Abstract
There is considerable confusion concerning the relationships among species of Sarcocystis found in donkeys and horses. Here, we describe a Sarcocystis species in Chinese donkeys (Equus asinus). Sarcocysts were found in 12 of 32 (37.5%) adult donkeys. By light microscopy, they were divided into two types, thin-walled and thick-walled. The thin-walled were macroscopic (up to 320 μm wide) and had short club-like protrusions (up to 2.7 μm long); the thick-walled were microscopic (up to 135 μm wide) and had villar protrusions (up to 5.4 μm long). Ultrastructures of the two types exhibited similar morphological characteristics, including bundled microtubules in the core of the villar protrusions penetrating diagonally into the ground substance, similar to wall type 11c. Three genetic markers, 18S rDNA, 28S rDNA, and mitochondrial cox1, obtained from the two morphotypes were sequenced and analyzed. The sequences of the three loci in the two morphotypes presented high intraspecific similarities of 97.2–99.5%, 97.8–99.6% and 99.0 − 99.9%, respectively. The most similar sequences in GenBank to the newly obtained 18S rDNA, 28S rDNA and cox1 sequences were those of Sarcocystis spp. in horses, with similarities of 90.0 − 97.5%, 94.7 − 95.1%, and 82.6 − 84.5%, respectively. Phylogenetic analysis using the three genetic markers indicated that the Sarcocystis sp. in donkeys formed an individual clade most closely related to a clade encompassing Sarcocystis spp. in horses. Further studies are needed for taxonomic identification of sarcocysts in donkeys because the Sarcocystis species in donkeys and horses are not successfully cross transmissible despite morphological similarities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The sarcocysts infecting donkeys (Equus asinus) were first described and named Sarcocystis asinus by Gadaev (1978) only based on the size, shape and staining of bradyzoites in smears. Subsequently, on the basis of cross-infection between horses and donkeys and the morphological similarity of the sarcocysts found in the two hosts, S. asinus was questioned, and the species of Sarcocystis infecting the two hosts came to be regarded as the same species (Matuschka 1983; Odening 1998; Dubey et al. 2015). Thus, the name S. bertrami is currently used for the sarcocysts diagnosed in the muscles of donkeys by most authors (Dubey et al. 2016; Passantino et al. 2019).
Currently, the sarcocysts found in horses are divided into two forms by Dubey et al. (2015): a macroscopic form (up to 15 mm long) with a thin cyst wall, named S. bertrami (synonym S. equicanis), and a microscopic form (up to 990 μm long) with a thick cyst wall, named S. fayeri. Ultrastructurally, the protrusions of S. bertrami sarcocysts are folded on the cyst wall, similar to type 11c, but those of S. fayeri sarcocysts appear almost vertical to the cyst wall, similar to type 11a. The morphological characteristics of horse sarcocysts have been observed to undergo changes in various stages of development, showing long protrusions (thick-walled) in the early phase and short protrusions (thin-walled) in the later phase (Fayer et al. 1983). Therefore, there is still considerable confusion concerning the relationship between S. bertrami/equicanis and S. fayeri (Odening et al. 1995; Ma et al. 2020).
In recent decades, molecular analysis based on nucleotide sequences has been recommended as a useful and efficient tool for delineating or identifying species of Sarcocystis from the same or different hosts. There are currently only limited donkey sarcocyst 18S rDNA and mitochondrial cox1 sequences deposited in GenBank, which were provided by Zeng et al. (2018). Based on the high similarity of mitochondrial cox1 sequences, these authors proposed S. bertrami (syn. S. fayeri) as the descriptor of the parasites of both horses and donkeys. However, the reliability of the meat samples used in their study was based mainly on the applied version of meat cutter (according to communication with the corresponding author of this paper, Dr. Yang). However, in local meat markets, meat sellers or butchers sometimes mix horse and donkey meat or replace donkey meat with horse meat to seek better returns because the price of donkey meat is higher than that of horse meat.
The morphological and molecular characteristics of S. bertrami sarcocysts isolated from horses in China have been investigated by our group previously (Ma et al. 2020). To clarify the relationship of the Sarcocystis species of donkeys and horses based on the molecular identification of meat samples, the aims of the present study were to investigate the morphological characteristics of donkey sarcocysts, to explore the relationship of the Sarcocystis species of donkeys and horses by molecular analysis, and to develop a method suitable for discrimination of the Sarcocystis species in the two animals via polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP).
Materials and methods
Collection and identification of meat samples
Donkey meat serves as a food source for humans and is commonly marketed in China. Muscle tissues from a total of 32 adult donkeys were purchased in a meat market in Kunming City located in Yunnan Province, China, in October 2019. Fresh muscle tissue (200 g) from each donkey was examined for sarcocysts.
To further confirm the species identification of the meat samples, 12 donkey meat and two horse meat (collected previously and stored at − 40 ℃) infected with sarcocysts were processed for molecular identification in the laboratory. DNA from each animal was extracted using a TIANamp Genomic DNA Kit (TIANGEN BIOTECH CO., LTD, Beijing, China) according to the manufacturer’s instructions. Mitochondrial cox1 was amplified with the primer pair ML1F/ML1R (5′-ACCACAAAGACATCGGCACT-3′/5′-CGTTTGGATGCGAATGCT-3′), designed using Oligo 5.0 software (National Biosciences, Inc., Plymouth, MN, USA) based on the highly conserved areas of mitochondrial cox1 sequences of Equus spp. deposited in GenBank. PCR assays were carried out in 25 μl reaction with 1X PCR buffer, 0.15 mmol MgCl2, 0.25 mmol dNTPs, 1 U Taq DNA polymerase (TakaRa, Dalian, China), 50–100 ng of DNA, and 25 pmol of each primer. The amplification program consisted of 35 cycles of 95 °C for 5 min, 54 °C for 30 s, and 72 °C for 5 min. The PCR products were sequenced on an ABI 3730XL automatic DNA sequencer (Applied Biosystems, Inc., Foster City, California).
Morphological examination of sarcocysts in donkeys
In the laboratory, 10 specimens of approximately 10 × 3 mm in size from each collected sample were pressed and squeezed between two glass slides and then inspected using a stereomicroscope. Thereafter, individual sarcocysts were extracted and isolated from skeletal muscular fibers using needles and processed for light microscopy (LM), transmission electron microscopy (TEM), and DNA analysis.
For TEM, four sarcocysts of each morphotype were fixed in 2.5% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.4) at 4 °C, postfixed in 1.0% osmium tetroxide in the same buffer, dehydrated in a graded alcohol series, and embedded in an Epon-Araldite mixture. Ultrathin sections were stained with uranyl acetate and lead citrate and then examined using a JEM100-CX transmission electron microscope (JEOL Ltd., Tokyo, Japan) at 100 kV.
Molecular characterization of sarcocysts in donkeys
For DNA analysis, six sarcocysts (three thin-walled cysts and three thick-walled cysts) obtained from donkey meat and two sarcocysts (one thin-walled cyst and one thick-walled cyst) isolated from the molecularly identified horse meat were subjected to genomic DNA extraction using a TIANamp Genomic DNA Kit. Three genetic markers, 18S rDNA, 28S rDNA, and mitochondrial cox1, were amplified from donkey sarcocysts with the primer sets S1/S4 (5′-CCATGCATGTCTAAGTATAAGC-3′/5′-TATCCCCATCACGATGCATAC-3′) (Fischer and Odening 1998), KL1/KL3 (5′-TACCCGCTGAACTTAAGC-3′/5′-CCACCAAGATCTGCACTAG-3′), KL4/KL5b (5′-AGCAGGACGGTGGTCATG-3′/5′-CTCAAGCTCAACAGGGTC-3′) and KL6a/KL2 (5′-GGATTGGCTCTGAGGG-3′/5′-ACTTAGAGGCGTTCAGTC-3′) (Mugridge et al. 1999), and SF1/SR9 (5′-ATGGCGTACAACAATCATAAAGAA-3′/5′-ATATCCATACCRCCATTGCCCAT-3′) (Gjerde 2013, 2014), respectively. PCR products were gel purified, cloned, sequenced, assembled, and analyzed using the methods detailed in a previous paper (Hu et al. 2016). Only mitochondrial cox1 was amplified from the sarcocysts obtained from horse meat using the SF1/SR9 primers for RFLP. To avoid increasing uncertainties, the 18S rDNA and mitochondrial cox1 sequences of donkey sarcocysts provided by Zeng et al. (2018) were not used in the present analysis.
To establish the PCR–RFLP strategy for discriminating the sarcocysts of donkeys and horses, the internal endonuclease cleavage sites in the mitochondrial cox1 nucleotide sequences of the sarcocysts of the two hosts were screened using Premier 5.0 software (Premier, Canada). Three mitochondrial cox1 nucleotide sequences (MH025631–MH025633) of S. bertrami from horses morphologically identified in our laboratory during a former investigation (Ma et al. 2020) were used for the detection of endonuclease cleavage sites. Based on the screening results, two restriction enzymes, EcoRI and HinfI, were selected to digest the amplified PCR products of mitochondrial cox1 from the thin-walled and thick-walled sarcocysts of the two hosts because of their ability to produce different numbers of fragments from the sarcocysts of donkey (three, of 196, 243, and 641 bp) and horse (two, of 416 and 644 bp). Aliquots of 10 μl of the resulting PCR products were double digested in 20 μl reactions with 1 μl EcoRI and 1 μl HinfI and 10 × Cutsmart buffer following the recommendations of the manufacturer (New England BioLabs). The digested products were analyzed via gel electrophoresis on 2% agarose gels stained with Goldenview at 100 V in 0.5 × TBE buffer.
Phylogenetic analyses were conducted separately on the nucleotide sequences of the 18S rDNA, 28S rDNA, and mitochondrial cox1 sequences by using MEGAX software (Kumar et al. 2018). The maximum likelihood (ML) trees of 18S rDNA, 28S rDNA and mitochondrial cox1 were generated with the Tamura 3-parameter, Hasegawa-Kishino-Yano, and Hasegawa-Kishino-Yano models, respectively, according to the Find Best DNA/Protein Models program integrated into MEGAX. The reliability of the maximum likelihood phylograms was tested via the bootstrap method using 1000 replications.
The 18S rDNA, 28S rDNA, and mitochondrial cox1 sequences of Sarcocystis spp. from different hosts were downloaded from GenBank and aligned using the ClustalW program implemented in MEGAX, using a gap opening penalty of 10/10 and a gap extension penalty of 0.1/0.2 as pairwise and multiple alignment parameters, respectively. The alignment was subsequently checked visually; some sequences were slightly truncated at both ends, so that all sequences started and ended at the same nucleotide positions. All sits were used. The final alignment of the 18S rDNA sequences consisted of 37 nucleotide sequences and 1218 aligned positions from eight taxa. Besnoitia besnoiti (DQ227418292304) and Toxoplasma gondii (U03070) were chosen as outgroups. The final alignment of the 28S rDNA sequences consisted of 19 nucleotide sequences and 3768 aligned positions from 14 taxa. Hammondia heydorni (AF159240), T. gondii (XR001974492), and B. besnoiti (XR001974492) were used as outgroup species to root the tree. The final alignment of mitochondrial cox1 sequences consisted of a total of 32 nucleotide sequences and 817 aligned positions from eight taxa. Hammondia heydorni (JX473251) and T. gondii (JX473253) were chosen as outgroups.
Results
Molecular identification of donkey and horse meat samples
The molecular identification based on mitochondrial cox1 sequences revealed that from 14 meat samples infected with sarcocysts 12 belonged to donkeys and two to horses. The 12 newly obtained mitochondrial cox1 sequences from donkeys were 1408 bp long and shared 99.7–100% identity (average 99.9%); the two newly obtained mitochondrial cox1 sequences from horses were 1408 bp and shared 100% identity. Therefore, only three nucleotide sequences from donkeys and one nucleotide sequence from the horse were deposited in GenBank, with accession numbers ON459761–ON459763 and ON459764, respectively. The identity between the sequences of donkeys and horses was 92.1–92.4% (average 92.3%). At this locus, the newly obtained nucleotide sequences from donkey and horse shared 97.8–100% (average 99.3%) and 98.2–100% (average 99.4%) identity, respectively, with those of E. asinus and E. caballus previously deposited in GenBank.
Morphological description of the detected sarcocysts in donkeys
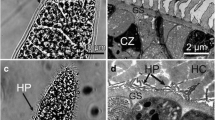
Sarcocysts were found in 12 of 32 (37.5%) adult donkeys. Using LM, the sarcocysts were divided into two types: thin-walled (Fig. 1a, b) and thick-walled (Fig. 1c). The thin-walled sarcocysts were macroscopic, measuring 2350–4856 × 110–320 μm [average = 2787 (± 442) × 210 (± 64) μm, (± SD); n = 20 isolated from five donkeys] in size, and exhibited numerous short club-like protrusions with lengths of 2.0 to 2.7 μm (mean = 2.4 ± 0.18 μm; n = 20 measurements from 10 sarcocysts); they were septate and contained bradyzoites measuring 14.5–17.4 × 3.5–5.0 μm [average = 15.8 (± 1.8) × 4.2 (± 0.4) μm, n = 20 measurements from three sarcocysts] in size. The thick-walled sarcocysts were microscopic, measuring 1200–3750 × 45–135 μm [average = 2213 (± 126) × 98 (± 21) μm, n = 20 isolated from four donkeys] in size and showed villar protrusions with lengths of 3.0 to 5.4 μm [mean = 4.2 (± 0.25) μm, n = 20 measurements from 10 sarcocysts]; they were septate and contained bradyzoites measuring 12.1–16.2 × 2.5–4.7 μm [average = 14.5 (± 1.2) × 4.1 (± 0.3) μm, n = 20 measurements from five sarcocysts] in size.
Morphological characteristics of Sarcocystis sp. sarcocysts isolated from skeletal muscles of donkeys. a Overview of a thin-walled sarcocyst (unstained, arrow) in muscular tissue (m) under a stereomicroscope. b Thin-walled sarcocyst (unstained) bound by short club-like protrusions (arrow) under light microscopy (LM). c Thick-walled sarcocyst (unstained) bound by villar protrusions (arrow) under LM. d Diagonal section of a thin-walled sarcocyst under transmission electron microscopy (TEM). The sarcocyst wall exhibits numerous villar protrusions (vp), which are often bent along the cyst surface. The vp contain bundled microtubules (mt) in their core, which penetrate diagonally into the ground substance (gs). Minute undulations (mu) present over the entire sarcocyst surface. e Longitudinal section of a thick-walled sarcocyst under TEM. The sarcocyst wall exhibits numerous vp, which are often sloping on the cyst surface. The vp contain bundled microtubules (mt) in the core, which penetrate diagonally into the ground substance (gs)
By TEM, the thin-walled and thick-walled sarcocysts exhibited similar morphological characteristics (Fig. 1d, e): the primary cyst wall had numerous villar protrusions with bundled microtubules in the core, which penetrated diagonally into the ground substances and sometimes reached the interior border of the ground substance. Minute undulations were present over the entire sarcocyst surface. A layer of ground substance was present beneath the protrusions. Overall, the cyst wall was similar to TEM type 11c classified by Dubey et al. (2015).
Molecular characterization of sarcocysts in donkeys
The three selected genes (18S rDNA, 28S rDNA and mitochondrial cox1) were successfully amplified from six individual sarcocysts (three thin-walled cysts and three thick-wall cysts) isolated from four donkeys. The three 18S rDNA sequences (accession numbers OM971696–OM971698) of the thin-walled sarcocysts were 1591–1614 bp long and shared 97.7–99.8% identity (average 98.4%). The three 18S rDNA sequences (OM971699–OM971701) of the thick-walled sarcocysts were 1589–1607 bp long and shared 97.7–98.5% identity (average 98.2%). The similarity between the two morphotypes was 97.2–99.5% (average 97.8%). The most similar sequences in GenBank to the newly obtained 18S rDNA sequences were those of Sarcocystis spp. obtained from horses in different regions, including S. bertrami (MH025625 − MH025628) from Chinese horses (95.6–97.5% identity, average 96.5%), S. fayeri (LC171838) from an Italian horse (95.2 − 97.1%, average 96.3%), S. fayeri (AB661437 − AB661447) from Japanese horses (90.8 − 97.4% identity, average 94.7%), S. fayeri (AB972440 − AB972443 and LC171831 − LC171837) from Canadian horses (90.0 − 97.1% identity, average 94.1%), and S. fayeri (MF614956) from an Egyptian horse (93.7 − 93.8%, average 93.8%).
The three 28S rDNA sequences (OM971683 − OM971685) obtained from thin-walled sarcocysts were 3441 − 3450 bp in length and shared 97.7 − 98.5% identity (average 98.1%). Only two 28S rDNA sequences (OM971686 and OM971687) of thick-walled sarcocysts were successfully assembled. They were 3445 and 3446 bp in length and shared 98.8% identity. The similarity between the two morphotypes was 97.8–99.6% (average 98.4%). The most similar sequences were those of S. bertrami (MH025629 − MH025630) from Chinese horses (94.7 − 95.1% identity, average 94.9%), followed by those of S. suihominis (MK867471 − MK867473) obtained from domestic pigs (90.0 − 91.2% identity, average 90.7%).
The three mitochondrial cox1 sequences (OM970235 − OM970237) of thin-walled sarcocysts were 1085 bp in length and shared 99.2 − 99.7% identity (average 99.4%). The three mitochondrial cox1 sequences (OM970238 − OM970240) of thick-walled sarcocysts were 1085 bp in length and shared 99.2 − 99.3% identity (average 99.3%). The identity between the two morphotypes was 99.0 − 99.9% (average 99.4%). The most similar sequences in GenBank were those of Sarcocystis spp. obtained from horses in different regions, including S. fayeri (LC171840 − LC171854) from Canadian horses (82.8 − 84.5% identity, average 83.9%), S. bertrami (MH025631 − MH025633) from Chinese horses (82.3 − 83.2% identity, average 82.9%), S. fayeri (LC171857) from an Italian horse (83.1–83.8% identity, average 83.4%), and S. fayeri (LC171855 and LC171856) from Japanese horses (82.7 − 83.4% identity, average 83.1%).
PCR–RFLP based on mitochondrial cox1 obtained from donkey and horse sarcocysts
The PCR-amplified products of mitochondrial cox1 from thin-walled and thick-walled sarcocysts in donkeys and horses were successfully digested by EcoRI and HinfI. This produced three fragments (196, 243, and 641 bp) and two fragments (416 and 644 bp) for the Sarcocystis sp. in donkeys and S. bertrami in horses, respectively (Fig. 2).
Results of PCR with primers SF1/SR9 and restriction enzyme digestion with EcoRI and HinfI for sarcocyst DNA from Sarcocystis bertrami from a naturally infected horse and Sarcocystis sp. isolated from a naturally infected donkey. M, molecular mass marker; Sb, Sarcocystis bertrami; Ss, Sarcocystis sp.; PI, PCR product of thin-walled sarcocyst; DPI, digestion of PCR product of thin-walled sarcocyst with EcoRI and HinfI; PII, PCR product of thick-walled sarcocyst; DPII, digestion of PCR product of thick-walled sarcocyst with EcoRI and HinfI
Phylogenetic analysis
Phylogenetic analysis based on the newly obtained 18S rDNA (Fig. 3), 28S rDNA (Fig. 4) and mitochondrial cox1 (Fig. 5) sequences confirmed their association with the Sarcocystis species, and the Sarcocystis sp. obtained from donkeys formed an individual clade most closely related to a clade encompassing S. bertrami and S. fayeri obtained from meat of horses originating from Chinese, Japanese, Canadian and Italian horses.
Phylogenetic tree based on 18S rDNA sequences. The tree was built using the maximum likelihood (ML) with the Tamura 3-parameter model. The analysis involved 37 nucleotide sequences (GenBank accession numbers behind the taxon names) and a total of 1218 aligned positions in the final dataset. The values between the branches represent bootstrap values per 1000 replicates, and values below 50% are not shown. The six new sequences of Sarcocystis sp. (OM971696–OM971701, shown in boldface) in donkeys formed an individual clade closely related to a clade encompassing S. bertrami and S. fayeri in horses
Phylogenetic tree based on 28S rDNA sequences. The tree was built using the maximum likelihood (ML) with the Hasegawa-Kishino-Yano model. The analysis involved 19 nucleotide sequences (GenBank accession numbers behind the taxon names) and a total of 3768 aligned positions in the final dataset. The values between the branches represent bootstrap values per 1000 replicates, and values below 50% are not shown. The five new sequences of Sarcocystis sp. (OM971683–OM971687, shown in boldface) in donkeys formed an individual clade closely related to a clade encompassing S. bertrami in horses
Phylogenetic tree based on mitochondrial cox1 sequences. The tree was built using the maximum likelihood (ML) with the Hasegawa-Kishino-Yano model. The analysis involved 32 nucleotide sequences (GenBank accession numbers behind the taxon names) and a total of 817 aligned positions in the final dataset. The values between the branches represent bootstrap values per 1000 replicates, and values below 50% are not shown. The six new sequences of Sarcocystis sp. (OM970235–OM970240, shown in boldface) obtained from donkeys formed an individual clade closely with S. bertrami and S. fayeri obtained from horses
Discussion
Sarcocystis is a common parasitic protozoan with a worldwide distribution found in a variety of mammals and birds, as well in domesticated food animals. Sarcocysts have been diagnosed in donkeys from the former USSR (Gadaev 1978), Austria (Hinaidy and Loupal 1982), Germany (Matuschka 1983), Morocco (Kirmse 1986), Egypt (Hilali and Nasser 1987; Dubey et al. 2016), China (Hu et al. 2001), and Italy (Passantino et al. 2019). In the present study, the prevalence of sarcocysts in the investigated Chinese donkeys was 37.5% (12/32), and it has been reported to be similar to 40% (8/20) in the former USSR (Gadaev 1978), higher than 22.0% (9/41) in Moroccan donkeys (Kirmse 1986) and 28.6% (40/140) in Italian donkeys (Passantino et al. 2019), but lower than 90.0% (18/20) in Egyptian donkeys (Hilali and Nasser 1987) and 92.3% (24/26) in Chinese donkeys surveyed by our group 20 years ago (Hu et al. 2001). The decrease in the prevalence of sarcocysts in Chinese donkeys may be due to the intensive culture of donkeys gradually replacing free-range farming, which reduces opportunities of the livestock meeting feces of domestic dogs.
In the present study, two morphotypes of sarcocysts (thin-walled cysts and thick-walled cysts) were observed in the muscle tissues of donkeys under LM. The thin-walled sarcocysts were macroscopic (up to 4856 long and 320 μm wide) and had short club-like protrusions (up to 2.7 μm long); the thick-walled sarcocysts were microscopic (up to 3750 μm long and 135 μm wide) and had villar protrusions (up to 5.4 μm long). The two morphotypes of sarcocysts were probably associated with the length of development time in the intermediate host tissues. Fayer et al. (1983) experimentally infected ponies with sporocysts collected from dogs that had been fed horsemeat containing visible sarcocysts. On day 127 post-infection (PI), the sarcocysts measured 50–360 × 12.6–30 μm. Some sarcocysts walls had long protrusions (4.5 μm), and others showed short protrusions (1 to 2 μm). However, on Days 157 and 184 PI, sarcocysts were up to 436.6 μm long, and only had short protrusions. Matuschka et al. (1986) performed a similar experimental infection and observed microscopic sarcocysts (< 1 mm long) and macroscopic sarcocysts (up to 2 mm) in ponies on day 378 PI. However, on day 1040 PI, only macroscopic sarcocysts were found, with sizes of up to 9 × 0.5 mm.
The ultrastructure of the sarcocyst wall is useful in evaluating the taxonomy of Sarcocystis species in a given host. Dubey et al. (2015) grouped sarcocysts into at least 42 types and several subtypes based on TEM morphological characteristics of sarcocyst wall. In our materials, the ultrastructures of the thin-walled and thick-walled sarcocysts presented characteristics of TEM type 11: the protrusions of the sarcocyst wall contained bundled microtubules in the core of the protrusions penetrated into the ground substance. Based on the inclination of protrusions over the sarcocyst surface, the TEM type of the two morphotypes of sarcocysts could be subdivided into TEM type 11c. All ultrastructural descriptions of sarcocysts obtained from donkeys and horses provided by different authors to date conform to the characteristics of TEM types 11a or 11c) (Table 1). Among them, sarcocysts from Egyptian donkeys are similar to TEM type 11c (Hilali and Nasser 1987; Dubey et al. 2016), and those from Italian donkeys are similar to TEM type 11a (Passantino et al. 2019). These sarcocysts were named Sarcocystis sp. (Hilali and Nasser 1987) or S. bertrami (Dubey et al. 2016; Passantino et al. 2019).
Morphologically similar sarcocysts frequently occur in different hosts, especially in closely related hosts, which sometimes creates controversy regarding species identification (Formisano et al. 2013; Dubey and Rosenthal 2013). Currently, PCR assays and sequencing procedures are considered much more practical, accurate, and reliable for the delineation and identification of Sarcocystis species than traditional methods based on morphological characteristics (Gjerde 2013). Therefore, a critical comparison of the molecular characteristics of Sarcocystis species in donkeys and horses should be performed to help reach a final conclusion (Dubey et al. 2016).
In the present study, three genetic markers, 18S rDNA, 28S rDNA, and mitochondrial cox1, were sequenced and analyzed in the two morphotypes of sarcocysts found in donkeys. The sequences of the three loci in the two types presented high intraspecific similarities of 97.2 − 99.5% (on average 97.8%), 97.8 − 99.6% (on average 98.4%), and 99.0 − 99.9% (on 99.4%), respectively. Therefore, combined with the similar morphological features observed under TEM, the two types of sarcocysts observed in donkeys are inferred to represent the same Sarcocystis species. The comparison of the newly obtained 18S rDNA, 28S rDNA, and mitochondrial cox1 sequences with those deposited in GenBank showed identities of 90.0 − 97.5% (average 94.7%), 94.7 − 95.1% (average 94.9%), and 82.6 − 84.5% (average 83.4%), respectively, with those of S. bertrami or S. fayeri obtained from horses. Phylogenetic analysis inferred from the three loci indicated that the Sarcocystis sp. in donkeys formed an individual clade separated from the clade encompassing S. bertami and S. fayeri in horse originated from different geographical areas. Additionally, the donkey sarcocyst could be successfully discriminated from the horse sarcocysts using PCR–RFLP based on the mitochondrial cox1 sequences of the two parasites. Therefore, the sarcocysts of donkeys should not belong to the species S. bertramior or S. fayeri found in horses.
Cross-infection is a criterion for revealing whether different intermediate hosts harbor the same parasite. To date, there has been only one reported attempt to perform the cross-infection of Sarcocystis between donkey and horse (Matuschka 1983). Tissues from 20 horses naturally infected with sarcocysts were fed to a dog, and those of 10 donkeys were fed to another dog. Both dogs excreted sporocysts. Experimental infections were carried out in 4 ponies (#1–4). One pony (#1) fed donkey-derived sporocysts became febrile on days 10 and 11 and 19 − 21, but no sarcocysts were detected in biopsies of the thigh muscles on days 44 and 59 PI. The same pony was then fed horse-derived sporocysts on day 117 and killed on day 138 PI, and sarcocysts were identified in the carcass. The other three ponies (#2 − 4) were fed horse-derived sporocysts and killed on days 197, 212, and 21 PI, respectively. Sarcocysts were detected in ponies #2 and 3, but no sarcocysts were found in pony #4. These results suggest the transmission of the parasite in horses and donkeys mainly based on the fever symptom of the donkey (#1) infected with horse-derived sporocysts. Therefore, the cross-infection of Sarcocystis between donkey and horse should be attempted, and the available molecular evidence also needs to be supplemented in the future.
In summary, two morphotypes of sarcocysts were observed in donkeys in China under LM. Based on TEM morphology and analysis of the three genetic markers, the two types of sarcocysts are attributed to one parasite species. Compared the newly obtained sequences with those of S. bertrami and S. fayeri in horses previously deposited in GenBank, the parasite in donkeys are distinct from S. bertrami and S. fayeri in horses. Gadaev (1978) first proposed the name S. asinus for the sarcocysts found in donkeys, but few cyst morphological characteristics and no molecular data provided. Considering the arguments of Sarcocystis spp. in horse and other equids (Dubey et al. 2015), here, the Sarcocystis sp. was used to name the sarcocysts found in donkeys to avoid increasing uncertainty.
Data availability
No other data and material are provided.
References
Cawthorn RJ, Clark M, Hudson R, Friesen D (1990) Histological and ultrastructural appearance of severe Sarcocystis fayeri infection in a malnourished horse. J Vet Diagn Invest 2:342–345. https://doi.org/10.1177/104063879000200418
Doflein F (1901) Sarcocystis bertrami n. sp. In: Die Protozoen als Parasiten und Krankheitserreger, nach biologischen Gesichtspunkten dargestellt. Gustav Fischer, Jena, pp 219–220
Dubey JP, Rosenthal BM (2013) Sarcocystis capracanis-associated encephalitis in sheep. Vet Parasitol 197:407–408. https://doi.org/10.1016/j.vetpar.2013.04.026
Dubey JP, Streitel RH, Stromberg PC, Toussant MJ (1977) Sarcocystis fayeri sp. n. from the horse. J Parasitol 63:443–447. https://doi.org/10.2307/3279997
Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R (2015) Sarcocystosis of animals and humans, 2nd edn. CRC Press, Boca Raton, pp 249–256
Dubey JP, Van Wilpe E, Verma SK, Hilali M (2016) Ultrastructure of Sarcocystis bertrami sarcocysts from a naturally infected donkey (Equus asinus) from Egypt. Parasitology 143:18–23. https://doi.org/10.1017/S0031182015001432
Fayer R, Hounsel C, Giles RC (1983) Chronic illness in a Sarcocystis infected pony. Vet Rec 113:216–217. https://doi.org/10.1136/vr.113.10.216
Fischer S, Odening K (1998) Characterization of bovine Sarcocystis species by analysis of their 18S ribosomal DNA sequences. J Parasitol 84:50–54. https://doi.org/10.2307/3284529
Formisano P, Aldridge B, Alony Y, Beekhuis L, Davies E, Del Pozo J, Dunn EK, Morrison L, Sargison N, Seguino A, Summers BA, Wilson D, Milne E, Beard PM (2013) Identification of Sarcocystis capracanis in cerebrospinal fluid from sheep with neurological disease. Vet Parasitol 193:252–255. https://doi.org/10.1016/j.vetpar.2012.12.016
Gadaev A (1978) On sarcocysts of ass (Equus asinus). Akad Nauk Uzbecks SSR 1:47–48
Gjerde B (2013) Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol 43:579–591. https://doi.org/10.1016/j.ijpara.2013.02.004
Gjerde B (2014) Sarcocystis species in red deer revisited: with a redescription of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. sp. based on mitochondrial cox1 sequences. Parasitology 141:441–452. https://doi.org/10.1017/S0031182013001819
Göbel E, Rommel M (1980) Light and electron microscopic study on cysts of Sarcocystis equicanis in the oesophageal musculature of horses. Berl Münch Tierärztl Wschr 93:41–47
Hinaidy HK, Loupal G (1982) Sarcocystis bertrami Doflein, 1901, a sarcosporidia of the horse. Equus Caballus. Zentralbl Veterinarmed B 29:681–701. https://doi.org/10.1111/J.1439-0450.1982.TB01269.X
Hilali M, Nasser AM (1987) Ultrastructure of Sarcocystis spp. from donkeys (Equus asinus) in Egypt. Vet Parasitol 23:179–183. https://doi.org/10.1016/0304-4017(87)90003-3
Hu JJ, Chen X, Zuo Y (2001) Ultrastructure of the cyst wall and experimental infectivity of Sarcocystis from donkeys. Chin J Vet Sci 21:145–148
Hu JJ, Liu TT, Liu Q, Esch GW, Chen JQ, Huang S, Wen T (2016) Prevalence, morphology, and molecular characteristics of Sarcocystis spp. in domestic goats (Capra hircus) from Kunming, China. Parasitol Res 115:3973–3981. https://doi.org/10.1007/s00436-016-5163-6
Kirmse P (1986) Sarcosporidioses in equines of Morocco. Br Vet J 142:70–72. https://doi.org/10.1016/0007-1935(86)90011-4
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Ma CL, Ye YL, Wen T, Huang ZM, Pan J, Hu JJ, Tao JP, Song JL (2020) Prevalence and morphological and molecular characteristics of Sarcocystis bertrami in horses in China. Parasite 27:1. https://doi.org/10.1051/parasite/2019078
Matuschka FR (1983) Infectivity of Sarcocystis from donkey for horse via sporocysts from dogs. Z Parasitenkd 69:299–304. https://doi.org/10.1007/BF00927871
Matuschka FR, Schnieder T, Daugschies A, Rommel M (1986) Cyclic transmission of Sarcocystis bertrami by the dog to the horse. Protistologica 22:231–234
Mugridge NB, Morrison DA, Heckeroth AR, Johnson AM, Tenter AM (1999) Phylogenetic analysis based on full-length large subunit ribosomal RNA gene sequence comparison reveals that Neospora caninum is more closely related to Hammondia heydorni than to Toxoplasma gondii. Int J Parasitol 29:1545–1556. https://doi.org/10.1016/S0020-7519(99)00150-2
Odening K (1998) The present state of species-systematics in Sarcocystis Lankester, 1882 (Protista, Sporozoa, Coccidia). Syst Parasitol 41:209–233. https://doi.org/10.1023/A:1006090232343
Odening K, Wesemeier HH, Walter G, Bockhardt I (1995) Ultrastructure of sarcocysts from equids. Acta Parasitol 40:12–20
Passantino G, Lia RP, Latrofa S, Annoscia G, Šlapeta J, Otranto D, Rossi R, Zizzo N (2019) Sarcocystis bertrami in skeletal muscles of donkeys (Equus africanus asinus) from Southern Italy. Vet Parasitol Reg Stud Rep 16:100283. https://doi.org/10.1016/j.vprsr.2019.100283
Rommel M, Geisel O (1975) Untersuchungen über die Verbreitung und de Lebenszyklus einer Sarkosporidienart des Pferdes (Sarcocystis equicanis n. spec.). Berl Münch Tierärztl Wschr 88:468–471
Saville WJA, Dubey JP, Oglesbee MJ, Sofaly CD, Marsh AE, Elitsur E, Vianna MC, Lindsay DS, Reed SM (2004) Experimental infection of ponies with Sarcocystis fayeri and differentiation from Sarcocystis neurona infections in horses. J Parasitol 90:1487–1491. https://doi.org/10.1645/GE-313
Tinling SP, Cardinet GH, Blythe LL, Cohen M, Vonderfecht SL (1980) A light and electron microscopic study of sarcocysts in a horse. J Parasitol 66:458–465. https://doi.org/10.2307/3280748
Zeng W, Sun L, Xiang Z, Li N, Zhang J, He Y, Li Q, Yang F, Song J, Morris J, Rosenthal BM, Sun L, Liu H, Yang Z (2018) Morphological and molecular characteristics of Sarcocystis bertrami from horses and donkeys in China. Vet Parasitol 252:89–94. https://doi.org/10.1016/j.vetpar.2018.01.024
Funding
This study was supported by the National Key R&D Program of China (grant 2017YFD0500400) and the Natural Sciences Foundation of China (Grant 31460557).
Author information
Authors and Affiliations
Contributions
Junjie Hu suggested the overall concept and design of the study and drafted the manuscript. Mingzhu Zhang, Kaiwen Wei, and Zhipeng Wu conducted specimen collection and molecular work. Jun Sun and Shuangsheng Deng performed observation of sarcocysts and data analysis. Jianping Tao provided suggestions for this manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval
The present study was approved by the Animal Ethics Committee of Yunnan University (permission number AECYU2018004).
Consent to participate
The authors declare that they have participated in this work.
Consent for publication
The authors declare that they know the content of this manuscript and agree to submit it to Parasitology Research.
Competing interests
The authors declare no competing interests.
Additional information
Section Editor: Kevin Tan
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, M., Wei, K., Wu, Z. et al. Morphological and molecular characterization of a Sarcocystis species infecting donkeys from China. Parasitol Res 121, 2917–2926 (2022). https://doi.org/10.1007/s00436-022-07616-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-022-07616-2