Abstract
Avian haemosporidians make up one of the most widely distributed and diverse vector borne parasite systems, found nearly worldwide in tropical and temperate areas. Despite the clear relationship between avian host fitness measures and infection, few studies have addressed the importance of source material selection when assessing these relationships. We show that source material, here blood and pectoral muscle, do not yield equivalent results when assessing prevalence and genetic diversity of haemosporidian genera. We find higher prevalence and genetic diversity are recovered from blood versus pectoral muscle for Haemoproteus. Contrastingly, we find that a higher prevalence of Plasmodium is detected from pectoral muscle, while higher genetic diversity is recovered from blood. Our results indicate that source material may bias parasite detection and be an important factor in study design, which is not only related to parasite infection, but by extension to the ecology and fitness of avian hosts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Haemosporidian detection studies inform a broad field of research, from host ecology and fitness to parasite evolution and diversity, yet differences in results across source material have thus far been inconclusively assessed. Avian haemosporidian ranges nearly worldwide and consists of the genera Haemoproteus, Leucocytozoon, and Plasmodium. Infection resulting in malaria, haemoproteosis, or leucocytozoonosis (caused by Plasmodium, Haemoproteus, and Leucocytozoon infections, respectively) can affect host fitness, from individual physical condition and reproductive success to reduced survival rates, as well as exhibiting a selective force on populations (Hamilton and Zuk 1982; Merino et al. 2000; Marzal et al. 2005; Knowles et al. 2010; Asghar et al. 2011; García-Longoria et al. 2014; Coon et al. 2016). They can also have devastating population- and community-level effects in non-endemic ranges, as evidenced by avian responses to malarial introductions on the Hawaiian Islands (van Riper et al. 1986). Avian haemosporidian research has grown as a model study system, and as resulting data has increased, we have begun to see the use of meta-studies to try and decipher broader evolutionary and ecological patterns (e.g., Clark et al. 2014; Outlaw et al. 2017). Overall, the majority of studies that have assessed avian haemosporidians have done so by targeting a small fragment of the mitochondrial DNA (mtDNA) cytochrome b (cytb) gene (479 bps), the standard barcode, and the measure of lineage diversity. There are currently 2381 unique lineages documented in MalAvi, the avian haemosporidian database, (MalAvi 2.2.9, December 17, 2016; Bensch et al. 2009), and descriptions of new variants occur regularly as the effects of malaria, haemoproteosis, and leucocytozoonosis on the ecology of their bird hosts are studied.
Avian haemosporidian parasite studies have predominantly sampled blood as the primary source material for parasite detection and analysis. Blood is most widely used due to its ease of collection, and the fact that birds are generally released unharmed, making it a more easily permitted and collected material. Previous work has found that Leucocytozoon detection did not vary when comparing blood collected from the brachial vein (circulation blood) to deep circulation blood (collected from the lungs), suggesting that location of blood draw will not influence detection (Holmstad et al. 2003). Further, various other tissue types (i.e., muscle, liver, and heart) are often used, as they may be widely accessible (i.e., already accessioned in natural history museum collections) or part of separate studies and collection efforts. Investigations most often utilize a single starting material due to study design, while other studies may use various source materials due to availability and logistics.
All haemosporidian genera undergo similar infection periods which include the following: prepatent, primary parasitemia (acute and chronic), latent stage, and secondary parasitemia (in response to relapse), within their vertebrate host. Vertebrate hosts, such as birds, are intermediate hosts and the location of asexual reproduction, while dipterans are the definitive host and location of sexual reproduction as well as the vector. However, there are distinct variations in life stages, patterns of development, and timing of transmission season across genera. All genera undergo development in both circulating blood and fixed tissues of hosts at various stages. Initial development for all genera occurs in fixed tissues (i.e., within parenchymal cells, spleen, liver, kidney heart, skeletal muscle, and endothelial lining of the capillaries) and is at the stage of trophozoites or meronts with the location of such being specific to genus and sometimes species (Valkiūnas 2005). At this stage (trophozoite or meront), either abortive development (i.e., dead end where parasite cannot develop into patent infection stages due to lack of host-parasite competence) occurs or the parasite continues to become a patent infection (Valkiūnas 2005). The patent circulating blood life stages are meronts and/or gametocytes depending on genera. Given these varying life-stages seen across haemosporidian genera, detection probability may be impacted across seasons and by host ecology (Mukhin et al. 2016).
Using single-species sampling, two previous studies have sought to address the differences in starting source material. Ramey et al. (2013) investigated Haemoproteus, Leucocytozoon, and Plasmodium in the Northern Pintail (Anas acuta) using paired blood and muscle source materials from 157 individuals. They found no significant difference between sources and primarily recovered Leucocytozoon. However, they did find higher haplotype diversity for Leucocytozoon in blood (five additional haplotypes) versus muscle. Svennson-coehlo et al. (2016) screened 46 White-shouldered Fire-eyes (Pyriglena leucoptera) for Haemoproteus and Plasmodium, implementing a comprehensive-paired sampling regime across of source materials (including blood, brain, heart, pectoral muscle, and liver) using several screening methods. Their investigation only recovered Plasmodium, with no significant differentiation when comparing across all source materials, although detection rates did vary: e.g., blood prevalence (28%) was lower than pectoral muscle prevalence (36%), while heart had the highest prevalence (39%). Detection in combined tissues (liver, heart, and pectoral muscle) as compared to blood was significant.
Here, we investigate whether blood and pectoral muscle source materials result in equivalent detections of avian haemosporidian prevalence and genetic diversity. We examined a taxonomically diverse suite of avian hosts from the tropical West African country of Benin, with sampling encompassing varied habitat types. Assessing equivalency across source materials for detection of haemosporidians may inform biases in previous avian haemosporidian research. Broad comparisons of parasite detections are important for understanding the role parasites play in host ecology. Given that parasites play a powerful selective force on populations understanding how to best address distributions informs or ability to detect them and make meaningful comparison across studies.
Methods
Avian sampling
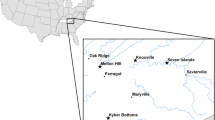
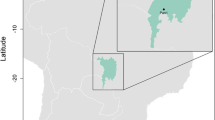
Birds were sampled between May and June of 2010, as part of a broader systematics and biodiversity survey, at six sampling localities across Benin (Parc W: Point Triplo [11.89828N, 2.41149E]; Parc W: Chutes de Koudou [11.68006N, 2.31689E]; Lake Toho [7.54305N, 2.60687E]; Dogo Forest [7.54305N, 2.60687E]; Lama Forest [6.96026N, 2.16830E]; and Abomey Calavi [6.42292N, 2.34996E]) (Fig. 1). These localities represent the diverse biogeographic regions and habitats found in Benin (see Linder et al. 2012). All localities have tropical savanna climates (Peel et al. 2007), with mean temperatures above 18 °C, a marked dry season and are differentiated by length and timing of wet seasons across Northern and Southern localities (Fig. 1) Northern localities, Parc W localities are characterized by a long dry season with a pronounced single wet season, occurring from June to September (http://worldclim.org; Hijmans et al. 2005). Sampling at these northern localities occurred between May 21st and 28th. All other southern localities (Dogo Forest, Lama Forest, Abomey Calavi, and Lake Toho) have two wet seasons, and sampling at these southern localities occurred from June 1st to 10th (http://worldclim.org; Hijmans et al. 2005).
Overall, 220 birds were captured via opportunistic mist-netting and sampled for blood and/or pectoral muscle tissue. Blood was sampled primarily directly from heart, after euthanasia. Blood was stored in Queen’s Lysis buffer, and tissues were stored in 20% DMSO salt saturated storage buffer. All voucher specimens collected are accessioned in the Biodiversity and Research Teaching Collections, at Texas A&M. A combined total of 421 samples from 220 individuals (blood = 213; pectoral muscle = 208) were collected. For analysis, we used only paired blood and pectoral muscle samples (i.e., both taken from the same bird specimen), totaling 199 pairs. Avian sampling from the 199 paired sample individuals consisted of 7 orders, 29 families, and 78 species, with most sampling representing Passeriformes (80%) (see Supplementary Table S1 for taxonomic information and sampling localities). Paired samples (N = 199) represent 119 individuals from Northern localities and 80 individuals from Southern localities. Based on skull ossification data taken for each of the 199 individual hosts, our bird samples consisted of 157 adults and 23 juveniles, while ossification was not noted for 19 individuals. Individuals were not partitioned by age given the small number of juvenile individuals collected. All collecting of specimens was done under protocols approved by the Institutional Animal Care and Use Committee at Texas A&M University. Birds were sacrificed via cardio-thoracic compression, as suggested by the Ornithological Council’s Guideline to the Use of Wild Birds in Research (Fair et al. 2010).
Molecular assessment of avian haemosporidians
We extracted whole genomic DNA from avian blood and pectoral muscle samples using the E.Z.N.A. Tissue extraction kit (Omega Bio-Tek, Norcross, GA) and standard protocols except for the final elution was 75 μl to increase overall DNA yield. Polymerase chain reaction (PCR) was used to identify haemosporidian infection by amplifying a fragment of the mtDNA cytb gene, and multiple primer pairs were used to amplify across known avian haemosporidian genetic diversity. Three primer pairs consisting of the same forward primer (UNIVF) and one of three distinct reverse primers—UNIVR1, UNIVR2, or UNIVR3—were used, yielding 505, 535, and 571 base pairs (bp) (Drovetski et al. 2014), respectively. These primers encompass the entire 479 bp that are the standard region that is collected in the MalAvi avian haemosporidian database (Bensch et al. 2009). Collectively, these primers amplify all three genera of avian haemosporidians: Haemoproteus (to include subgenera Parahaemoproteus and Haemoproteus), Leucocytozoon, and Plasmodium. PCR amplification was carried out in 18.75 μl reaction containing 1× GoTaq Flexi buffer, 2.5 mM MgCl2, 0.2 mM of dNTP, 0.19 μl of each primer, and 0.9375 μ of GoTaq Flexi DNA polymerase (Promega Madison, Wisconsin, USA) with 1 μl of DNA template. Each sample (both blood and pectoral muscle) was tested via PCR for each primer pair, and up to three times per primer pair. If a positive amplicon was not detected and successfully sequenced during this process, we considered the sample to negative for haemosporidians. All PCRs included four positive controls as well as two negative controls. Automated sequencing was performed bi-directionally using BigDye (Applied Biosystems, Foster City, CA, USA), and products were run out using an ABI 3730 at Beckman Coulter Genomics (Danvers, MA, USA).
Sequences were verified and aligned by eye using Geneious 6.1.8 (http://www.geneious.com; Kearse et al. 2012). Multiple infections were determined by the presence of double peaks on both chromatograms at one or more base positions. For sequences with double peaks, we re-PCR’d and re-sequenced two to three times to verify these double peaks. Sequences with < 3 double peaks were treated as single infections, due to a high probability; these are not multiple infections but instead the result of sequencing error (Szymanski and Lovette 2005). Verified multiple infections were further examined using the heterozygous plug-in in Geneious 6.1.8. Criteria to better qualify a multiple peak were > 70% peak similarity, a base calling confidence mean score of > 25 at each multiple peak site, and visual assessment of the strand quality. We implemented conservative criteria to remove the possibility of sequencing errors being erroneously called a multiple infection peak. Sequences with multiple peaks having met the above criteria (N = 5) and were then assigned the appropriate IUPAC nucleotide ambiguity code for double infections.
All data were assessed phylogenetically and identified to genus by use of the MalAvi blast (Version 2.2.8, Bensch et al. 2009) and NCBI BLAST (Altschul et al. 1990) functions. Double infection data were phased using the Phase 2.1 (Stephens et al. 2001) implementation in DnaSP 5.10.1 (Librado and Rozas 2009) for reconstruction of single infection haplotypes. Phase input datasets were separated by genus and included all single infection data from our data along with top MalAvi blast (Version 2.2.8, Bensch et al. 2009) and NCBI BLAST matches. Phase was run for each genus with a 1000 iteration burn-in, 10 thinning intervals, and 1000 iterations. Phasing of data reconstructs haplotypes from diploid gametic alleles and therefore may have flaws related to its ability to reliably assess multiple infections haplotypes. Phased data (N = 10) were then added to the final sequence dataset.
Statistical analyses
All statistical analyses were performed in R 3.3.2 (R Core Team 2016). Datasets were assessed for normality using Shapiro-Wilks test and assessed using quantile-quantile plots. Data were partitioned in five ways for further analysis. Partitioning allowed us to determine an overall infection rate as a percentage of host individuals detected positive (successful pcr detection and definitive sequencing), as well as to determine the overall detection rates for each avian haemosporidian genus. In dataset A, we assessed all host individuals (N = 199) as binary (detected versus not detected) for avian haemosporidian detection, regardless of source material. In dataset B, all host individuals were assessed separately for each tissue source (i.e., for statistical purposes, blood and pectoral muscle were treated as dependent variables), and all samples were coded as binary (detected versus not detected) for comparison. We used a McNemar’s chi-squared test as implemented in the exact 2 × 2 package of R (Fay 2010) for paired sample data to detect equality of detection rates across starting source materials (blood versus pectoral muscle). In dataset C, all host individuals were treated as paired samples (blood versus pectoral muscle), and we analyzed counts of the number of detections found in blood versus pectoral muscle, using a dependent t test to determine mean difference of blood versus pectoral muscle detections. We included count data for host individuals determined as positive for detection, because we found one to three distinct lineages of avian haemosporidian per positive individual (given blood and pectoral muscle sources and three distinct primer sets, allowing a total of six possible detections per individual plus possible double infections). In dataset D, all host individuals (N = 199) were assessed for each parasite genus separately, and then within genus by blood versus pectoral muscle. We then examined genus specific subsets using McNemar’s chi-squared test and binary coding of detection. Each genus was examined independently, as detectability may vary between blood and pectoral muscle due to variations across genera life cycles. In dataset E, we examined all recovered avian haemospordian lineages (E1) from positive samples (detected and sequenced, N = 155 recovered from 103 host individuals), allowing us to assess the lineages recovered and their source material while removing any bias from individuals not infected. We separated lineages by genus (Haemoproteus (N = 55); Leucocytozoon (N = 11); Plasmodium (N = 89)) and starting source material. We then utilized McNemar’s chi-squared test to determine within genus equality of detection for blood and pectoral muscle source materials. We reduced the number of infections produced from double infections, all of which were in the genus Haemoproteus and counted these as single infections (E2) to correct for the possible error in PHASE assignment (reduced E2 Haemoproteus N = 50).
Genetic diversity analysis
We also assessed dataset E3 to examine genetic diversity recovered across blood and pectoral muscle starting materials. The final sequence dataset (same as dataset E1) was separated by genus (as above: Haemoproteus (N = 55), Leucocytozoon (N = 11), and Plasmodium (N = 89) and assessed for haplotype diversity using PopART 1.7 (http://popart.otago.ac.nz) implementing the minimum spanning for networks (Bandelt et al. 1999). Further, each sequence was coded for trait data as being recovered from blood, pectoral muscle, or both to determine differences in genetic diversity as resolved by each starting source material. Lineages recovered have been deposited in Genbank under accession numbers (MG018625 - MG018709) and the MalAvi database.
Results
Statistical analyses
For dataset A, detections at the individual host level (N = 199, blood and pectoral muscle combined) resulted in 103 individuals with one or more detections of avian haemosporidians, an overall 51.8% infection rate (Haemoproteus 22.6%, Leucocytozoon 6.5%, Plasmodium 34.2%) (Table 1). For dataset B, 69 host individuals recovered detections in both blood and pectoral muscle material, 14 individuals had detections in blood alone, and 20 individuals had detections in pectoral muscle alone. Paired binary detections (detected/not detected) did not differ across starting source material (McNemar’s X 2 (1) = 1.059, P = 0.303, Table 1, B). For dataset C, total counts of detections were equal (P = 1.0) in the dependent samples t test for paired count data (N = 199), with blood and pectoral muscle each having 105 detections (both have M = 0.528, Table 1, C).
For dataset D, there was some detection differentiation evident for source material when assessed by genus (N = 199, paired binary infection state, Table 1, D). Haemoproteus was detected in both blood and pectoral muscle for 20 host individuals, while 11 detections where found in blood alone, and 7 were detected in pectoral muscle alone. Of the 199 hosts with paired sampling, 161 individuals resulted in no Haemoproteus detections, and within Haemoproteus, there was no significant differentiation across source material (P = 0.346; Table 1, D). We detected Leucocytozoon in both blood and pectoral muscle for eight host individuals, while four detections were observed in blood alone, and one detection was in pectoral muscle alone, and 186 host individuals recovered no Leucocytozoon detections. As in Haemoproteus, we found no significant difference in Leucocytozoon detections across source material (P = 0.18; Table 1, D). We detected Plasmodium in both blood and pectoral muscle for 26 host individuals, while 12 detections were found in blood alone, 25 detections were found in pectoral muscle alone, and 136 host individuals resulted in no Plasmodium detections. Unlike Haemoproteus and Leucocytozoon, we found significant inequality in detection between blood and pectoral muscle in Plasmodium (P = 0.032; Table 1).
For dataset E1, where recovered lineages (detected and sequenced) across host individuals and their source material were separated by genus, we recovered 157 sequences, with an average of 0.789 infections per individual (range of one to four infections per individual). For Haemoproteus (N = 55), 22 infections were recovered from blood alone, while 9 infections were recovered from pectoral muscle alone, and 24 identical lineages were recovered from both source materials. This resulted in a highly significant differentiation of detections dependent upon source material for Haemoproteus (P = 0.019; Table 1, E1). However, if we removed the double infection data and treat those as single infections (E2), with 18 detections in blood only and 9 in pectoral muscle only, we still recover a marginally significant difference in detections for blood and pectoral muscle (P = 0.083; Table 1, E2). We found few unique Leucocytozoon detections (N = 11, Table 1, E1) with five lineage detections found in blood alone and two detections found in pectoral muscle alone, along with six identical lineage detections recovered in the same individual from both source materials. Resulting differences were not significant (P = 0.257; Table 1, E1). Lastly, for Plasmodium (N = 89), we recovered 23 lineages in blood alone, 38 lineages were recovered in pectoral muscle alone, and 28 identical detections were recovered in blood and pectoral muscle. Detection differences were marginally significant (P = 0.055; Table 1, E1).
Genetic diversity
For dataset E3, using a network analysis for each genus, we recovered a total of 37 unique haplotypes for Haemoproteus from 57 sequences mtDNA cytb sequences, 18 of which were recovered in both blood and pectoral muscle host samples (Fig. 2). However, 11 Haemoproteus haplotypes were unique to blood, and 4 were unique to pectoral muscle, resulting in a 33.3% loss of recovered genetic diversity if sampling pectoral muscle alone for Haemoproteus, and a 19.4% loss of genetic diversity if sampling blood alone. For Leucocytozoon, we detected a total of 11 unique haplotypes (Fig. 2) from 13 mtDNA cytb sequences, where 6 haplotypes were recovered in blood and pectoral muscle paired samples. We found two haplotypes unique to blood and one unique to pectoral muscle, resulting in a 22.2% loss of genetic diversity if sampling pectoral muscle alone, and a 11.1% loss in diversity if sampling blood alone. Finally, we recovered 31 unique haplotypes for Plasmodium from 87 mtDNA cytb sequences (Fig. 2). While 18 were shared between blood and pectoral muscle, 10 were unique to blood comprising 32.2% of genetic diversity recovered, and only three were unique to pectoral muscle comprising 9.7% of the diversity. This resulted in a 32.2% loss in recovered genetic diversity if sampling pectoral muscle alone, and a 9.7% loss of recovered genetic diversity if sampling blood alone for Plasmodium.
Discussion
Our study is the first to compare haemosporidian detection across source materials, using a broad sampling of avian species. Although we found no differences in detection when we assessed all avian haemosporidian genera together, we found clear differences when examining each genus independently. Our results recovered more detections, as well as higher genetic diversity, in Haemoproteus from blood source materials as compared to pectoral muscle. For Plasmodium, we found higher detections in pectoral muscle while recovering higher genetic diversity from blood source material. We found few Leucocytozoon detections across both blood and pectoral muscle starting materials. When taken together, our results suggest that detection numbers and captured genetic diversity are not equivalent across pectoral muscle and blood starting material for each genus (Table 1).
Leucocytozoon prevalence in our study is similar to previous work in nearby Cameroon and Gabon (Beadell et al. 2009). The localities previously sampled in Cameroon and Gabon included sites with tropical savanna climate (like Benin) but were predominantly tropical monsoon climates with higher overall precipitation with less variance in temperatures. Leucocytozoon vectors (Simuliidae: blackflies) are present in Benin (species inventories have been conducted for most countries with highest interest in medically relevant species), but the vectors have not been assessed for abundance or biogeographically across much of Africa (Adler and Crosskey 2015). Blackfly presence is closely tied to rivers and other bodies of water and therefore is spatially restricted (Sutcliffe 1986). Species richness has been associated largely with temperature and stream discharge, and it has been suggested that they are rarer in tropical climates (Young et al. 1993; Ya’cob et al. 2016). Therefore, we attribute the Leucocytozoon prevalence in our study to the timing of sampling (early in the rainy season) and the relative climatic characteristics of the sampling regions and associated vector abundance.
The differences in life stage development and transmission seasonality vary as we assess each genus, and within these differentiations, we believe lay three possible explanations for the patterns in detections and haplotype diversity across genera and source material. First, the seasonality of transmission period and the associated length of these infection periods vary across parasite host genera and even within closely related parasites lineages, indicating that parasite lineages may exhibit evolutionary adaptions to transmission strategy (Valkiūnas et al. 2004; Pérez-Rodríguez et al. 2015). Avian hosts spend more time in chronic or latent infection periods as compared to prepatent or acute infection periods, hence leading to high detections in pectoral muscle (Valkiūnas 2005). However, acute parasitemia is generally under-sampled due to reduced host activity (50% less activity e.g., highly reduced flight, foraging, and breeding activity, as compared to uninfected birds) (Valkiūnas 2005; Mukhin et al. 2016). Birds that can activate a sufficient immune response move from acute to the chronic phase (low parasitemia) and then to latent infections; otherwise, mortality from acute infection likely occurs (Valkiūnas 2005). Infected birds with low-level chronic infections are the most detectable via mist netting given the relatively benign symptoms, which do not hinder normal physiological maintenance movements of birds (i.e., foraging, nesting, breeding). These low-level infections are detectable in both blood and fixed tissue and thereby resulted in high detection rates in both blood and muscle. Additionally, the chronic stage is longer than other stages (months for Haemoproteus and Leucocytozoon; up to a year for Plasmodium), making this stage the most detectable in wild populations (Valkiūnas 2005). Moreover, older birds have been shown to have higher parasite prevalence than younger birds, possibly due to increased hormone levels and the associated incidence of relapse combined with the exposure to new seasonal infections (Greiner and Mundy 1979; Deviche et al. 2001).
Second, life stages may impact haemosporidian detection probability across source starting material. The location (i.e., in blood, pectoral muscle, or other organs) of the parasite across life stages is known from experimental infection studies (Fallis and Bennett 1960; Hepler et al. 1966; Khan and Fallis 1970; Atkinson et al. 1986; Atkinson et al. 1988; Zehtindjiev et al. 2008; Valkiūnas et al. 2015), although these stages are described from a small proportion of species from each haemosporidian genus (Valkiūnas 2005; Bensch et al. 2009). So, while informative, we lack a broad knowledge regarding how consistent life stage locations (and durations in each) are across the high diversity of lineages recovered, particularly if the transmission strategies show plasticity as previously seen in discrete lineage clades (Pérez-Rodríguez et al. 2015). Regardless, we expect overall lower detections in blood for birds unless specifically targeting the timing of active transmission periods (i.e., primary parasitemia: acute and chronic), when meronts and gametocytes are found in the circulating blood. Our high detection rates in pectoral muscle may be due to parasite recovery from the tissue stages of infections (i.e., located in the endothelial lining of capillary cells or skeletal muscle) which may be confounded by possible carryover of circulating blood in muscle (Valkiūnas 2005). Further, some of the muscle tissue only detections may have captured abortive infections (i.e., dead end infections) (Markus 2011). Abortive development has been found to result in morbidity and acute disease as a result of the damage from meronts to internal organs. Consequently, this has most commonly been detected in exotic captive species, being difficult to capture and assess in wild populations (Donovan et al. 2008; Olias et al. 2011; Cannell et al. 2013). Diagnosing abortive development requires necropsy and histology examinations, though we can examine molecular sequences for signals of abortive development by looking at host associations along with haemosporidian genera host specificity. We only found one likely detection of abortive development, where a Striated Heron (Butorides striata) recovered a lineage most homologous to a Haemoproteus lineage previously detected in the Common Blackbird (Turdus merula).
Third, resident birds (i.e., non-migratory or narrowly intra-continental migrants) in tropical climates engage in diverse breeding strategies. Timing of tropical breeding is mostly related to precipitation patterns (i.e., rainy season occurrence and the resulting humid period) and subtle photoperiodic cues, though some avian species have continuous or opportunistic breeding strategies which are often related to non-seasonal rainfall (Hau et al. 2008). Relapses of infection have been associated with gonadal, stress, and pineal hormones which are activated by photoperiod and associated breeding cues, though no conclusive mechanism has been discovered (Desser et al. 1968; Applegate and Beaudoin 1970; Valkiūnas et al. 2004; Cornelius et al. 2014). Additionally, if temperature and relative humidity are appropriate for parasite development and vectors are available, transmission may be occurring year-round (Hasselquist et al. 2007; Sorensen et al. 2016). However, extreme dry periods characterizing the tropical savanna climate of our Benin sampling localities make year-round transmission unlikely to occur. Furthermore, parasite persistence is sustained via these relapses, which then enables the infection of nestlings and juveniles (Bennett and Cameron 1974; Greiner and Mundy 1979; Deviche et al. 2001; Dunn et al. 2016). Yet, sampling for this study took place before the rainy season in the northern sampling localities, so presumably breeding for most birds was not yet initiated and consequently chronic, latent, and relapse infections are most likely detectable during this period. Sampling for southern localities took place in the middle of the first of two annual rainy seasons suggesting some birds had commenced breeding for the season and that relapses as well as new primary parasitemia periods are likely to be detected in these birds.
Previous source material comparison studies found no significant differences for haemosporidian parasite detection (Ramey et al. 2013; Svennson-coehlo et al. 2016). These investigations were limited to single host species studies, recovery of a single primary haemosporidian genus (Leucocytozoon and Plasmodium, respectively), and the concordant host-parasite relationship bias. Given that not all avian hosts are equally susceptible to Haemoproteus, Leucocytozoon, and Plasmodium, broader comparisons and extrapolations can therefore not be made without including greater host taxon sampling. Avian host susceptibility also varies across each parasite genus, with Haemoproteus and Leucocytozoon species having higher host specificity (phylogenetically host family restricted) as compared to Plasmodium which exhibits much lower host specificity (Bennett et al. 1982; Atkinson and Van Riper 1991; Beadell et al. 2009; Lutz et al. 2015). Host susceptibility is also affected by different vector associations, as each parasite genus is vectored by different dipteran groups whose presence and abundance have differing ecological constraints and timing of emergence (Atkinson and van Riper 1991; Valkiūnas 2005). Therefore, our results indicate that each avian haemosporidian genus should be assessed independently. Blood starting source material was the most informative for Haemoproteus, providing a higher number of detections and greater genetic diversity. Both blood and pectoral muscle were informative for Plasmodium, with pectoral muscle providing more detections and blood providing higher genetic diversity. When examining just one source material, there may be a potential loss of genetic diversity and possible prevalence underestimation, particularly when pectoral muscle is the single source material. We realize that many studies may not have the opportunity to collect tissues other than blood, but we encourage those associated with museums to employ broader collections of source materials when performing broad diversity studies of hosts (as was the impetus for our sampling in Benin).
We conclude by suggesting that when investigations of avian haemosporidians are initiated, study design needs to take into consideration three factors. First, the genus (genera) of interest should inform the choice of sampling material. Second, the timing of sampling has two important facets, as seasonality of transmission is dependent on availability and abundance of vectors, and the periods shortly after host breeding season or rainy season are likely best for capturing the chronic infection stage. Clearly, more vector assessment is needed for all haemosporidian vectors across Africa (Dipterans: Ceratopogonidae, Culicidae, Hippoboscidae, and Simuliidae) in order to disentangle parasite transmission strategies (i.e., changes in length of chronic periods) from limitations of vector seasonality. Third, the climate of the sampling location (i.e., tropical versus temperate) as climate is informative for seasonality of hosts and vectors. The heterogeneity in detection we found across source materials at the genus level indicates the importance of source material selection for parasite studies of not only parasite diversity and ecology, but for deeper understanding of ecological patterns using comparative studies.
References
Adler PH, Crosskey RW (2015) World blackflies (Diptera: Simuliidae): a comprehensive revision of the taxonomic and geographical inventory. 123 pp. Available from: http://www.clemson.edu/cafls/biomia/pdfs/blackflyinventory.pdf
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Applegate JE, Beaudoin RL (1970) Mechanism of spring relapse in avian malaria: effect of gonadotropin and corticosterone. J Wildl Dis 6:443–447
Asghar M, Hasselquist D, Bensch S (2011) Are chronic avian haemosporidian infections costly in wild birds? Avian Biol 42:530–537
Atkinson CT, Forrester DJ, Greiner EC (1988) Pathogenicity of Haemoproteus meleagridis (Haemosporina: Haemoproteidae) in experimentally infected domestic turkeys. J Parasitol 74:228–239
Atkinson CT, Greiner EC, Forrester DJ (1986) Pre-erythrocytic development and associated host responses to Haemoproteus meleagridis (Haemosporina: Haemoproteidae) in experimentally infected domestic Turkeys. J Protozool 33:375–381
Atkinson CT, Van Riper C (1991) Pathogenicity and epizootiology of avian haematozoa: Plasmodium, Haemoproteus, and Leucocytozoon. In: Loye JE, Zuk M (eds) Bird-parasite interactions: ecology, evolution, and behavior. Oxford University Press, Oxford, UK, pp 19–48
Bandelt H, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Beadell JS, Covas R, Gebhard C, Ishtiaq F, Melo M, Schmidt BK, Perkins SL, Graves GR, Fleischer RC (2009) Host associations and evolutionary relationships of avian blood parasites from West Africa. Int J Parasitol 39:257–266
Bennett GF, Cameron M (1974) Seasonal prevalence of avian hematozoa in passeriform birds of Atlantic Canada. Can J Zoo 52:1259–1264
Bennett GF, Whiteway M, Woodworth-Lynas C (1982) A host-parasite catalogue of the avian haematozoa. Memorial University of Newfoundland Occasional Papers in Biology, no. 5, St. John's, Newfoundland
Bensch S, Hellgren O, Pérez-Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour 9:1353–1358
Cannell BL, Krasnec KV, Campbell K, Jones HI, Miller RD, Stephens N (2013) The pathology and pathogenicity of a novel Haemoproteus spp. infection in wild Little Penguins (Eudyptula minor). Vet Parasitol 197:74–84
Clark NJ, Clegg SM, Lima MR (2014) A review of global diversity in avian Haemosporidians (Plasmodium and Haemoproteus: Haemosporida): new insights from molecular data. Int J Parasitol 44:329–338
Coon CAC, García-Longoria L, Martin LB, Magallanes S, de Lope F, Marzal A (2016) Malaria infection negatively affects feather growth rate in the house sparrow Passer domesticus. J Avian Biol 47:779–787
Cornelius JM, Zylberberg M, Breuner CW, Gleiss AC, Hahn TP (2014) Assessing the role of reproduction and stress in the spring emergence of haematozoan parasites in birds. J Exp Biol 217:841–849
Desser SS, Fallis AM, Garnham PCC (1968) Relapses in ducks chronically infected with Leucocytozoon simondi and Parahaemoproteus nettionis. Can J Zool 46:281–285
Deviche P, Greiner AC, Manteca X (2001) Seasonal and age-related changes in blood parasite prevalence in dark- eyed juncos (Junco hyemalis, Aves, Passeriformes). J Exp Zool 289:456–466
Donovan TA, Schrenzel M, Tucker TA, Pessier AP, Stalis IH (2008) Hepatic hemorrhage, hemocoelom, and sudden death due to Haemoproteus infection in passerine birds: eleven cases. J Vet Diagn Investig 20:304–313
Drovetski SV, Aghayan SA, Mata VA, Lopes RJ, Mode NA, Harvey JA, Voelker G (2014) Does the niche breadth or trade-off hypothesis explain the abundance-occupancy relationship in avian Haemosporidia? Mol Ecol 23:3322–3329
Dunn JC, Stockdale JE, Bradford EL, Mccubbin A, Morris AJ, Grice PV, Goodman J, Hamer KC (2016) High rates of infection by blood parasites during the nestling phase in UK Columbids with notes on ecological associations. Parasitology:1–7
Fair J, Paul E, Jones J (eds) (2010) Guidelines to the use of wild birds in research. Ornithological Council, Washington, D.C. URL www.nmnh.si.edu/BIRDNET/guide
Fallis AM, Bennett GF (1960) Description of Haemoproteus canachites n. sp. (Sporozoa: Haemoproteidae) and sporogony in Culicoides (Diptera: Ceraptogonidae). Can J Zool 38:455–464
Fay MP (2010) Two-sided exact tests and matching confidence intervals for discrete data. R Journal 2:53–58
García-Longoria L, Garamszegi LZ, Møller AP (2014) Host escape behavior and blood parasite infections in birds. Behav Ecol 00:1–11
Greiner EC, Mundy PJ (1979) Hematozoa from Southern African vultures, with a description of Haemoproteus janovyi sp. n. J Parasitol 65:147–153
Hamilton WD, Zuk M (1982) Heritable true fitness and bright birds: a role for parasites? Science 218:384–387
Hasselquist D, Östman Ö, Waldenström J, Bensch S (2007) Temporal patterns of occurrence and transmission of the blood parasite Haemoproteus payevskyi in the great reed warbler Acrocephalus arundinaceus. J Ornithol 148:401–409
Hau M, Perfito N, Moore IT (2008) Timing of breeding in tropical birds: mechanisms and evolutionary implications. Ornitol Neotrop 19:39–59
Hepler PK, Huff CG, Helmuth S (1966) The fine structure of the exoerythrocytic stages of Plasmodium fallax. J Cell Biol 30:333–358
Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A (2005) Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 25:1965–1978
Holmstad PR, Anwar A, Iezhova T, Skorping A (2003) Standard sampling techniques underestimate prevalence of avian hematozoa in willow ptarmigan (Lagopus lagopus). J Wildl Dis 39:354–358
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Khan RA, Fallis AM (1970) Relapses in birds infected with species of Leucocytozoon. Can J Zool 48:451–455
Knowles SCL, Palinauskas V, Sheldon BC (2010) Chronic malaria infections increase family inequalities and reduce parental fitness: experimental evidence from a wild bird population. J Evol Biol 23:557–569
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Linder HP, de Klerk HM, Born J, Burgess ND, Fjeldså J, Rahbek C (2012) The partitioning of Africa: statistically defined biogeographical regions in sub-Saharan Africa. J Biogeogr 39:1189–1205
Lutz HL, Hochachka WM, Engel JI, Bell JA, Tkach VV, Bates JM, Hackett SJ, Weckstein JD (2015) Parasite prevalence corresponds to host life history in a diverse assemblage of Afrotropical birds and haemosporidian parasites. PLoS One 10:e0121254
Markus MB (2011) The hypnozoite concept, with particular reference to malaria. Parasitol Res 108:247–252
Marzal A, de Lope F, Navarro C, Møller AP (2005) Malarial parasites decrease reproductive success: an experimental study in a passerine bird. Oecologia 142:541–545
Merino S, Moreno J, Sanz JJ, Arriero E (2000) Are avian blood parasites pathogenic in the wild? A medication experiment in blue tits (Parus caeruleus). Proc Biol Sci 267:2507–2510
Mukhin A, Palinauskas V, Platonova E, Kobylkov D, Vakoliuk I, Valkiunas G (2016) The strategy to survive primary malaria infection: an experimental study on behavioural changes in parasitized birds. PLoS One 11:1–15
Olias P, Wegelin M, Zenker W, Freter S, Gruber AD, Klopfleisch R (2011) Avian malaria deaths in parrots, Europe. Emerg Infect Dis 17:950–952
Outlaw DC, Harvey JA, Drovetski SV, Voelker G (2017) Diversity and distribution of avian haemosporidians in sub-Saharan Africa: an inter-regional biogeographic overview. Parasitology 144:394–402
Peel MC, Finlayson BC, Mcmahon TA (2007) Updated world map of the Köppen-Geiger climate classification. Hydrol Earth Syst Sci 4:439–473
Pérez-Rodríguez A, de la Hera I, Bensch S, Pérez-Tris J (2015) Evolution of seasonal transmission patterns in avian blood-borne parasites. Int J Parasitol 45:605–611
R Core Team (2016) R: a language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria. URL https://www.R-project.org/
Ramey AM, Fleskes JP, Schmutz JA, Yabsley MJ (2013) Evaluation of blood and wing pectoral muscle tissues for molecular detection and characterization of hematozoa infections in northern pintails (Anas acuta) wintering in California. Int J Parasitol Parasites Wildl 2:102–109
Sorensen MC, Asghar M, Bensch S, Fairhurst GD, Jenni-Eiermann S, Spottiswoode CN (2016) A rare study from the wintering grounds provides insight into the costs of malaria infection for migratory birds. J Avian Biol 47:1–8
Stephens M, Smith N, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Sutcliffe JF (1986) Black fly host location: a review. Can J Zool 64:1041–1053
Svensson-coelho M, Silva GT, Santos SS, Miranda LS, Ricklefs RE, Miyaki CY, Maldonado-Coelho M (2016) Lower detection probability of avian plasmodium in blood compared to other tissues. J Parasit Dis 102:559–561
Szymanski MM, Lovette IJ (2005) High lineage diversity and host sharing of malarial parasites in a local avian assemblage. J Parasitol 91:768–774
Valkiūnas G (2005) Avian malaria parasites and other haemosporidia. CRC Press. Boca Raton, FL, Boca Raton
Valkiūnas G, Bairlein F, Iezhova TA, Dolnik OV (2004) Factors affecting the relapse of Haemoproteus belopolskyi infections and the parasitaemia of Trypanosoma spp. in a naturally infected European songbird, the blackcap, Sylvia atricapilla. Parasitol Res 93:218–222
Valkiūnas G, Iezhova TA, Palinauskas V, Ilgūnas M, Bernotienė R (2015) The evidence for rapid gametocyte viability changes in the course of parasitemia in Haemoproteus parasites. Parasitol Res 114:2903–2909
van Riper IIIC, van Riper SG, Goff ML, Laird M (1986) The Epizootiology and ecological significance of malaria in Hawaiian land birds. Ecol Monogr 56:327
Ya'cob Z, Takaoka H, Pramual P, Low VL, Sofian-Azirun M (2016) Breeding habitat preference of preimaginal black flies (Diptera: Simuliidae) in peninsular Malaysia. Acta Trop 153:57–63
Young BE, Garvin MC, McDonald DB (1993) Blood parasites in birds from Monteverde, Costa Rica. J Wildl Dis 29:555–560
Zehtindjiev P, Ilieva M, Westerdahl H, Hansson B, Valkiūnas G, Bensch S (2008) Dynamics of parasitemia of malaria parasites in a naturally and experimentally infected migratory songbird, the great reed warbler Acrocephalus arundinaceus. Exp Parasitol 119:99–110
Acknowledgements
We are grateful to the country of Benin for the permission to perform this research. We would like to thank Alphonse Adite for the invaluable assistance in obtaining permits and field collecting and Robert Adite for his driving prowess. We thank Dr. Toby J. Hibbitts and Jerry W. Huntley for the field collections of samples. We are grateful to Carter Atkinson for proposing valuable comments and suggestions that improved the manuscript. We would also like to thank Jerry Huntley and Jonathan Puritz for the much need insightful comments on an earlier draft of the paper. We also thank Danielle Walkup for the helpful discussions on the material along the way. This is publication number 1552 of the Biodiversity, Research, and Teaching Collections at Texas A&M University. This research received no specific grant from any funding agency, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Online Resources 1
Host species taxonomic information including, locality sampled (Chutes de Koudou (C), Point Triplo (P), Dogo Forest (D) Lama Forest (L), Lake Toho (T), Abomey Calavi (A), number of individuals sampled (N), samples greater then 1 per locality listed, and frequency of detection across Haemoproteus (H), Leucocytozoon (L), Plasmodium (P), bioregion sampled in, novel lineages recovered, MalAvi lineages recovered (XLSX 44 kb)
Rights and permissions
About this article
Cite this article
Harvey, J.A., Voelker, G. Avian haemosporidian detection across source materials: prevalence and genetic diversity. Parasitol Res 116, 3361–3371 (2017). https://doi.org/10.1007/s00436-017-5654-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-017-5654-0