Abstract
Background
Colorectal cancer (CRC) is one of the most common cancers in the world. MicroRNAs play important roles in the progression of CRC. This study aimed to investigate the role of miR-206 and its novel mechanism in the invasion and metastasis of CRC.
Methodology
Real-time RT-PCR or Western blotting was used to detect the expressions of miR-206, FMNL2 and c-MET in CRC cell lines and tissues. Luciferase reporter assays were conducted to detect the associations between miR-206 and 3′UTRs of FMNL2 and c-MET. A series of loss-of-function and gain-of-function assays were performed to evaluate the effect of miR-206 on the proliferation, invasion and metastasis of CRC cells.
Results
miR-206 was significantly down-regulated in CRC tissues and correlated closely with differentiation, lymphatic metastasis and serosal invasion. miR-206 suppressed CRC cell proliferation by arresting CRC cells in the G1/G0 phase and accelerating apoptosis. miR-206 also inhibited cell invasion and lung metastasis in CRC cells. Mechanically, FMNL2 and c-MET were identified as direct targets of miR-206. And FMNL2 rescued the suppression of miR-206 in the proliferation and invasion of CRC cells.
Conclusions
This study revealed functional and mechanistic links between miR-206 and oncogene FMNL2 and c-MET in the progression of CRC. miR-206 functioned as a tumor suppressor in the progression of CRC by targeting FMNL2 and c-MET. Restoration of miR-206 expression may represent a promising therapeutic approach for targeting malignant CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) has become one of the most common types of malignant tumors in the world, which shows high morbidity and mortality (Siegel et al. 2013). Although there are many diagnostic and therapeutic strategies for CRC, owing to a complicated network with multiple steps and multiple molecular processes of initiation of CRC (Migliore et al. 2011; Weitz et al. 2005; Harris and McCormick 2010), the survival of CRC patients is still very low. Thus, it is necessary to understand the molecular mechanisms which regulate the initiation and progression of CRC, and explore the novel molecular targets which may help to generate more effective therapies.
Recently, microRNAs (miRNAs), a class of small regulatory RNA molecules which are highly conserved in the regulation of a diversity of basic biological processes (Kloosterman and Plasterk 2006; He and Hannon 2004), have been reported as new biomarkers of CRC (Bonfrate et al. 2013). Through posttranscriptional regulation of the expression of target mRNA transcripts by binding to their 3′-untranslated region (UTR), miRNAs can dysregulate the development and progression of CRC (Hrasovec and Glavac 2012; Bartel 2009). Several studies about miRNA expression have been conducted in CRC, and a series of miRNAs including miR-20, miR-21, miR-17-5p, miR-224, miR-181b and miR-137 have been proved to be important biomarkers related to the pathogenesis of CRC (Hrasovec and Glavac 2012; Aslam et al. 2009; Cummins et al. 2006; Liao et al. 2013, 2014). Although miRNAs have been the subject of extensive research in recent years, the molecular regulatory mechanisms of miRNAs and their effects on cancer are not well understood.
miR-206 has been found to be lost in several cancers, such as hepatocellular cancer, gastric cancer, breast cancer and oral squamous cell carcinoma (Liu et al. 2014; Yang et al. 2013; Kondo et al. 2008; Lin et al. 2014). It functions as a pleiotropic modulator of cancer cell proliferation, invasion, lymphangiogenesis and metastasis (Kondo et al. 2008; Haas et al. 2011; Alteri et al. 2013; Song et al. 2009). The known targets of miR-206 in tumor include KRAS, cyclin D, CDK4, Notch3 and Cdc42 (Lin et al. 2014; Alteri et al. 2013; Song et al. 2009; Gagan et al. 2012; Liu et al. 2010). Although one paper has reported that miR-206 as a tumor suppressor in CRC (Wang et al. 2015), the role and precise mechanisms of miR-206 in the progression of CRC have not been characterized.
In this study, we identify miR-206 to be significantly down-regulated in CRC tissues. We reveal that miR-206 is a novel negative regulator in the progression of CRC by targeting FMNL2. Thus, we raise the intriguing possibility that miR-206 may be an attractive candidate for miRNA-based anticancer therapies.
Materials and methods
Construction of plasmids and transfection
MiR-206 lentivirus-expressing vector pEZX-MR01/miR-206 containing the enhanced green fluorescent protein (EGFP) gene (GeneCopier, Rockville city, MD) was transfected into lentiviral packaging cell lines 293T. Then, 1 mL of viral supernatant containing 4 Ag of polybrene was added into CRC cell lines for stable transduction. After 14 days, puromycin-resistant cell pools were established. pGC FU-GFP-LVFMNL2-CT lentiviral-expressing vector constructed in our previous study contains functional FMNL2-CT fragment without the 3′UTR region, which was used to show FMNL2 as a positive regulator of CRC invasion and metastasis (Liang et al. 2013). To obtain the miR-206/FMNL2 co-expressing cells, 3 μL of FMNL2-CT lentivirus-expressing vector concentrated solution was added into miR-206-expressing SW620 and LOVO cells. Five to approximately 8 μg/mL of polybrene then was mixed in those cells. After 72 h, Western blotting was performed to detect the expression of FMNL2. Cells were transfected with miR-206 inhibitor or negative control (NC) by using Lipofectamine 2000 (Invitrogen, Foster city, CA).
Proliferation, plate colony formation, cell cycle, apoptosis, cell migration and cell invasion assays in vitro
1 × 103 Cells were seeded into 96-well plates. The number of viable cells was determined by Cell Counting Kit-8 (CCK-8) (Dojindo, Kumamoto, Japan) from 1 to 7 days. Briefly, 10 mL CCK-8 solution was added, and absorbance at 490 nm was measured after 2 h of incubation at 37 °C. Each cell group was plated in three duplicate wells.
The plate colony formation, cell cycle, apoptosis, migration and invasion of transfected CRC cells were determined as previously described (Liang et al. 2013; Ren et al. 2014).
Animal models
For in vivo tumor growth assay, xenograft tumors were generated by subcutaneous injection of 1 × 107 cells. Tumors were measured with calipers to estimate volume from day 5 to day 28 after injection. For tail vein metastasis assay, a total of 1 × 106 cells were injected into the tail veins of nude mice. After 40 days, mice were killed, and lung tissues were dissected and subjected to histological examination. Metastatic tumors were detected by H&E staining and quantified by counting metastatic lesions in each section. Images were taken by Olympus DP72 upright microscope and were outputted by DP2-BSW software.
Luciferase activity assay
For the binding of miR-206 to 3′UTR segments of FMNL2 and MET, the 3′UTR segments of FMNL2 and MET genes were amplified by PCR and inserted into the vector. A mutant construct in two miR-206 binding sites of FMNL2 3′UTR region also was generated using Quick Change Site-Directed Mutagenesis Kit (Agilent, Roseville City, CA). Co-transfections of FMNL2 3′UTR or mut-FMNL2 3′UTR plasmid with miR-206 lentivirus vector into the cells were accomplished by using Lipofectamine 2000 (Invitrogen). Luciferase activity was measured 48 h after transfection by the Dual-Luciferase Reporter Assay System (Promega). Each assay was repeated in six independent experiments.
Results
miR-206 is down-regulated in CRC tissues
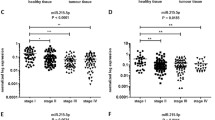
To explore the expression pattern of miR-206 in CRC tissues, we detected the expression of miR-206 in 40 paired samples of CRC patients by real-time RT-PCR. The result showed that the expression level of miR-206 in CRC tissues was significantly lower than in adjacent normal tissues (Fig. 1a, b, P = 0.007) and decreased along with the progression of T classifications (Fig. 1c, P < 0.01), N classifications (Fig. 1d, P < 0.01) and distant metastasis (Fig. 1e, P < 0.01). Clinicopathological analyses showed that miR-206 expression was correlated strongly with differentiation (P = 0.000, Table 1), lymphatic metastasis (P = 0.014, Table 1) and serosal invasion (P = 0.046, Table 1). We also measured miR-206 expression in six CRC cell lines by real-time RT-PCR. miR-206 expression was relatively lower in highly aggressive metastatic SW620 and LOVO cells, and gradually increased in lowly metastatic LS174T, SW480, HT29 and HCT116 cells (P < 0.05, Fig. 1f). These results indicate that miR-206 may function as a tumor suppressor in CRC.
miR-206 is down-regulated in CRC tissues. a Expression of miR-206 in 40 paired human CRC tissues and the corresponding normal mucosa by real-time PCR. Each bar represents the mean of three independent experiments. b Real-time PCR analysis of miR-206 expression in CRC tissues compared to the corresponding normal mucosa. P = 0.007. c Correlation between miR-206 expression and T classification (T1–T4) in 40 cases of CRC tissues and normal intestine epithelial tissue (Normal, n = 40). d Correlation between miR-206 expression and N classification (N0–N2) in 40 cases of CRC tissues. e Correlation between miR-206 expression and distant metastasis. f Real-time RT-PCR analysis of miR-206 in six CRC cell lines. *P < 0.05
Effect of miR-206 on cell proliferation of CRC in vitro and in vivo
To elucidate candidate effects of miR-206 in the progression of CRC, miR-206 was over-expressed in LOVO and SW620 cells (Fig. 2a) and silenced in HCT116 and SW480 cells (Fig. 2b). Alterations in cell proliferation, cell cycle transition and apoptosis induced by miR-206 were then examined. CCK-8 assays revealed that over-expression of miR-206 significantly decreased the growth rate of SW620 and LOVO cells at the sixth and seventh days (Fig. 2c, P < 0.05), while depletion of miR-206 in SW480 and HCT116 cells showed the opposite effects (Fig. 2d, P < 0.05). Similar results were observed in colony formation assays (Fig. 2e–h). To explore the possible mechanisms of miR-206’s function in cell proliferation, we determined the distribution of cells within the stages of the cell cycle by flow cytometry. Ectopic miR-206 in LOVO cells induced a significant increase in the percentage of cells in the G1/G0 peak and a decrease in the percentage of cells in the S and G2/M peaks (Fig. 3a). However, miR-206 knockdown in SW480 cells led to an increase in the percentage of cells in the S and G2/M peaks (Fig. 3b). We also measured the effect of miR-206 on apoptosis in CRC cells. Ectopic miR-206 in LOVO and SW620 cells obviously increased the rate of apoptosis (Fig. 3c), while depletion of miR-206 in HCT116 cells had the reverse effect (Fig. 3d). These results suggested that miR-206 could inhibit cell proliferation by arresting the tumor cells at the G1/G0 phase and accelerating apoptosis.
Effect of miR-206 on the proliferation of CRC cells in vitro. a Analyses of miR-206 expression in miR-206-expressing SW620 and LOVO cells by real-time PCR. b Analyses of miR-206 expression in miR-206-depleting SW480 and HCT116 cells by real-time PCR. c Effect of ectopic miR-206 on cell proliferation in vitro by CCK-8 assay. d Effect of miR-206 knockdown on cell proliferation in vitro by CCK-8 assay. e, g Effect of ectopic miR-206 on cell proliferation in vitro by plate colony formation assay. f, h Effect of miR-206 knockdown on cell proliferation in vitro by plate colony formation assay. Error bars represent mean ± SD from three independent experiments. *P < 0.05
Effect of miR-206 on CRC cell cycle transition, apoptosis in vitro and tumor growth in vivo. a, b Effects of miR-206 over-expression and knockdown on cell cycle transitions by flow cytometry. c, d Effects of miR-206 over-expression and knockdown on cell apoptosis by flow cytometry. e Analyses of miR-206 expression in miR-206 stable expressing LOVO cells by real-time PCR. f Effects of miR-206 over-expression on tumor growth in nude mice (n = 6). g Histopathological analyses of xenograft tumors. The tumor sections were stained with H&E or subjected to IHC staining using an antibody against Ki-67, and scale bars represent 20 μm. Error bars represent mean ± SD from three independent experiments. *P < 0.05
To further confirm the suppression of miR-206 in cell proliferation, we established stable miR-206-expressing LOVO cells (Fig. 3e) and injected them into the subcutaneous site of mice. Expectedly, over-expression of miR-206 significantly inhibited tumor growth in vivo at the sixth and seventh days (Fig. 3f, P < 0.01). Furthermore, miR-206-over-expressed tumors displayed decreased proliferation indices (calculated by Ki-67 expression) compared to control groups (Fig. 3g, P < 0.01). Taken together, these results indicate that miR-206 suppresses the proliferation of CRC cells.
Effect of miR-206 on CRC invasion and metastasis in vitro and in vivo
We next assessed the effect of miR-206 on the invasion and metastasis of CRC cells. Results of Boyden chamber assay showed that over-expression of miR-206 in SW620 and LOVO cells markedly decreased the invasive abilities (P < 0.05, Fig. 4a, c), while suppression of miR-206 in SW480 and HCT116 cells showed the opposite effect (Fig. 4b, d, P < 0.05). Moreover, miR-206-expressing subcutaneous tumors were well encapsulated and noninvasive, while control tumors displayed evidence of local invasion (Fig. 4e). To test the effect of miR-206 on CRC metastasis in vivo, we injected miR-206-expressing LOVO cells into the tail vein of nude mice to examine the role of miR-206 in lung colonization. MiR-206 strikingly inhibited the capacity of LOVO cells to seed lung metastases (Fig. 4f). The number of metastatic lesions in miR-206-expressing group was significantly less than in control group (P < 0.05, Fig. 4g). These results demonstrate that miR-206 profoundly decreases invasion and metastasis of CRC cells in vitro and in vivo.
Effect of miR-206 on the invasion and metastasis of CRC cells in vitro and in vivo. a, c Effects of miR-206 over-expression on cell invasion in vitro. Morphological comparison of cells penetrating the artificial basement membrane is also shown. b, d Effects of miR-206 knockdown on cell invasion in vitro. Morphological comparison of cells penetrating the artificial basement membrane is also shown. e Effect of miR-206 over-expression on cell invasion in vivo. The tumor sections were stained with H&E. Scale bars represent 20 μm. f, g Effects of miR-206 over-expression on pulmonary metastases in vivo. The tumor sections were stained with H&E. Scale bars represent 50 μm. The number of metastatic nodules in individual mice was counted under the microscope. Error bars represent mean ± SD from three independent experiments. *P < 0.05
miR-206 directly targets FMNL2 and c-MET
To explore the potential mechanisms underlying miR-206-mediated CRC cell behaviors, three common bioinformatic algorithms (TargetScan, Pictar, and miRANDA) were used to predict the mRNA targets of miR-206. Based on the representation of miR-206 sites in their 3′UTRs, >200 mRNAs were predicted to be regulated by miR-206. Among those mRNAs, FMNL2 and c-MET were predicted by all three databases and associated with tumor invasion and metastasis (Fig. 5a). We cloned the wild-type (WT) 3′UTRs of FMNL2 and c-MET into luciferase constructs. Reporter assays revealed that miR-206 repressed the luciferase activities of WT FMNL2 and c-MET 3′UTR (Fig. 5b, c). We also introduced mutant (mut) FMNL2 3′UTR into luciferase constructs and found that miR-206 had no effect on the activity of mut FMNL2 3′UTR (Fig. 5a, b). In addition, ectopic miR-206 in SW620 and LOVO cells reduced the protein levels of FMNL2 and c-MET, while knockdown of miR-206 in SW480 and HCT116 cells led to increased expressions of FMNL2 and c-MET (Fig. 5d and Figure S1A-B). miR-206 also inhibited the mRNA levels of FMNL2 and c-MET (Fig. 5e, f). These results demonstrate that miR-206 could directly target FMNL2 and c-MET in CRC cells.
miR-206 directly targets FMNL2 and c-MET. a Predicted binding sites of miR-206 in wild-type 3′ UTRs of FMNL2 and c- MET, and mutant 3′ UTRs of FMNL2. b Luciferase activities of wild-type and mutant 3′UTR-FMNL2-luc constructs in SW480 and HCT116 cells after transfection of miR-206. c Luciferase activities of wild-type 3′UTR-c-MET-luc construct in SW480 and HCT116 cells after transfection of miR-206. d Western blotting analyses of FMNL2 and c-MET expressions in miR-206-expressing SW620, LOVO and HCT116 cells. e Real-time PCR analyses of FMNL2 mRNA level in miR-206-over-expressing or miR-206-depleting cells. f Real-time PCR analyses of c-MET mRNA level in miR-206-over-expressed or miR-206-depleted cells. *P < 0.01
FMNL2 is necessary for miR-206-induced cell proliferation and invasion of CRC cells
FMNL2 is regarded as a positive regulator of cell motility and metastasis in CRC (Liang et al. 2013). And c-MET has been reported to be a target of miR-1/206 in the development of rhabdomyosarcoma (Yan et al. 2009). Thus, we speculated that FMNL2 might act as the vital effector of miR-206 in the progression of CRC. To determine whether cancer cell phenotypes associated with miR-206 expression could be reversed via restoration of FMNL2, we stably transduced miR-206-expressing LOVO cells with FMNL2 construct lacking the 3′UTR regions and confirmed its over-expression (Fig. 6a and Figure S1C). Results of CCK-8 assays showed that FMNL2 reversed at least partially miR-206-mediated repression of proliferation in vitro at the sixth and seventh days (Fig. 6b, P < 0.05). Moreover, reintroduction of FMNL2 rescued miR-206-imposed cell cycle transition and apoptosis (Fig. 6c, d). In addition, the constitutive expression of FMNL2 rescued miR-206-induced invasion of LOVO cells (P < 0.05, Fig. 6e). These data make it obvious that miR-206 inhibits proliferation and invasion by targeting FMNL2. Finally, to further examine whether the above findings could be supported by observations in human primary CRC tissues, we detected the expressions of miR-206, FMNL2 and c-MET (Fig. 6f, g and Figure S1C) in the normal and tumor tissues from 20 CRC patients by Q-PCR and Western blotting. The results showed that miR-206 expression was negatively correlated with the expressions of FMNL2 (Fig. 6h, P = 0.0021, r = −0.6449) and c-MET (Fig. 6i, P = 0.0377, r = −0.4674). However, there is no significant correlation between the expressions of FMNL2 and c-MET (Fig. 6j, P = 0.100, r = 0.3768).
FMNL2 is necessary for miR-206-induced cell proliferation and invasion in CRC cells. a Western blotting analysis of FMNL2 expression in LOVO/miR-206 and LOVO/miR-206/FMNL2 cells. b Effect of miR-206 and miR-206/FMNL2 on cell proliferation of CRC cell in vitro by CCK-8 arrays. c Effect of miR-206 and miR-206/FMNL2 on cell cycle transition of CRC cell in vitro by flow cytometry. d Effect of miR-206 and miR-206/FMNL2 on cell apoptosis of CRC cell in vitro by flow cytometry. e Effect of miR-206 and miR-206/FMNL2 on the invasion of CRC cell in vitro. Morphological comparison of cells penetrating the artificial basement membrane was also shown. Error bars represent mean ± SD from three independent experiments. f Real-time PCR analysis of miR-206, FMNL2 and c-MET expressions in the normal and tumor tissues from 20 CRC patients. g Western blotting analysis of FMNL2 and c-MET expressions in the normal and tumor tissues from 20 CRC patients; β-actin was used as loading control. h Spearman’s correlation analyses between relative miR-206 expression and relative mRNA expression levels of FMNL2 or c-MET in the normal and tumor tissues from 20 CRC patients. *P < 0.05
Discussion
Here we show that down-regulation of miR-206 in CRC is important for cancer cells to sustain their proliferative, invasive and metastatic capacities. We detected the expression of miR-206 in 40 paired CRC tissues and 6 CRC cell lines and found that miR-206 was down-regulated in CRC tissues and CRC cell lines with highly metastatic capacities. Moreover, its expression was correlated strongly with T, N, M classifications, differentiation, lymphatic metastasis and serosal invasion (P < 0.05). Previously, miR-206 has been found to be deregulated in several tumors such as hepatocellular cancer, gastric cancer and breast cancer (Liu et al. 2014; Yang et al. 2013; Kondo et al. 2008). In cancer cells, it is plausible that miR-206 is silenced by epigenetic modifications (Geisler et al. 2013). A recent study reveals that miR-206 is negatively regulated by nuclear factor erythroid-2-related factor 2, a transcription factor, which is critical for regulating tumor growth through glucose metabolism reprogramming (Singh et al. 2013). In CRC, the exact mechanism that leads to the suppression of miR-206 needs further investigation.
Accumulating data demonstrate that miR-206 functions as a tumor suppressor in the carcinogenesis and aggressiveness of several cancers. For example, miR-206 inhibits cellular proliferation and invasion of estrogen receptor α-positive ovarian cancer cells (Pan et al. 2008). Previous studies in rhabdomyosarcomas (MacQuarrie et al. 2012; Missiaglia et al. 2010), hepatocellular carcinoma (Liu et al. 2014) and endometrioid adenocarcinoma (Chen et al. 2012) show that miR-206 is involved in the negative control of cell cycle. miR-206 induces G1 arrest in melanoma by inhibition of CDK4 and cyclin D (Gagan et al. 2012). miR-206 has been shown to target Notch3 and activate apoptosis in HeLa cells (Song et al. 2009). miR-206 acts at steps that suppress metastasis in gastric cancer by targeting several metastasis regulatory genes (Alteri et al. 2013). Consistent with the previous reports, our study showed that miR-206 negatively regulated the proliferation of CRC cells by inducing cell cycle arrest and promoting apoptosis. Moreover, miR-206 suppressed in vitro invasion as well as in vivo metastasis in CRC cells. One of the possible mechanisms for the suppression of miR-206 in tumor invasion and migration is that miR-206 partly regulates actin cytoskeleton remodeling such as filopodia formation (Zhang et al. 2013). Taken together, the above results provide evidence that miR-206 can function as a suppressor in the progression of CRC.
Having established the expression and role of miR-206 in CRC, we subsequently conducted three publicly available bioinformatic algorithms (TargetScan, Pictar and miRANDA) to search possible direct targets of miR-206. Our results identified that FMNL2 and c-MET were targets of miR-206 in CRC cells with high predictive value. c-MET is a hepatocyte growth factor (HGF) receptor with tyrosine kinase activity, and aberrant activation of receptor tyrosine kinase c-Met/HGF pathway is shown to be associated with cell proliferation, invasion and poor prognosis in several tumor types (Taulli et al. 2006). However, c-MET is a known target of miR-206 in the development of rhabdomyosarcoma (Yan et al. 2009). So, we focused on FMNL2 as the vital effector of miR-206 in CRC. In our previous study, FMNL2 was screened as a potential metastasis-associated gene by comparing the gene expression profiles of highly metastatic M5, SW620 cells and low metastatic SW480 cells (Huang et al. 2003). Over-expression of FMNL2 in CRC tissues was associated with invasion and lymphatic metastasis (Zhu et al. 2008). We also documented that FMNL2 promoted CRC cell proliferation, motility, invasion and metastasis (Yan et al. 2009) and enhanced the invasive potential of CRC by inducing epithelial–mesenchymal transition (Li et al. 2010). Other research groups propose that FMNL2 drives amoeboid invasive cell motility downstream of RhoC (Kitzing et al. 2010) and FMNL2 co-localizes with F-actin dots at the tips of cellular protrusions in melanoma cells (Block et al. 2012). In our study, the rescue experiments showed that the tumor-suppressive role of miR-206 in the proliferation and invasion of CRC cells was at least partially via down-regulation of FMNL2.
In summary, our findings suggest that miR-206 is down-regulated in CRC and may affect proliferation, invasion and metastasis of CRC cells by inhibiting FMNL2. Restoration of miR-206 might become a promising therapeutic approach for targeting malignant CRC. Thus, our data demonstrate a potential mechanistic connection between miR-206 dysregulation and the progression of human CRC and may shed light on therapeutic strategies for CRC prevention and treatment.
References
Alteri A, De Vito F, Messina G et al (2013) Cyclin D1 is a major target of miR-206 in cell differentiation and transformation. Cell Cycle 12(24):3781–3790
Aslam MI, Taylor K, Pringle JH, Jameson JS (2009) MicroRNAs are novel biomarkers of colorectal cancer. Br J Surg 96:702–710
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Block J, Breitsprecher D, Kühn S et al (2012) FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr Biol 22(11):1005–1012
Bonfrate L, Altomare DF, Di Lena M et al (2013) MicroRNA in colorectal cancer: new perspectives for diagnosis, prognosis and treatment. J Gastrointestin Liver Dis 22:311–320
Chen X, Yan Q, Li S, Zhou L, Yang H et al (2012) Expression of the tumor suppressor miR-206 is associated with cellular proliferative inhibition and impairs invasion in ERalpha-positive endometrioid adenocarcinoma. Cancer Lett 314(1):41–53
Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA Jr, Sjoblom T et al (2006) The colorectal microRNAome. Proc Natl Acad Sci USA 103:3687–3692
Gagan J, Dey BK, Layer R, Yan Z, Dutta A (2012) Notch3 and Mef2c proteins are mutually antagonistic via Mkp1 protein and miR-1/206 MicroRNAs in differentiating myoblasts. J Biol Chem 287(48):40360–40370
Geisler A, Schön C, Größl T et al (2013) Application of mutated miR-206 target sites enables skeletal muscle-specific silencing of transgene expression of cardiotropic AAV9 vectors. Mol Ther 21(5):924–933
Haas JD, Nistala K, Petermann F et al (2011) Expression of miRNAs miR-133b and miR-206 in the Il17a/f locus is co-regulated with IL-17 production in αβ and γσ T cells. PLoS One 6(5):e20171
Harris TJ, McCormick F (2010) The molecular pathology of cancer. Nat Rev Clin Oncol 7:251–265
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531
Hrasovec S, Glavac D (2012) MicroRNAs as novel biomarkers in colorectal cancer. Front Genet 3:180
Huang XZ, Sun Q, Ding YQ et al (2003) Mining microarray gene expression data of metastatic colorectal cancer by literature profiling. Di Yi Jun Yi Da Xue Xue Bao 23:1195–1197
Kitzing TM, Wang Y, Pertz O et al (2010) Formin-like 2 drives amoeboid invasive cell motility downstream of RhoC. Oncogene 29:2441–2448
Kloosterman WP, Plasterk RH (2006) The diverse functions of micro-RNAs in animal development and disease. Dev Cell 11:441–450
Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H (2008) miR-206 expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res 68(13):5004–5008
Li Y, Zhu X, Zeng Y et al (2010) FMNL2 enhances invasion of colorectal carcinoma by inducing epithelial-mesenchymal transition. Mol Cancer Res 8:1579–1590
Liang L, Li X, Zhang X, Lv Z, He G et al (2013) MicroRNA-137, an HMGA1 target, suppresses colorectal cancer cell invasion and metastasis in mice by directly targeting FMNL2. Gastroenterology 144:624–635
Liao WT, Li TT, Wang ZG, Wang SY, He MR et al (2013) microRNA-224 Promotes cell proliferation and tumor growth in human colorectal cancer by repressing PHLPP1 and PHLPP2. Clin Cancer Res 19:4662–4672
Liao WT, Ye YP, Zhang NJ, Li TT, Wang SY et al (2014) MicroRNA-30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2. J Pathol 232(4):415–427
Lin F, Yao L, Xiao J et al (2014) MiR-206 functions as a tumor suppressor and directly targets K-Ras in human oral squamous cell carcinoma. Onco Targets Ther 7:1583–1591
Liu H, Cao YD, Ye WX, Sun YY (2010) Effect of microRNA-206 on cytoskeleton remodelling by down-regulating Cdc42 in MDA-MB-231 cells. Tumori 96:751–755
Liu W, Xu C, Wan H, Liu C et al (2014) MicroRNA-206 overexpression promotes apoptosis, induces cell cycle arrest and inhibits the migration of human hepatocellular carcinoma HepG2 cells. Int J Mol Med 34(2):420–428
MacQuarrie KL, Yao Z et al (2012) miR-206 integrates multiple components of differentiation pathways to control the transition from growth to differentiation in rhabdomyosarcoma cells. Skelet Muscle 2:7
Migliore L, Migheli F, Spisni R et al (2011) Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol 2011:792362
Missiaglia E, Shepherd CJ, Patel S, Thway K (2010) MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br J Cancer 102(12):1769–1777
Pan Q, Luo X, Chegini N (2008) Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids. J Cell Mol Med 12(1):227–240
Ren XL, Zhu XH, Li XM et al (2014) Down-regulation of BTG3 promotes cell proliferation, migration and invasion and predicts survival in gastric cancer. J Cancer Res Clin Oncol 141(3):397–405
Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63:11–30
Singh A, Happel C, Soumen K et al (2013) Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J Clin Invest 123(7):2921–2934
Song GS, Zhang YX, Wang L (2009) MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J Biol Chem 284(46):31921–31927
Taulli R, Scuoppo C, Bersani F et al (2006) Validation of met as a therapeutic target in alveolar and embryonal rhabdomyosarcoma. Cancer Res 66(9):4742–4749
Wang XW, Xi XQ, Wu J et al (2015) MicroRNA-206 attenuates tumor proliferation and migration involving the downregulation of NOTCH3 in colorectal cancer. Oncol Rep 33(3):1402–1410
Weitz J, Koch M, Debus J, Hohler T, Galle PR, Buchler MW (2005) Colorectal cancer. Lancet 365:153–165
Yan D, Dong XDE, Chen X et al (2009) MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem 284(43):29596–29604
Yang Q, Zhang C, Huang B, Li H, Zhang R, Huang Y, Wang J (2013) Downregulation of microRNA-206 is a potent prognostic marker for patients with gastric cancer. Eur J Gastroenterol Hepatol 25(8):953–957
Zhang X, Xu J, Wang J et al (2013) Reduction of MicroRNA-206 contributes to the development of bronchopulmonary dysplasia through up-regulation of fibronectin 1. PLoS One 8(9):e74750
Zhu XL, Liang L, Ding YQ (2008) Expression of FMNL2 and its relation to the metastatic potential of human colorectal cancer cells. Nan Fang Yi Ke Da Xue Xue Bao 28:1775–1778
Acknowledgments
Dr. Ren X.L., He G.Y., Li X.M., Yi L.Z. and Lu G.F. carried out experiments. Dr. Xin S.N., Wu P.X. and Li Y.L. took on the statistical analysis. Ren X.L., He G.Y. and Li X.M. contributed equally to this work. Professor Liao WT gave assistance in collecting tissue samples or animal experiments. Professor Ding Y.Q. and Liang L. conceived experiments and analyzed data. All authors were involved in writing the paper and had final approval of the submitted and published versions. We thank Professor Geevan and Reddy for editing the English writing.
Funding
This work was supported by the National Natural Science Foundation of China (81272759, 81172382, 81472313), National Basic Research Program of China (973 Program, 2015CB554002), the Science and Technology Planning Project of Guangdong Province (S2013010014544), Major projects of science and technology of Guangzhou (201300000056) and the Key Project of National Natural Science Fund (Guangdong Province NSFC-Joint Fund, U1201226).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All protocols for animal studies were reviewed and approved by the Institutional Animal Care and Use Committee of Southern Medical University and have been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
Conflict of interest
All the authors declare that they have no conflict of interest.
Additional information
X. L. Ren, G. Y. He and X. M. Li have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ren, X.L., He, G.Y., Li, X.M. et al. MicroRNA-206 functions as a tumor suppressor in colorectal cancer by targeting FMNL2. J Cancer Res Clin Oncol 142, 581–592 (2016). https://doi.org/10.1007/s00432-015-2053-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-015-2053-8