Abstract
Main conclusion
The nuclear localized TaWZY1-2 helps plants resist abiotic stress by preserving the cell’s ability to remove reactive oxygen species and decrease lipid oxidation under such conditions.
Abstract
In light of the unpredictable environmental conditions in which food crops grow, precise strategies must be developed by crops to effectively cope with abiotic stress and minimize damage over their lifespan. A key component in this endeavor is the group II of late embryogenesis abundant (LEA) proteins, known as dehydrins, which play crucial roles in enhancing the tolerance of plants to abiotic stress. Tawzy1-2 is a dehydrin-encoding gene which is constitutively expressed in various tissues of wheat. However, the biological function of TaWZY1-2 is not yet fully understood. In this study, TaWZY1-2 was isolated and identified in the wheat genome, and its functional role in conferring tolerance to abiotic stresses was detected in both prokaryotic and eukaryotic cells. Results showed that TaWZY1-2 is a nuclear localized hydrophilic protein that accumulates in response to multiple stresses. Escherichia coli cells expressing TaWZY1-2 showed enhanced tolerance to multiple stress conditions. Overexpression of TaWZY1-2 in Nicotiania benthamiana improved growth, germination and survival rate of the transgenic plants exposed to four kinds of abiotic stress conditions. Our results show that Tawzy1-2 transgenic plants exhibit improved capability in clearing reactive oxygen species and reducing lipid degradation, thereby enhancing their resistance to abiotic stress. This demonstrates a significant role of TaWZY1-2 in mitigating abiotic stress-induced damage. Consequently, these findings not only establish a basis for future investigation into the functional mechanism of TaWZY1-2 but also contribute to the expansion of functional diversity within the dehydrin protein family. Moreover, they identify potential candidate genes for crop optimization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Food crops are essential for human survival, with wheat being one of the most crucial resources that supports 60% of the world’s population (Gupta 2011). However, current crop yield trajectories indicate that it may not be sufficient to sustain the global population beyond 2050 (Ray et al. 2013). This inadequacy is exacerbated by the various stress factors that crops face in their natural environment, including drought, high salinity, and extreme temperatures. These environmental pressures have a detrimental effect on the development of wheat and are beyond human control. Consequently, achieving a transformation in crop quality through leveraging the innate adaptive traits of crops to mitigate external environmental variations becomes imperative (Bailey-Serres et al. 2019).
Plants, when exposed to environmental stress, undergo serious cellular damage, which can result in plant death (Cleal et al. 2021). In response, plants have evolved defense mechanisms over long periods of evolution to counter potential damage, such as upregulating various protective proteins. Among these proteins, late embryogenesis abundant (LEA) proteins play a particularly significant role. Recent research has significantly advanced our understanding of these proteins, providing compelling evidence for their effectiveness in enhancing resistance to adverse conditions. Hence, an increasing focus on studying the characteristics of LEA proteins is contributing to a more evidence-based comprehension of their role in conferring resilience to crops in adverse conditions (Priya et al. 2019).
Dehydrins (DHNs), belonging to group II of LEA proteins, exhibit a high proportion of hydrophilic amino acids. When exposed to a membrane-mimetic environment using sodium dodecyl sulfate micelles, some dehydrins adopt an alpha-helical conformation. These proteins are characterized as heat-stable and typically contain an amino acid sequence with at least one K-segment (XKXGXXD/EKIKD/EKXPG) rich in lysine, which modulates the structural and biochemical properties of wild potato FSK3 dehydrin (Szabala 2023). Furthermore, many dehydrins comprise S-segments consisting of five to seven serine residues, the phosphorylation of which can recruit proteins with nuclear localization signals to the nucleus (Goday et al. 1994). Moreover, a Y-segment (V/TD/E/QYGNP) with a conserved tyrosine at the N-terminal end, which can be substituted with phenylalanine or histidine, is also present. Some dehydrins feature the F-segment, containing one or two phenylalanine residues (Strimbeck 2017). The organization of these segments is modular, leading to the division of dehydrins into seven subclasses—YnSKn, FnSKn, FnK, YnKn, SKn, Kn, and KnS—based on the presence and order of these segments (Smith and Graether 2022).
Dehydrins are multifunctional proteins that are thought to protect the cellular machinery from stress-induced damage and confer stress tolerance to a variety of plants (Liu et al. 2017). For in vivo studies, the overexpression of dehydrin in cells often enhances the stress tolerance of plants, a number of studies demonstrated that overexpression of dehydrins in transgenic plants enhanced stress tolerance as a result of dehydrin multiple protective functions on distinct biomacromolecules, including membrane lipids, proteins, and nucleic acids (Graether and Boddington 2014). Dehydrin has the ability to lower the transition temperature of different membrane components, as well as the relative humidity needed for the transition to the HII phase. This property enables the membrane to retain its fluidity under lower environmental temperatures. Consequently, dehydrin acts as an inhibitor for the fusion of two liposomes by preventing membrane exchange (Eriksson et al. 2011; Clarke et al. 2015).
Dehydrin is known to have the crucial function of protecting intracellular macromolecules from stress and environmental changes. In this regard, lactate dehydrogenase (LDH) is frequently used as a marker to evaluate the protective effect of dehydrins on enzyme activity due to its extreme sensitivity to low temperatures (Houde et al. 1995; Bravo et al. 2003; Reyes et al. 2008). In vitro experiments demonstrated that the purified DHN24, SbDHN2, and WZY2 dehydrins were effective in shielding LDH from freeze-induced denaturation (Szabala 2023; Yang et al. 2015; Halder et al. 2016). Furthermore, they were found to prevent the excessive aggregation of malate dehydrogenase and alcohol dehydrogenase, indicating their potential role in safeguarding the activity of other intracellular protein molecules under various stress conditions (Kovacs et al. 2008; Reyes et al. 2008; Clarke et al. 2015). This protective function of dehydrin is considered one of the key factors in cellular stress resistance. However, it is noteworthy that CuCOR15, a KnS dehydrin, was shown to have a specific interaction with a 200 bp fragment of DNA (Hara et al. 2009). Similarly, VrDHN1, a dehydrin with the YSK2 type from Vitis riparia, also exhibited DNA binding characteristics (Boddington and Graether 2019), suggesting that dehydrins may be involved in protecting DNA or regulating gene expression.
It is important to note that environmental stress not only disrupts the stability of cell membranes and intracellular molecules, but also elevates the levels of intracellular reactive oxygen species (ROS), particularly hydroxyl radicals and singlet oxygen, which can cause severe damage in biological systems (Gechev et al. 2006; Del Rio 2015). Previous research has indicated that dehydrin plays a role in the physiological antioxidant processes in plants. For instance, the pepper (Capsicum annuum L.) dehydrin gene CaDHN3 has been shown to enhance tolerance to salt and drought stresses by reducing the accumulation of ROS (Meng et al. 2021). Similarly, in maize, the KS-type dehydrin has been found to improve the tolerance of transgenic tobacco to oxidative stress (Liu et al. 2019). Studies have demonstrated that SbDHN2 exhibits a strong binding affinity for metal ions through histidine-rich motifs, thereby preventing the generation of ROS (Halder et al. 2016). In addition, when DHN-5 was transformed into Arabidopsis, the transgenic plants showed greater salinity tolerance than the wild type (WT), with DHN-5 mediating the response to salt stress by regulating proline metabolism and scavenging ROS (Saibi et al. 2015). These findings collectively indicate that dehydrin may play a role in the cellular signaling pathway associated with the clearance of ROS, although its precise functions in plants are yet to be fully understood.
In previous studies, the 789-bp open reading frame (ORF) sequence of dehydrin Tawzy1-2 (GenBank Accession no. EU124658.1), encoding a 262-residue protein, was isolated from Triticum aestivum cultivar Zhengyin 1, a prominent winter wheat cultivar that originated from Italy in 1965 and is currently extensively cultivated in numerous provinces of China. Zhengyin 1 is known to exhibit heightened sensitivity to drought stress compared to other strains. The promoter sequence of Tawzy1-2 was scrutinized, as these sequences often contain crucial biological information relevant to their function (Zhu et al. 2014). In order to investigate the stress-related biological functions of TaWZY1-2, we conducted an analysis of the expression pattern of Tawzy1-2 in diverse tissues and under various abiotic stress conditions. The biological function of TaWZY1-2 was also assessed using prokaryotic expression and transgenic technology in both E. coli and N. benthamiana. These assessments provided the groundwork for future research on stress resistance types and signaling pathways of TaWZY1-2, which is pivotal for uncovering the regulatory mechanisms of TaWZY1-2 and dehydrins during stress conditions.
Materials and methods
Plant materials and stress treatments
The seeds of Zhengyin 1 utilized in this study were obtained from the College of Life Sciences/State Key Laboratory of Crop Stress Biology for Arid Areas at Northwest A & F University in Yangling, China, where they have been carefully preserved. Prior to planting, the seeds were initially germinated on two layers of moist filter paper following an overnight soaking in distilled water to facilitate the germination process. Once germinated, the plump seeds were then planted in soil and subsequently cultivated under controlled conditions, with a 16-h light and 8-h dark cycle, at a temperature of 28 °C.
Amplification of cDNA sequence of TaWZY1-2
The first step of the experiment involved the extraction of total RNA using Trizol reagent (Invitrogen), following the manufacturer’s instructions. The extracted RNA was then assessed for both quality and quantity. This assessment included UV spectrophotometer analysis at 260 nm and 280 nm using Nanodrop ND-1000 as well as electrophoresis of a 5 μL sample on a 1% agarose gel at a constant current of 100 V for 45 min. Subsequently, 500 ng of the total RNA was utilized to generate first-strand cDNAs according to the manufacturer’s instructions of PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China).
Next, the ORF sequence of Tawzy1-2 was isolated using specific primers listed in Table 1 through PCR amplification from the obtained cDNA. The PCR program was initiated at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 40 s, and a final extension at 72 °C for 10 min, terminating at 4 °C. The resulting PCR products were verified by sequencing and subsequently used for further studies.
Sequence and phylogenetic tree analysis of TaWZY1-2
The information about TaWZY1-2 was analyzed by multiple software. The MEME suite (http://meme-suite.org/) was used to determine the conserved motifs. Hydrophobicity analysis was conducted by http://www.expasy.ch/tools/protscale.html. The amino acid sequences of TaWZY1-2 in different plants were analyzed using ClustalX software with the default settings. The phylogenetic tree was built by MEGA 7.0 software with NJ (Neighbor-Joining Algorithm) method based on the following parameters: bootstrap analysis with 1000 replicates.
Expression profiles of wheat Tawzy1-2 gene under various stress conditions
Uniformly sized seedlings at the two-leaf growth stage were subjected to simulated drought (20% PEG6000, w/v), salinity (500 mM NaCl), suboptimal temperature (4 °C), and high-temperature stress (42 °C), under a 16/8 light/dark at 28 °C. For drought and salinity stress, the seedlings were watered with 20% PEG6000 and 500 mM NaCl solution, respectively, and the double filter paper was kept moist. For the temperature stress, seedlings were treated with 4 °C for low-temperature stress and placed under 42 °C for high-temperature stress. Plants grown under normal conditions were used as controls. For each treatment, 10 plants were selected to carry out the assay. All samples were collected every 12 h until 72 h after treatment initiation, immediately frozen in liquid nitrogen, and stored at – 80 °C.
Total RNA was extracted using Trizol reagent (Invitrogen) according to manufacturer instructions. cDNA was synthesized with a PrimeScriptTM RT reagent kit with gDNA Eraser (TaKaRa) following the manufacturer’s instructions. The qRT-PCR reactions were carried out with SYBR Green PCR Master Mix in an iCycler iQ5 system (BioRad, Hercules, CA, USA) using the following program: 95 °C for 3 min; 95 °C for 15 s; 60 °C for 1 min; 40 cycles; 95 °C for 15 s; 60 °C for 5 s and 95 °C at the end. The cycle threshold (Ct) 2−∆∆Ct method was used for analyzing the transcriptional levels. The elongation factor gene (GenBank accession no. GR302794) served as the reference gene. Primers used were given in Table 1. Each reaction was replicated three times and relative-fold expression was analyzed.
Preparation of protoplasts
Lysozyme was added into the bacterial solution with OD600 value of 0.5, held in a water bath at 36 °C for 30 min, regular sampling was taken, and the formation of protoplasts was observed by microscopy. When more than 95% of the cells became spherical protoplasts, the suspension was centrifuged at 2616 g for 10 min, the supernatant discarded, and washed with hyperosmolar buffer to remove the enzymes. The protoplasts were then suspended in a 5 mL hypertonic buffer. The remaining bacteria count was determined immediately.
For the preparation of tobacco protoplasts, the infiltrated leaves were digested using an enzyme solution containing cellulase and macerozyme R-10 to obtain protoplasts according to the referred protocol (Yoo et al. 2007).
Subcellular localization of TaWZY1-2 in Nicotiana benthamiana
pTF486 is a reformed vector with the CaMV 35S promoter and GFP gene sequence (Wang et al. 2023). The cDNA sequence of Tawzy1-2 with no stop codon, containing the restriction enzyme site Sal I at the 5′ end and the Nco I at the 3′ end, was inserted between the CaMV 35S promoter and the green fluorescence protein (GFP) sequence by the enzymatic digestion and ligation reactions (primers are shown in Table 1) (Fig. 7a). Positive recombinant vectors were transferred into Agrobacterium tumefaciens GV3101 and infiltrated into three fully stretched blades of N. benthamiana of 4-week-old using a needleless syringe. Protoplasts of N. benthamiana were prepared and observed under an inverted confocal microscope (Nikon A1R, Tokyo, Japan). The GFP fusions proteins were excited with a 488 nm argon laser and detected using a 505–530 nm band-pass emission filter, and the mCherry channel was excited using a 561 nm laser and detected using a custom-made 595–620 nm band-pass emission filter to examine the chloroplast auto-fluorescence, to ensure the cell state. Experiments were replicated three times.
Stress tolerance assays with E. coli transformants
The ORF of Tawzy1-2 was amplified with the forward primer containing an EcoR I site (underlined), and the reverse primers contained a Hind III site (underlined) (Table 1). The PCR products were digested with EcoR I and Hind III and ligated into the expression vector pET28a (Novagen, Madison, WI, USA), which had been digested with the same enzymes. The recombinant plasmid was transformed into E. coli strain BL21 (DE3) according to Novagen’s protocol.
To investigate the effect of TaWZY1-2 on the viability of E. coli under abiotic stress, the transformants harboring pET28a-Tawzy1-2 or the pET28a vector were subject to the stress treatments described below. They were first transferred to liquid Luria–Bertani (LB) medium containing 50 μg/mL kanamycin and 1.0 mM isopropyl-β-d-thiogalactoside (IPTG) at 37 °C until an OD600 of 1.0 was achieved. Then, 1 mL culture was inoculated into 50 mL fresh LB medium containing 50 μg/mL kanamycin and 1.0 mM IPTG. Samples were cultured at 37 °C in liquid LB medium containing 800 mM mannitol to simulate osmotic stress or 500 mM NaCl for high-salinity stress or at 8 °C to simulate suboptimal temperature stress and at 45 °C for heat stress. Culture aliquots of 3 mL were removed for measurement of OD600. The experiment was replicated three independent times in triplicate for each sample. Recombinant vector construction primers are shown in Table 1.
Generation of transgenic tobacco plants
The Tawzy1-2 gene was amplified using primers (TG-F, TG-R) containing Xba I and Sac I restriction enzyme sites, as indicated in Table 1. The cDNA was then digested with Xba I and Sac I, purified, and ligated to the linearized pBI121 binary vector (Clontech), which contains the powerful Cauliflower mosaic virus (CaMV) 35S promoter and the kanamycin resistance gene. Afterward, the recombinant plasmid was transferred into Agrobacterium tumefaciens (GV3101) competent cells, and positive strains were utilized to infect tobacco leaves, which were cut into small pieces (2 × 2 cm), leading to the formation of callus through tissue culture on solid MS medium. The transformation of Nicotiania benthamiana was carried out using the leaf disc method (Li et al. 2008). In addition, the resultant buds were placed in MS medium containing 100 mg/L kanamycin to select positive plants. Subsequently, these plants were grown for 2 months, after which they were transferred to soil until the seeds were harvested.
Following this, Tawzy1-2-transgenic T1 seeds were planted on solid MS medium containing 50 μg/mL kanamycin, and selected seedlings were transplanted into soil and grown under a 16 h photoperiod at 25 °C to compile the seeds.
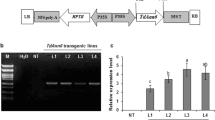
The total RNA was extracted from leaves of the transgenic T2 generation and WT tobacco plants to serve as the first-strand cDNA synthesis template by a reverse transcriptase system (Takara). Semi-qPCR was then performed to assess the expression level of Tawzy1-2, with the β-actin gene (GenBank Accession no. JQ256516.1) of N. benthamiana serving as a loading control, utilizing the primers specified in Table 1.
Evaluation of abiotic stress tolerance and physiological parameters in transgenic tobacco
Three T2 transgenic lines were chosen for abiotic stress-tolerance assays. WT seeds, which were generated at the same time as the transgenic lines, were used as controls. The transgenic seeds and WT seeds were placed on separate areas of solid MS medium with sucrose containing 100 or 200 mM mannitol to simulate osmotic stress, or 100 or 200 mM NaCl to simulate salinity stress. In addition, seeds on normal solid MS medium were incubated at 10 and 45 °C to simulate suboptimal- and high-temperature stress, respectively. After 14 days of growth, the phenotypic differences were analyzed. For the germination rate analysis, the number of germinated seeds on different media was counted daily. The survival rate was evaluated according to the ratio of seedlings surviving at 14 days after stress treatment initiation. Experiments were replicated at least three times for each treatment.
Measurement of H2O2 and ROS contents
The E. coli cell homogenate was obtained by ultrasonication, and H2O2 content was determined using the method reported by Chakrabarty and Datta (2008). For tobacco, samples were ground with 0.1% TCA (w/v), followed by centrifugation at 12,000 g for 15 min. Subsequently, 0.5 mL of the supernatant was combined with 2 mL of 1 M KI and 0.5 mL of 100 mM potassium sulfate buffer, and then kept in the dark for 1 h. The absorption value of the E. coli and tobacco samples was determined at 390 nm, with each detection being repeated more than three times. To determine ROS production in E. coli containing pET28a or pET28a-Tawzy1-2 vector, and the leaves of both treated and control transgenic N. benthamiana plants, we used the reactive oxygen species assay kit (Yeasen, Shanghai, China). The cell walls of E. coli and tobacco cells were removed by lysozyme and macerozyme R-10, respectively, to facilitate the entry of the fluorescent dye DCFH-DA into the cell. Following the instruction of the ROS kit, the fluorescence signal was detected using a fluorescence microplate reader (Varioskan LUX; Thermo Fisher, Waltham, MA, USA) at the maximum excitation wavelength of 480 nm and the maximum emission wavelength of 525 nm. The intensity of the fluorescence signal is proportional to the content of ROS. In addition, leaves detached from untreated wild-type and transgenic plants were used as mock controls, and the measurements were repeated at least three times.
Antioxidant-related enzyme activity measurement assay
Enzyme activities including ascorbate peroxidase (APX), peroxidase (POD), superoxide dismutase (SOD), and malondialdehyde (MDA) content were measured in 4-week-old transgenic and WT seedlings grown in the presence of osmotic, salinity, and low- or high-temperature stress for 48 h. APX, POD, and SOD were measured according to the methods described by Leclercq et al. (2012). The measurement of MDA was performed using the thiobarbituric acid (TBA) reaction (Yang et al. 2008).
Data analysis
Statistical analysis among stress treatments and controls was performed using t tests. Data shown are means ± SE. Analysis of variance (ANOVA) with a correction for multiple comparisons or t tests was used to assess differences among groups using the GraphPad Prism software (ver. 8.0; GraphPad Software).
Results
Gene structure and conserved motifs of TaWZY1-2
The ORF sequence of Tawzy1-2 is 789-bp encoding a 262-residue protein with a calculated molecular mass of 28 kDa and is rich in glutamic acid (16.41%), lysine (14.12%), alanine (9.16%) and glycine (9.16%) (Figs. 1, 2a). The amino acid sequence analysis of TaWZY1-2 showed that it contained the conversed domain of one serine-rich S-fragment (SSSSSSSSS, residues 81 to 89) (the blue region in Fig. 1) and three lysine-rich K-fragments (KRKKKKGLKEKLQGKLPG, residues 105 to 122, EEEKKGFLEKIKEKLPG, residues 173 to 189, and KEKKGLLGKIMDKLPG, residues 226 to 241) (the orange region in Fig. 1), belonging to the SK3 type dehydrin (Fig. 1). Its Grand Average of Hydrophobicity (GRAVY) is – 1.051, which indicated that TaWZY1-2 is highly hydrophilic with only one hydrophobic region (vertical axis score > 1) according to the analysis result of ProtScale software on its hydrophobic content (Fig. 2b).
Multiple protein sequence alignments revealed that TaWZY1-2 had significant homology with the previously reported bread wheat proteins WCOR410b and WCOR410c, with amino acid sequence similarities of 94% and 97%. The similarity of DHN and LEA proteins in durum wheat reached 89% and 94%, respectively, and the similarity of DHN, DHN8, and PAF93 proteins in barley also reached 89%, 90%, and 90% (Fig. 3).
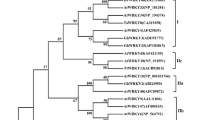
Phylogenetic analysis of TaWZY1-2
The phylogenetic relationship between TaWZY1-2 and members of the dehydrin protein family from different dicotyledonous, monocotyledonous or bryophyte species, was investigated using software MEGA 7.0, NJ (Neighbor-Joining Algorithm) and bootstrap value 1000 were used to construct the phylogenetic tree in different plants (C. intybus, L. sativa, Q. saponaria, E. salsugineum, C. sativus, C. melo, M. charantia, C. pepo, C. moschata, C. maxima, M. prunifolia, M. domestica, M. sylvestris, R. chinensis, A. thaliana, A. suecica, A. lyrata, C. rubella, C. sinensis, A. annua, Z. mays, S. italica, S. bicolor, S. purpurea, H. vulgare, T. turgidum, T. aestivum, T. dicoccoides, T. urartu, L. rigidum, L. perenne, O. sativa, B. distachyon, R. pubera, C. nucifera, H. orientalis, Z. officinale, A. shenzhenica, P. patens). As the results show in Fig. 4, members of this family are widely distributed in different spermatophyte, ranging from bryophyte to angiosperms. The phylogenetic tree is organized into three subfamilies. At the base of the tree, the first subfamily consists of bryophytes, exhibiting a relatively primitive evolutionary degree from a topological standpoint. The second subfamily, representing monocotyledonous plants, clusters together on a main branch, suggesting closely related species sequence, such as two wheat sequences located adjacent to each other. Furthermore, various species are positioned on different sub-branches within this group. The third main branch is comprised of dicotyledonous plants, which are further divided into two sub-branches. The first sub-branch is typified by Arabidopsis thaliana, while the second sub-branch is represented by pumpkin.
Phylogenetic tree of dehydrin TaWZY1-2 among the dicotyledoneae (C. intybus, L. sativa, Q. saponaria, E. salsugineum, C. sativus, C. melo, M. charantia, C. pepo, C. moschata, C. maxima, M. prunifolia, M. domestica, M. sylvestris, R. chinensis, A. thaliana, A. suecica, A. lyrata, C. rubella, C. sinensis, A. annua), monocotyledoneae (Z. mays, S. italica, S. bicolor, S. purpurea, H. vulgare, T. turgidum, T. aestivum, T. dicoccoides, T. urartu, L. rigidum, L. perenne, O. sativa, B. distachyon, R. pubera, C. nucifera, H. orientalis, Z. officinale, A. shenzhenica), and bryophytes (P. patens) based on the full-length protein sequences using the neighbor-joining method. Different groups are indicated by different colors
Expression patterns of Tawzy1-2 in wheat tissues and under various stress conditions
To investigate the expression pattern of Tawzy1-2 in different tissues, total RNA was extracted from wheat roots, stems, leaves, and seeds without any stress treatment and utilized as templates for real-time quantitative PCR. Tawzy1-2 transcripts were found to be most abundant in wheat seeds and roots, followed by stems and leaves, as illustrated in Fig. 5. These results clearly indicate that Tawzy1-2 is preferentially expressed in seeds.
Subsequently, the transcript levels in wheat seedlings were analyzed using real-time quantitative PCR assay to assess the involvement of Tawzy1-2 in abiotic stress responses such as drought, cold, heat, and salinity. The results displayed a steady increase in the expression level of Tawzy1-2 in wheat seedlings treated with 20% (W/V) PEG6000, peaking at 60 h and reaching almost ninefold higher expression compared to the unstressed condition (Fig. 6a). Similarly, under salinity stress, Tawzy1-2 exhibited a slow increase in expression within the first 36 h, followed by a larger upregulation after 48 h, reaching its peak at 60 h (Fig. 6b). The response to high-temperature stress mirrored that under salinity stress during the initial 24 h, and then showed significant upregulation at 36 h, followed by a decrease at 48 h, before reaching fourfold higher expression at 60–72 h (Fig. 6c). In contrast, under cold stress, Tawzy1-2 displayed a rapid increase in expression, reaching a peak at 36 h, with a 14-fold increase from the initial level (Fig. 6d). These data collectively indicate that Tawzy1-2 transcript levels were effectively induced by drought, salinity, and extreme temperature stress conditions.
Transcript level of wheat Tawzy1-2 in response to different abiotic stress conditions. Expression pattern of the Tawzy1-2 gene upon exposure of wheat seedlings to a 20% (w/v) PEG 6000, b 500 mM NaCl, c high temperature (45 °C) and d low temperature (4 °C). The elongation factor gene was employed as an internal control. Data are means ± SD from three independent experiments (n = 3; biological replicates)
Subcellular localization of TaWZY1-2
The subcellular localization of TaWZY1-2 was determined by constructing a recombinant vector with GFP protein fused at the C-terminal and expressing it in tobacco leaves. Upon excitation with 488 nm light from a confocal scanning laser microscope, the green fluorescence of the TaWZY1-2-GFP fusion protein was observed to localize in the nucleus. Conversely, in control cells containing only GFP protein, the green fluorescence signal exhibited diffuse distribution throughout the entire cell without distinct organelle localization (Fig. 7b).
Functional analysis of TaWZY1-2 on E. coli viability under stress conditions
To assess the function of TaWZY1-2 in prokaryotic cells under abiotic stress conditions, E. coli BL21 (DE3) strains overexpressing TaWZY1-2 protein and pET28a empty vector were cultured under five different culture conditions (37, 45, and 8 °C, with 800 mM mannitol or 500 mM NaCl) simulating heat, cold, drought, and salinity stress, respectively. BL21/pET28a and BL21/TaWZY1-2 strains cultured at 37 °C were used as controls.
Growth curves were generated for BL21/pET28a and BL21/TaWZY1-2 exposed to abiotic stress conditions. Under optimum growth conditions (Fig. 8a), both BL21/pET28a and BL21/TaWZY1-2 showed similar growth trends, reaching a final OD600 value of 2.2 after 24 h. However, under osmotic, high salinity, and extreme temperature stresses, their growth curves diverged, with BL21/TaWZY1-2 displaying better growth tendencies than the BL21/pET28a group. Particularly, under salt stress, the OD600 value for BL21/TaWZY1-2 reached 1.0 at 14 h, compared to 0.36 for BL21/pET28a, indicating significant growth advantage for BL21/TaWZY1-2 (Fig. 8c). Furthermore, BL21/TaWZY1-2 exhibited greater resistance to extreme temperatures than BL21/pET28a. At 8 °C, BL21/pET28a and BL21/TaWZY1-2 both exhibited growth signs after 60 h, with OD600 values reaching 1.6 and 1.8, respectively, after 348 h. Despite the extremely low temperature, BL21/TaWZY1-2 displayed superior growth momentum compared to BL21/pET28a (Fig. 8d). The difference between BL21/pET28a and BL21/TaWZY1-2 under high-temperature stress was weaker, and the low-temperature stress had more impact on the growth of E. coli than high-temperature stress (Fig. 8d and e). Overall, overexpression of TaWZY1-2 enhanced the tolerance of E. coli strains to osmotic, salinity, and extreme temperature stress conditions.
Growth curves of E. coli cultures transformed with TaWZY1-2 or control pET28a exposed to the indicated abiotic stress conditions. E. coli strains grown under standard culture conditions (a), in medium supplemented with 800 mM mannitol (b) or 500 mM NaCl (c), and at 8 °C (d) or 45 °C (e). OD600 was measured as an indicator of cell density. Each stress assay was performed three times and the significance of differences was analyzed using Student’s t test (*P < 0.05 or **P < 0.01)
In addition to the differences in growth curves, the intracellular H2O2 and ROS contents of BL21/TaWZY1-2 and BL21/pET28a were also investigated. The H2O2 and ROS contents of logarithmic strains under different culture conditions were determined, and the results indicated that the contents of H2O2 and ROS in BL21/TaWZY1-2 cells were lower than those in the control group under different culture conditions (Fig. 9).
Analysis of H2O2 and ROS accumulation in E. coli transformant strains of BL21/TaWZY1-2 and BL21/pET28a in response to abiotic stresses. H2O2 (a) and ROS (b) contents in BL21/TaWZY1-2 and BL21/pET28a under abiotic stresses. Each reaction was repeated three times. Statistically significant differences were analyzed using Student’s t test (*P < 0.05 or **P < 0.01)
Tawzy1-2 overexpression enhances abiotic stress tolerance in N. benthamiana
Selected for stress assays were three transgenic lines with high Tawzy1-2 expression levels: OE1, OE2, and OE3. When subjected to drought, salinity, or extreme temperature stress conditions, these transgenic tobacco seedlings displayed different phenotypes compared to their sibling WT counterparts. No distinction was observed in control seedlings grown under normal conditions; however, the growth advantage of the transgenic strains was significantly enhanced when exposed to drought or salinity stress. Notably, a higher proportion of transgenic seeds germinated, indicating a stronger growth capacity compared to the few WT seeds that germinated. Similarly, under 200 mM NaCl stress, the growth of both WT and transgenic seedlings was substantially suppressed, yet 11 transgenic seeds surpassed the germination of only 6 out of 30 WT seeds (Fig. 10b and e). Under suboptimal temperature, all four strains exhibited limited growth, yet the transgenic plants outperformed the WT, with only slight phenotypic differences observed between the two under high-temperature stress (Fig. 11a, b, and d).
Drought and salinity tolerance of wild type (WT) and Tawzy1-2-overexpressing N. benthamiana plants. a Semi-quantitative PCR for identification of transgenic tobacco. b The phenotype of WT and overexpression (OE) lines grown on MS medium containing the indicated concentrations of mannitol and NaCl. c, d Root length of WT and OE lines on MS medium under normal conditions or in the presence of 200 mM mannitol or 200 mM NaCl. e Germination rate and f survival rate of WT and OE lines cultured on MS medium under normal conditions or in the presence of 200 mM mannitol or 200 mM NaCl. The significance of differences was determined by one-way ANOVA (*P < 0.05 or **P < 0.01)
Cold and heat tolerance of WT and Tawzy1-2-overexpressing N. benthamiana plants. a Phenotype of the WT and OE lines on MS medium under low- and high-temperature treatments. b, c Root length of WT and OE lines on MS medium under normal conditions, or at 10 or 45 °C. d Germination rate and e survival rate of WT and OE lines on MS medium under normal conditions, or at 10 or 45 °C. The significance of all differences was determined by one-way ANOVA (*P < 0.05 or **P < 0.01)
Under normal conditions, seedling root length was similar in the four strains. However, following exposure to abiotic stress, the average root length of transgenic tobacco seedlings was notably greater than that of WT seedlings, particularly under higher levels of osmotic and salinity stress (Figs. 10c, d and 11b, c). In the absence of abiotic stress, the four groups of experimental tobacco seeds exhibited a similar survival rate. However, following exposure to drought, salinity, suboptimal temperature, or high-temperature stress, the survival rate of transgenic seeds was significantly greater than that of WT seeds, with the difference being somewhat less pronounced under high-temperature stress (Fig. 11d and e).
Determination of antioxidant-related physiological parameter
To investigate whether Tawzy1-2 expression conferred an elevated tolerance to oxidative stress in the transgenic plants, detached leaves from the wild-type and the transgenic plants subjected to NaCl, drought, and extreme temperatures were used to measure the accumulation of H2O2 and ROS contents. It is widely recognized that excess intracellular ROS is detrimental to cells, and environmental stress is a known pathway of intracellular ROS accumulation (Gechev et al. 2006). Results showed that under normal growth conditions, the H2O2 and ROS contents of the wild-type and the transgenic plants were similar. However, following treatment with NaCl, drought and extreme temperatures, the contents of H2O2 and ROS were significantly lower in the transgenic plants compared to the wild-type plants, indicating a considerable decrease in the level of ROS (Fig. 12). These data suggest that overexpression of TaWZY1-2 either inhibits ROS production or enhances the efficient scavenging of excess ROS, leading to lower levels of ROS production in the transgenic plants.
Analysis of H2O2 and ROS accumulation in WT and OE plants in response to abiotic stresses. H2O2 (a) and ROS (b) content in WT and OE plants under abiotic stresses. Each reaction was repeated three times. Statistically significant differences were analyzed using Student’s t test (*P < 0.05 or **P < 0.01)
Furthermore, the activities of APX, POD, and SOD, as well as MDA content in Tawzy1-2 transgenic and wild-type (WT) seedlings, were determined following exposure to osmotic, salinity, or extreme temperature stress. The results indicated that all three enzyme activities showed varying degrees of improvement in transgenic tobacco compared to the WT, with the activity of APX being notably increased. In addition, the MDA content in transgenic plants was significantly lower than in WT plants, further demonstrating the enhanced tolerance of the transgenic plants to oxidative stress (Fig. 13).
Antioxidant enzyme activities and MDA content in WT and Tawzy1-2-overexpressing plants upon exposure to stress. The activity of APX (a), POD (b) and SOD (c), and MDA content (d) in WT and OE transgenic lines under osmotic, salinity, and low and high-temperature stress. Experiments were repeated three times with 10 repetitions for each treatment group. Statistical analysis was done with one-way ANOVA (*P < 0.05, **P < 0.001)
Discussion
Abiotic stress, such as drought, salinity, and extreme temperature, poses significant challenges to the growth and development of plants in natural environments (Munns and Millar 2023). One of the proteins that plays a crucial role in mitigating these challenges is dehydrin. TaWZY1-2, as a member of the dehydrin protein family, is anticipated to be involved in plant stress tolerance reactions due to the presence of various stress-related response elements in its promoter sequence (Zhu et al. 2014). The isolation of Tawzy1-2 from Triticum aestivum cultivar Zhengyin 1 revealed its substantial expression in wheat seeds, consistent with its role as a highly expressed protein in late embryonic development. In addition, Tawzy1-2 was found to be induced under various abiotic stress conditions, such as drought, high salinity, and extreme temperatures, confirming the earlier predictions regarding its promoter sequence. Therefore, these findings underscore the importance of TaWZY1-2 in the plant’s response to abiotic stress and highlight its potential as a target for further investigation into plant stress tolerance mechanisms.
The multifunctional role of dehydrin is evident not only in responding to various abiotic stressors, but also in the diverse biological functions linked to stress resistance across prokaryotic and eukaryotic cells (Hoi et al. 2012; Pochon et al. 2013; Becker et al. 2020; Zhang et al. 2022). To investigate the specific in vivo biological functions of TaWZY1-2, TaWZY1-2 overexpressing E. coli and Tawzy1-2-transgenic tobacco were generated. Subsequently, a range of physiological indicators including E. coli growth curve, tobacco phenotype, root length, germination rate, and antioxidant-related enzyme activity were measured. It was thereby confirmed that the overexpression of TaWZY1-2 significantly enhanced the vitality of E. coli and tobacco under abiotic stress conditions.
Plants produce reactive oxygen species (ROS) as a byproduct of aerobic metabolism, and to maintain cellular equilibrium, it is crucial to regulate the production and elimination of ROS. Adverse environmental conditions can lead to an excessive accumulation of ROS, resulting in cellular damage (Ahmad et al. 2008). Nonetheless, plants possess enzymatic antioxidant defense systems to protect cells from oxidative damage by scavenging ROS (Alscher et al. 1997). Some dehydrins have been found to mitigate the accumulation of ROS and the resultant damage induced by ROS-promoting treatments (Kumar et al. 2014; Halder et al. 2018). Furthermore, the scavenging ability of plants is evidenced by the higher activity of antioxidant enzymes (APX, POD, and SOD) and lower ROS content in Tawzy1-2-transgenic tobacco as compared to the wild type. This confirmation directly verifies the role of TaWZY1-2 in aiding plants to eliminate intracellular free radicals under unfavorable conditions. This scavenging ability is also observed in prokaryotic cells. Notably, lipid degradation frequently occurs when plants are under stress (Goggin et al. 2022). Malondialdehyde (MDA), the final product of lipid oxidative decomposition, serves as an important physiological indicator of membrane lipid peroxidation and the plant’s response to stress conditions (Liang et al. 2023). The lower content of MDA in Tawzy1-2-transgenic tobacco demonstrates the function of TaWZY1-2 in reducing lipid degradation under abiotic stress conditions.
In the context of plant stress, TaWZY1-2 has been identified as a potential enhancer of the antioxidant capacity of plants. The crucial question, however, pertains to the mechanism through which TaWZY1-2 is involved in the regulation of intracellular ROS clearance and lipid metabolism. Considering its nuclear localization and the propensity to form homodimers (Wang et al. 2020), it is plausible that TaWZY1-2 could interact with nucleic acids, partake in gene regulation, and influence ROS clearance and gene expression in lipid metabolism-related signaling pathways to carry out its biological function. This is supported by the findings of related studies, wherein homologous proteins, such as PtrDHN-3 from Populus trichocarpa and CaDHN4 and CaDHN5, have been shown to modulate the enzyme activity of SOD and POD, as well as impact the expression of corresponding genes when manipulated in Arabidopsis thaliana under stress conditions (Zhou et al. 2022). The knock-out of CaDHN4 and CaDHN5 resulted in inhibited expression of manganese superoxide dismutase (MnSOD) and peroxidase (POD) genes, leading to severe wilting and ROS accumulation in the leaves of Arabidopsis, elucidating the involvement of DHNs in the regulation of antioxidant-related gene expression to counteract peroxidation induced by stress (Zhang et al. 2019; Luo et al. 2019). Ongoing research endeavors are dedicated to delving into the intricacies of the underlying mechanisms and signaling pathways responsible for the biochemical functions of TaWZY1-2.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- APX:
-
Ascorbate peroxidase
- DHN:
-
Dehydrin
- GFP:
-
Green fluorescence protein
- LEA:
-
Late embryogenesis abundant
- MDA:
-
Malondialdehyde
- OE:
-
Overexpressing
- ORF:
-
Open reading frame
- POD:
-
Peroxidase
- ROS:
-
Reactive oxygen species
- SOD:
-
Superoxide dismutase
References
Ahmad P, Sarwat M, Sharma SJ (2008) Reactive oxygen species, antioxidants and signaling in plants. J Plant Biol 51(3):167–173. https://doi.org/10.1007/BF03030694
Alscher RG, Donahue JL, Cramer CL (1997) Reactive oxygen species and antioxidants: relationships in green cells. Physiol Plant 100(2):224–233. https://doi.org/10.1111/j.1399-3054.1997.tb04778.x
Bailey-Serres J, Parker JE, Ainsworth EA, Oldroyd GED, Schroeder JI (2019) Genetic strategies for improving crop yields. Nature 575(7781):109–118. https://doi.org/10.1038/s41586-019-1679-0
Becker B, Feng XH, Yin YB, Holzinger A (2020) Desiccation tolerance in streptophyte algae and the algae to land plant transition: evolution of LEA and MIP protein families within the Viridiplantae. J Exp Bot 71(11):3270–3278. https://doi.org/10.1093/jxb/eraa105
Boddington KF, Graether SP (2019) Binding of a Vitis riparia dehydrin to DNA. Plant Sci 287:110172. https://doi.org/10.1016/j.plantsci.2019.110172
Bravo LA, Gallardo J, Navarrete A, Olave N, Martínez J, Alberdi M, Close TJ, Corcuera LJ (2003) Cryoprotective activity of a cold-induced dehydrin purified from barley. Physiol Plant 118(2):262–269. https://doi.org/10.1034/j.1399-3054.2003.00060.x
Chakrabarty D, Datta SK (2008) Micropropagation of gerbera: lipid peroxidation and antioxidant enzyme activities during acclimatization process. Acta Physiol Plant 30(3):325–331. https://doi.org/10.1007/s11738-007-0125-3
Clarke MW, Boddington KF, Warnica JM, Atkinson J, McKenna S, Madge J, Barker CH, Graether SP (2015) Structural and functional insights into the cryoprotection of membranes by the intrinsically disordered dehydrins. J Biol Chem 290(45):26900–26913. https://doi.org/10.1074/jbc.M115.678219
Cleal CJ, Pardoe HS, Stolle E (2021) Plant evolution, floral diversity and the response of plants to environmental stress in deep time. Palaeogeogr Palaeocl 584(8):110674–110675. https://doi.org/10.1016/j.palaeo.2021.110674
Del Rio LA (2015) ROS and RNS in plant physiology: an overview. J Exp Bot 66(10):2827–2837. https://doi.org/10.1093/jxb/erv099
Eriksson SK, Kutzer M, Procek J, Grobner G, Harryson P (2011) Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell 23(6):2391–2404. https://doi.org/10.1105/tpc.111.085183
Gechev TS, Breusegem FV, Stone JM, Denev I, Laloi C (2006) Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays 28(11):1091–1101. https://doi.org/10.1002/bies.20493
Goday A, Jensen AB, Culianez M, Alba MM, Figueras M, Serratosa J, Torrent M, Pages M (1994) The maize abscisic acid-responsive protein Rab17 is located in the nucleus and interacts with nuclear localization signals. Plant Cell 6(3):351–360. https://doi.org/10.2307/3869755
Goggin FL, Shah J, Gillaspy G (2022) Editorial: Lipid metabolism and membrane structure in plant biotic interactions. Front Plant Sci 13:1096268. https://doi.org/10.3389/fpls.2022.1096268
Graether SP, Boddington KF (2014) Disorder and function: a review of the dehydrin protein family. Front Plant Sci 5:00576. https://doi.org/10.3389/fpls.2014.00576
Gupta RK (2011) Food security, genetically modified crops and environment. In: International Conference on Environmental Science and Development, Mumbai, India
Halder T, Agarwal T, Ray S (2016) Isolation, cloning, and characterization of a novel Sorghum dehydrin (SbDhn2) protein. Protoplasma 253(6):1475–1488. https://doi.org/10.1007/s00709-015-0901-7
Halder T, Upadhyaya G, Basak C, Das A, Chakraborty C, Ray S (2018) Dehydrins impart protection against oxidative stress in transgenic tobacco plants. Front Plant Sci 9:00136. https://doi.org/10.3389/fpls.2018.00136
Hara M, Shinoda Y, Tanaka Y, Kuboi T (2009) DNA binding of citrus dehydrin promoted by zinc ion. Plant Cell Environ 32(5):532–541. https://doi.org/10.1111/j.1365-3040.2009.01947.x
Hoi JWS, Beau R, Latge JP (2012) A novel dehydrin-like protein from Aspergillus fumigatus regulates freezing tolerance. Fungal Genet Biol 49(3):210–216. https://doi.org/10.1016/j.fgb.2012.01.005
Houde M, Daniel C, Lachapelle M, Allard F, Laliberté S, Sarhan F (1995) Immunolocalization of freezing-tolerance-associated proteins in the cytoplasm and nucleoplasm of wheat crown tissues. Plant J 8(4):583–593. https://doi.org/10.1046/j.1365-313X.1995.8040583.x
Kovacs D, Kalmar E, Torok Z, Tompa P (2008) Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol 147(1):381–390. https://doi.org/10.1104/pp.108.118208
Kumar M, Lee SC, Kim JY, Kim SJ, Aye S, Kim SR (2014) Over-expression of dehydrin gene, OsDhn1, improves drought and salt stress tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). J Plant Biol 57(6):383–393. https://doi.org/10.1007/s12374-014-0487-1
Leclercq J, Martin F, Sanier C, Clement-Vidal A, Fabre D, Oliver G, Lardet L, Ayar A, Peyramard M, Montoro P (2012) Over-expression of a cytosolic isoform of the HbCuZnSOD gene in Hevea brasiliensis changes its response to a water deficit. Plant Mol Biol 80(3):255–272. https://doi.org/10.1007/s11103-012-9942-x
Li D, Shen J, Wu T, Xu Y, Zong X, Shu H (2008) Overexpression of the apple alcohol acyltransferase gene alters the profile of volatile blends in transgenic tobacco leaves. Physiol Plant 134(3):394–402. https://doi.org/10.1111/j.1399-3054.2008.01152.x
Liang Y, Huang Y, Liu C, Chen K, Li M (2023) Functions and interaction of plant lipid signalling under abiotic stresses. Plant Biol (stuttg) 25(3):361–378. https://doi.org/10.1111/plb.13507
Liu Y, Song Q, Li D, Yang X, Li D (2017) Multifunctional role of plant dehydrins in response to environmental stresses. Front Plant Sci 8:01018. https://doi.org/10.3389/fpls.2017.01018
Liu Y, Li D, Song Q, Zhang T, Li D, Yang X (2019) The maize late embryogenesis abundant protein ZmDHN13 positively regulates copper tolerance in transgenic yeast and tobacco. Crop J 7(3):403–410. https://doi.org/10.1016/j.cj.2018.09.001
Luo D, Hou X, Zhang Y, Meng Y, Zhang H, Liu S, Wang X, Chen R (2019) CaDHN5, a dehydrin gene from pepper, plays an important role in salt and osmotic stress responses. Int J Mol Sci 20(8):1989. https://doi.org/10.3390/ijms20081989
Meng Y, Zhang H, Pan X, Chen N, Chen R (2021) CaDHN3, a pepper (Capsicum annuum L.) dehydrin gene enhances the tolerance against salt and drought stresses by reducing ROS accumulation. Int J Mol Sci 22(6):3205. https://doi.org/10.3390/ijms22063205
Munns R, Millar AH (2023) Seven plant capacities to adapt to abiotic stress. J Exp Bot 74(15):4308–4323. https://doi.org/10.1093/jxb/erad179
Pochon S, Simoneau P, Pigne S, Balidas S, Bataille-Simoneau N, Campion C, Jaspard E, Calmes B, Hamon B, Berruyer R, Juchaux M, Guillemette T (2013) Dehydrin-like proteins in the necrotrophic fungus Alternaria brassicicola have a role in plant pathogenesis and stress response. PLoS ONE 8(10):e75143. https://doi.org/10.1371/journal.pone.0075143
Priya M, Dhanker OP, Siddique KHM, Hanumantharao B, Nair RM, Pandey S, Singh S, Varshney RK, Prasad PVV, Nayyar HJT, Genetics A (2019) Drought and heat stress-related proteins: an update about their functional relevance in imparting stress tolerance in agricultural crops. Theor Appl Genet 132:1607–1638. https://doi.org/10.1007/s00122-019-03331-2
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8(6):e66428. https://doi.org/10.1371/journal.pone.0066428
Reyes JL, Campos F, Wei H, Arora R, Yang Y, Karlson DT, Covarrubias AA (2008) Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ 31(12):1781–1790. https://doi.org/10.1111/j.1365-3040.2008.01879.x
Saibi W, Feki K, Ben Mahmoud R, Brini F (2015) Durum wheat dehydrin (DHN-5) confers salinity tolerance to transgenic Arabidopsis plants through the regulation of proline metabolism and ROS scavenging system. Planta 242(5):1187–1194. https://doi.org/10.1007/s00425-015-2351-z
Smith MA, Graether SP (2022) The disordered dehydrin and its role in plant protection: a biochemical perspective. Biomoleculars 12(2):294. https://doi.org/10.3390/biom12020294
Strimbeck GR (2017) Hiding in plain sight: the F segment and other conserved features of seed plant SKn dehydrins. Planta 245(5):1061–1066. https://doi.org/10.1007/s00425-017-2679-7
Szabala BM (2023) The cationic nature of lysine-rich segments modulates the structural and biochemical properties of wild potato FSK(3) dehydrin. Plant Physiol Biochem 194:480–488. https://doi.org/10.1016/j.plaphy.2022.11.039
Wang X, Yu Z, Liu H, Zhang Y, Bai Z, Zhang L (2020) Effect of K-/S- segments on subcellular localization and dimerization of wheat dehydrin WZY1-2. Plant Signal Behav 15:12. https://doi.org/10.1080/15592324.2020.1827583
Wang X, Liu H, Yu Z, Zhu W, Zhang L, Wang B (2023) Characterization of wheat Wrab18 gene promoter and expression analysis under abiotic stress. Mol Biol Rep 50(7):5777–5789. https://doi.org/10.1007/s11033-023-08485-3
Yang L, Tang R, Zhu J, Liu H, Mueller-Roeber B, Xia H, Zhang H (2008) Enhancement of stress tolerance in transgenic tobacco plants constitutively expressing AtIpk2beta, an inositol polyphosphate 6-/3-kinase from Arabidopsis thaliana. Plant Mol Biol 66(4):329–343. https://doi.org/10.1007/s11103-007-9267-3
Yang W, Zhang L, Lv H, Li H, Zhang Y, Xu Y, Yu J (2015) The K-segments of wheat dehydrin WZY2 are essential for its protective functions under temperature stress. Front Plant Sci 6:00406. https://doi.org/10.3389/fpls.2015.00406
Yoo SD, Cho YH, Sheen J (2007) Mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2(7):1565–1572. https://doi.org/10.1038/nprot.2007.199
Zhang H, Liu S, Ma J, Wang X, Haq S, Meng Y, Zhang Y, Chen R (2019) CaDHN4, a salt and cold stress-responsive dehydrin gene from pepper decreases abscisic acid sensitivity in Arabidopsis. Int J Mol Sci 21:26. https://doi.org/10.3390/ijms21010026
Zhang L, Tian J, Ye L, Liao K, Han J, Wang S, Cao J, Ye Z, Xu J (2022) Transcriptomic analysis reveals the mechanism of low/high temperature resistance in an outstanding diet alga Nannochloropsis oceanica. Aquac Rep 27:101365. https://doi.org/10.1016/j.aqrep.2022.101365
Zhou M, Peng N, Yang C, Wang C (2022) The poplar (Populus trichocarpa) dehydrin gene PtrDHN-3 enhances tolerance to salt stress in Arabidopsis. Plants-Basel 11(20):2700. https://doi.org/10.3390/plants11202700
Zhu W, Zhang D, Lu X, Zhang L, Yu Z, Lv H, Zhang H (2014) Characterisation of an SKn-type dehydrin promoter from wheat and its responsiveness to various abiotic and biotic stresses. Plant Mol Biol Rep 32(3):664–678. https://doi.org/10.1007/s11105-013-0681-1
Acknowledgements
We thank Dr. Zhenqing Bai, and members of the Zhang laboratory for their kind help during the preparation of this manuscript. This work was supported by funding from the National Natural Science Foundation of China (31671608) and the State Key Laboratory of Crop Stress Biology for Arid Areas (CSBA2015007).
Funding
National Natural Science Foundation of China, 31671608, Linsheng Zhang, the State Key Laboratory of Crop Stress Biology for Arid Areas, CSBA2015007, Linsheng Zhang.
Author information
Authors and Affiliations
Contributions
The experimental procedures for this research were carried out by XW, who proposed the ideas and designed the experiments, and HL, who collaborated in performing the laboratory experiments. YL was responsible for the analysis of the data, while LZ conducted the enzyme assays and provided the necessary germplasm resources. Upon completion, XW drafted the manuscript and subsequently revised it, with BW participating in the revision process and also contributing to the transient expression assay. Ultimately, all the authors were involved in reading and approving the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
We declare that we have no significant competing financial, professional, or personal interests that might have influenced the performance or presentation of the work described in this manuscript.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, X., Liu, H., Li, Y. et al. Heterologous overexpression of Tawzy1-2 gene encoding an SK3 dehydrin enhances multiple abiotic stress tolerance in Escherichia coli and Nicotiania benthamiana. Planta 259, 39 (2024). https://doi.org/10.1007/s00425-023-04328-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-023-04328-4