103, MF596217–MF596237, KX156838, MF596217–MF596237, KX156838, KF601301, KY947306, and KY947307) found in Arctic foxes (Vulpes lagopus), red foxes (V. vulpes), and gray wolves (C. lupus) in Europe and/or the USA.
At the 28S rDNA locus, the sequence obtained from the S. caninum cyst was most similar to those of S. arctica (KF601312, KY947308, KY947309, KY609323, MF596240–MF596260, and KX022104–KX022107) found in Arctic foxes, red foxes, and gray wolves from Europe and/or the USA (99.9–100% sequence identity).
Based on the sequence from mitochondrial cox1, the S. caninum in our samples was 99.4–100% similar to S. arctica (MF596286–MF596306, KX022112–KX022115, KY947304, KY947305, and KF601318–KF601321) from Arctic foxes, red foxes, and gray wolves found in Europe and/or the USA.
The sequence of ITS1 obtained from the S. caninum cyst shared 99.4% of its identity with that of Sarcocystis sp. (JX993923) found in a dog from the USA, and 99.6–100% of its identity with those of S. arctica (MF596262–MF596282, KF601306, KF601308, KY947310, KY947311, and KX022108–KX022111) from Arctic foxes, red foxes, and gray wolves found in Europe and/or the USA.
The rpoB sequence obtained from the S. caninum cyst shared 99.8% of its identity with that of S. caninum (KC191641) found in a dog from the USA, and 99.7–100% of its identity with those of S. arctica (MF596311–MF596331) found in red foxes from Europe.
Phylogenetic analysis based on the 28S rDNA (Fig. 2) or mitochondrial cox1 (Fig. 3) sequences confirmed the relationship of the S. caninum found in our samples with Sarcocystis spp. from different hosts. In the phylogenetic tree constructed based on the 28S rDNA sequences, S. caninum was in the same clade as S. arctica from the Arctic fox (KF601312), red fox (MF596260), and gray wolf (KX022106), and this clade was within a group that comprised various Sarcocystis spp., including those found in raptorial birds (definitive hosts), such as S. glareoli (AF044251) and S. microti (AF044252) from voles and S. calchasi (FJ232949) from the domestic pigeon (Columba livia). In the phylogenetic tree constructed based on the mitochondrial cox1 sequences, S. caninum formed a clade with S. arctica from the Arctic fox (KF601321), red fox (MF596306), and gray wolf (KX022114), and this clade was within a group comprising Sarcocystis spp. that employ raptorial birds as definitive hosts, including S. lari (MF596283) from the great black-backed gull (Larus marinus), S. calchasi (KU220952) from the domestic pigeon, S. halieti (MF946583) from the white-tailed sea eagle (Haliaeetus albicilla), and Sarcocystis sp. (KY348756) from Cooper’s hawk (Accipiter cooperii).
Phylogenetic tree of selected members of Sarcocystidae based on 28S rDNA sequences and inferred using the maximum parsimony (MP) method with the tree bisection-regrafting algorithm (TBR). The analysis involved 23 nucleotide sequences (GenBank accession numbers shown) and a total of 1697 aligned positions in 21 taxa within the final dataset. The values between the branches represent the percent bootstrap values per 1000 replicates; the values below 50% were not shown. Sarcocystis caninum (shown in bold) formed a clade with S. arctica within a group comprising Sarcocystis spp. that use raptorial birds as definitive hosts
Phylogenetic tree for selected members of Sarcocystidae based on mitochondrial cox1 sequences and inferred using the maximum parsimony (MP) method with the tree-bisection-regrafting algorithm (TBR). The analysis involved 35 nucleotide sequences (GenBank accession numbers shown) and a total of 958 aligned positions in 33 taxa within the final dataset. The values between the branches represent the percent bootstrap values per 1000 replicates; the values below 50% were not shown. Sarcocystis caninum (shown in bold) formed a clade with S. arctica within a group comprising Sarcocystis spp. that use raptorial birds as definitive hosts
Discussion
Muscular sarcocystosis is a rare infection in dogs; this study is the first report of muscular Sarcocystis infection in dogs from China. Between 1964 and 1991, incidental Sarcocystis infections transmitted from unknown species were recorded in four dogs from the USA, India, and Kenya (Sahasrabudhe and Shah 1966; Hill et al. 1988; Blagburn et al. 1989; Bwangamoi et al. 1993). Recently, severe myositis has been reported in five dogs: two from Canada (Chapman et al. 2005; Dubey et al. 2015), two from the USA (Sykes et al. 2011; Dubey et al. 2015), and one from Finland (Hagner et al. 2018); all of these were associated with two new species of Sarcocystis, S. caninum, and S. svanai (Dubey et al. 2015). It is noteworthy that the morphological characterization of S. caninum based on LM and TEM examination closely resembles that of S. arctica isolated from different canid intermediate hosts, such as the Arctic fox (Gjerde and Schulze 2014; Cerqueira-Cézar et al. 2017), red fox (Pavlásek and Máca 2017; Kirillova et al. 2018), and gray wolf (Calero-Bernal et al. 2016). In the present study, sarcocysts from both infected dogs were similar to those of S. caninum and closely resembled the “type 9c” sarcocysts as classified by Dubey et al. (2016); no further clinical information was available for either dog.
The ultrastructure of sarcocysts has traditionally been used as a reliable indicator for the characterization of different Sarcocystis species present within the same host (Dubey et al. 2016). However, for the identification of morphologically similar sarcocysts in different but closely related intermediate hosts, sequence analysis using different genetic markers have proved to be useful (Gjerde 2013). Here, five loci (18S rDNA, 28S rDNA, mitochondrial cox1, ITS-1, and rpoB) in DNA samples from S. caninum sarcocysts were sequenced and analyzed in order to ascertain the relationship of the sarcocysts with S. arctica from different intermediate hosts. Among these sequences, those from the 28S rDNA and mitochondrial cox1 loci were the first evidence of the presence of Sarcocystis spp. in dog muscle tissue to be added to GenBank. In our samples, the five loci appeared to be high conserved (100% identity) among the four different isolates of S. caninum. Molecularly, the 18S rDNA and rpoB sequences shared 100 and 99.8% of their identity, respectively, with those obtained from S. caninum in the dogs from the USA. Additionally, the S. caninum sequences from the 18S rDNA, 28S rDNA, mitochondrial cox1, ITS-1, and rpoB loci, which have previously demonstrated varying levels of interspecies sequence variation (Wendte et al. 2010; Gjerde 2013), shared similarities of 99.9–100%, 99.0–100%, 99.4–100%, 99.6–100%, and 99.7–100%, respectively, with those from S. arctica found in the gray wolf, Arctic fox, and red fox from the USA and/or Europe. Thus, based on the similarities in morphology and between the five loci, the S. caninum found in dogs and the S. arctica found in the Arctic fox, red fox, or gray wolf may in fact be from the same species of Sarcocystis. However, this will need to be evaluated using cross transmission experiments in controlled conditions in the future.
The definitive hosts of S. caninum and S. arctica are not yet clear. Phylogenetic analysis based on the sequences of genetic markers has proved to be a useful method to infer the possible definitive hosts of Sarcocystis species (Hu et al. 2017). Here, the phylogenetic tree constructed based on the sequences from 28S rDNA and mitochondrial cox1 indicated that the clade formed by S. caninum and S. arctica was within a group comprising Sarcocystis spp. found in raptorial birds, which serve as the definitive hosts. Therefore, we surmise that S. caninum and S. arctica might use a bird as their definitive hosts, and this may be the reason that muscular sarcocystosis has been found to occur only sporadically in dogs.
References
Barta JR, Martin DS, Liberator PA, Dashkevicz M, Anderson JW, Feighner SD, Elbrecht A, Perkins-Barrow A, Jenkins MC, Danforth HD, Ruff MD, Profous-Juchelka H (1997) Phylogenetic relationships among eight Eimeria species infecting domestic fowl inferred using complete small subunit ribosomal DNA sequences. J Parasitol 83:262–271
Blagburn BL, Braund KG, Amling KA, Toivio-Kinnucan M (1989) Muscular Sarcocystis in a dog. Proc Helminthol Soc Wash 56:207–210
Bwangamoi O, Ngatia TA, Richardson JD (1993) Sarcocystis-like organisms in musculature of a domestic dog (Canis familiaris) and wild dogs (Lycaon pictus) in Kenya. Vet Parasitol 49:201–205
Calero-Bernal R, Cerqueira-Cézar CK, Verma SK, Mowery J, Carmena D, Beckmen K, Dubey JP (2016) Sarcocystis arctica (Apicomplexa: Sarcocystidae): ultrastructural description and its new host record, the Alaskan wolf (Canis lupus). Parasitol Res 115:2893–2897. https://doi.org/10.1007/s00436-016-5067-5
Cerqueira-Cézar CK, Thompson PC, Verma SK, Mowery J, Calero-Bernal R, Antunes Murata FH, Sinnett DR, Van Hemert C, Rosenthal BM, Dubey JP (2017) Morphological and molecular characterization of Sarcocystis arctica-like sarcocysts from the Arctic fox (Vulpes lagopus) from Alaska, USA. Parasitol Res 116:1871–1878. https://doi.org/10.1007/s00436-017-5462-6
Chapman J, Mense M, Dubey JP (2005) Clinical muscular sarcocystosis in a dog. J Parasitol 91:187–190. https://doi.org/10.1645/GE-406R
Dubey JP, Sykes JE, Shelton GD, Sharp N, Verma SK, Calero-Bernal R, Viviano J, Sundar N, Khan A, Grigg ME (2015) Sarcocystis caninum and Sarcocystis svanai n. spp. (Apicomplexa: Sarcocystidae) associated with severe myositis and hepatitis in the domestic dog (Canis familiaris). J Eukaryot Microbiol 62:307–317. https://doi.org/10.1111/jeu.12182
Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R (2016) Sarcocystosis of animals and humans, 2nd edn. CRC Press, Boca Raton
Fenger CK, Granstrom DE, Langemeier JL, Stamper S, Donahue JM, Patterson JS, Gajadhar AA, Marteniuk JV, Xiaomin Z, Dubey JP (1995) Identification of opossums (Didelphis virginiana) as the putative definitive host of Sarcocystis neurona. J Parasitol 81:916–919
Fischer S, Odening K (1998) Characterization of bovine Sarcocystis species by analysis of their 18S ribosomal DNA sequences. J Parasitol 84:50–54
Gjerde B (2013) Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol 43:579–591. https://doi.org/10.1016/j.ijpara.2013.02.004
Gjerde B (2014a) Sarcocystis species in red deer revisited: with a re-description of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. Sp. based on mitochondrial cox1 sequences. Parasitology 141:441–452. https://doi.org/10.1017/S0031182013001819
Gjerde B (2014b) Molecular characterisation of Sarcocystis rileyi from a common eider (Somateria mollissima) in Norway. Parasitol Res 113:3501–3509. https://doi.org/10.1007/s00436-014-4062-y
Gjerde B, Schulze J (2014) Muscular sarcocystosis in two arctic foxes (Vulpes lagopus) due to Sarcocystis arctica n. sp.: sarcocyst morphology, molecular characteristics and phylogeny. Parasitol Res 113:811–821. https://doi.org/10.1007/s00436-013-3711-x
Hagner K, Jokinen TS, Lavikainen A, Sukura A (2018) Acute fulminant necrotizing myopathy in a dog caused by co-infection with ultrastructural Sarcocystis caninum and Sarcocystis svanai-like apicomplexan protozoa. Vet Parasitol 252:153–156. https://doi.org/10.1016/j.vetpar.2018.02.011
Hill JE, Chapman WL Jr, Prestwood AK (1988) Intramuscular Sarcocystis sp. in two cats and a dog. J Parasitol 74:724–727
Hu JJ, Huang S, Wen T, Esch GW, Liang Y, Li HL (2017) Morphology, molecular characteristics, and demonstration of a definitive host for Sarcocystis rommeli from cattle (Bos taurus) in China. J Parasitol 103:471–476. https://doi.org/10.1645/16-187
Kirillova V, Prakas P, Calero-Bernal R, Gavarāne I, Fernández-García JL, Martínez-González M, Rudaitytė-Lukošienė E, Martínez-Estéllez MÁH, Butkauskas D, Kirjušina M (2018) Identification and genetic characterization of Sarcocystis arctica and Sarcocystis lutrae in red foxes (Vulpes vulpes) from Baltic States and Spain. Parasit Vectors 11:173. https://doi.org/10.1186/s13071-018-2694-y
Mugridge NB, Morrison DA, Johnson AM, Luton K, Dubey JP, Votýpka J, Tenter AM (1999) Phylogenetic relationships of the genus Frenkelia: a review of its history and new knowledge gained from comparison of large subunit ribosomal ribonucleic acid gene sequences. Int J Parasitol 29:957–972
Pavlásek I, Máca O (2017) Morphological and molecular identification of Sarcocystis arctica sarcocysts in three red foxes (Vulpes vulpes) from the Czech Republic. Parasitol Int 66:603–605. https://doi.org/10.1016/j.parint.2017.05.003
Sahasrabudhe VK, Shah HL (1966) The occurrence of Sarcocystis sp. in the dog. J Protozool 13:531–531
Sykes JE, Dubey JP, Lindsay LL, Prato P, Lappin MR, Guo LT, Mizisin AP, Shelton GD (2011) Severe myositis associated with Sarcocystis spp. infection in 2 dogs. J Vet Intern Med 25:1277–1283. https://doi.org/10.1111/j.1939-1676.2011.00828.x
Wendte JM, Miller MA, Nandra AK, Peat SM, Crosbie PR, Conrad PA, Grigg ME (2010) Limited genetic diversity among Sarcocystis neurona strains infecting southern sea otters precludes distinction between marine and terrestrial isolates. Vet Parasitol 169:37–44. https://doi.org/10.1016/j.vetpar.2009.12.020
Acknowledgements
This work was supported by the National Key R&D Program of China (grant 2017YFD0500400), the Natural Sciences Foundation of China (grant 31460557), and the Southeast Asia Biodiversity Research Institute, Chinese Academy of Science (grant Y4zk111B01).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Section Editor: Daniel K Howe
Rights and permissions
About this article
Cite this article
Ye, Y., Liang, Y., Hu, J. et al. First isolation of Sarcocystis caninum sarcocysts from two domestic dogs (Canis familiaris) from China. Parasitol Res 117, 3613–3618 (2018). https://doi.org/10.1007/s00436-018-6060-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-018-6060-y