Abstract
In Sicily, small differences exist between wild and cultivated rosemary biotypes; VOCs and genetic profiles may be a useful tool to distinguish them. A germplasm collection of Rosmarinus officinalis L. was harvested from 15 locations in Sicily. Eleven wild and four cultivated populations were collected and, due to the surveyed area covered, they can be considered as a representative panel of Sicilian genetic background of the species. Ex situ plant collection was transferred to the field cultivation in homogeneous conditions for characterizing through a multidisciplinary approach. The study included morphological traits observations (growth habitus, flower color, number and size of leaves, length and number of internodes), VOC profiles using HS-SPME, genome size by flow cytometry analysis, and genetic characterization by means of DNA and nuclear microsatellite (nSSR) investigation. To detect any pattern within- and among-populations variability, all morphological and chemical data were submitted to ANOVA, while clustering and structure population analysis were carried out using genetic profiles. The present work allowed us to distinguish rather well between wild and cultivated genotypes and to underline the biodiversity richness among rosemary Sicilian germplasm, never highlighted, useful for future breeding programs addressed to exploit this important resource.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rosemary (Rosmarinus officinalis L.) is a xeromorphic, evergreen shrub belonging to Lamiaceae, including wild and cultivated forms distributed throughout the Mediterranean area, classified in three subspecies: R. officinalis subsp. officinalis, R. officinalis subsp. palaui (Bolòs and Molinier) Malag., native to Maiorca and Minorca, and R. officinalis subsp. valentinus Ferrer, Guillén and Gómez Nav., recently described in the coastal area around Valencia, in South-Eastern Spain (Ferrer-Gallego et al. 2014). Rosemary is commonly used for culinary and ornamental purposes since ancient times (Mateu-Andrés et al. 2013), and being rich in bioactive compounds, it has many important medicinal and functional properties, ranging from antibacterial to antidiabetic, anti-inflammatory, antitumor and antioxidant (Sánchez-Camargo and Herrero 2017; Andrade et al. 2018). Moreover, rosemary is also a source of natural compounds with allelopathic potential (Alipour and Saharkhiz 2016; Atak et al. 2016) as many other Mediterranean species (Mamoci et al. 2011; Araniti et al. 2013, 2014; Mercati et al. 2019). Three Rosmarinus species grow wild in the Mediterranean area: (1) R. officinalis, widespread throughout the Basin; (2) R. eriocalix Jord. and Fourr., present in the South-Eastern of Spain, Morocco, Algeria and Libya; and (3) R. tomentosus Hub.-Mor. and Maire, native to the coastal area between Granada and Malaga, in Southern Spain. Several hybrids were also found, including Rosmarinus×lavandulaceus De Noé (R. eriocalix×R. officinalis) and R. x mendizabalii Sagredo ex Rosúa (R. officinalis×R. tomentosus) (Rosúa 1981; Morales 2010; Euro+Med 2018). More recently, a new classification included the three species within the genus Salvia, with the denominations Salvia rosmarinus Schleid., Salvia jordanii J.B.Walker, and Salvia granatensis B.T. Drew, respectively (Drew et al. 2017).
In Italy, R. officinalis is the only native plant of the genus (Pignatti 1982), occurring with a variety of growth habits, morphological traits, flower colors, and aromatic features (Nunziata et al. 2019). In Sicily, wild populations of R. officinalis may be found in a specific phytocoenosis (Rosmarinetea officinalis) located in rocky ridges and eroded slopes of carbonate nature mostly along the North-Eastern sea coast, from which they sometimes extend into the inland (Gianguzzi et al. 2015). The interested area is one of the 52 glacial refugia identified within the Mediterranean basin, and, together with Sardinia, Corsica and Balearic Islands, represents one of the 10 regional hotspots of plant biodiversity (Tyrrhenian islands; Médail and Quézel 1999; Médail and Diadema 2009).
The need to favor the safeguard and the crop exploitation of wild Sicilian rosemary is a critical point, due to two major aspects. The first is related to the concrete risk that wild Sicilian populations may be further reduced due to the increased harvesting for domestic self-supply, addressed to food or self-medical purposes. Under ecological balance conditions, the collection from wild or semi-wild populations is usually able to cope with the demand from market, provided it is limited and steady. However, the increase in demand, due to the enhancement of researches that enlarge the exploitation opportunities for the species, often leads to the impossibility to cope with it by means of a simple increase of collection from wild populations. The increasing interest of industry towards wild plants has in some cases contributed to a decline in natural populations, and many species all around the world are presently at risk of extinction. Such depletion model, described in the early 90s (Homma 1992, 1996), has been extensively validated for many spontaneous populations belonging to different species. In such conditions, especially for slowly growing species and in the absence of specialized cultivations, wild populations may severely decline (Lamrani Alaoui and Hassikou 2018). This issue has a great importance for many species native to the rainy forests of Amazonia, but it is also relevant for many Mediterranean plants, since depletion in natural stands was claimed already for some wild population of Spanish Arnica, Gentian, and others (Schippmann et al. 2002). Indeed, an extensive decrease of rosemary wild populations due to the excessive pressure of gathering practices has been already described in Sardinia (Mulas and Mulas 2005), and could become a concrete possibility also in Sicily. A medium-large scale cultivation of the plants that bear a major interest for industrial purposes, such as rosemary, could be an important step to safeguard their natural populations.
The second reason for addressing efforts in the exploitation of Sicilian rosemary germplasm is due to a lack of homogeneity in the marketed material. Even when plant material is supplied by means of nurseries and multiplication centers, limited attention is paid to its genetic characterization with the aim to avoid a large heterogeneity. The lack of genetic knowledge about rosemary germplasm hampers breeding programs for an efficient exploitation of this species.
The available literature offers a great deal of references about rosemary’s morphological variability. Notwithstanding, in contrast to other medicinal and aromatic plants, an official descriptors list for rosemary is not available as far, making it difficult to compare literature data collected from different environments. To date, two different descriptor lists were proposed by the Italian Council for Research in Agriculture (CREA 2013) and the International Union for the Protection of new Varieties of Plants (UPOV 2000). Although they are substantially different in the approach to data measurements and in the importance assigned to each character, both proposals discriminate varieties mainly for ornamental purposes, insofar as the UPOV list sets as reference varieties the two ornamental Barbecue and Blue Lagoon (Hatch 2013).
In addition to morphological and agronomic traits, several efforts were addressed to explore rosemary chemical variability. Based on their essential oil profile, three main chemotypes of rosemary were identified: cineoliferum (with a high occurrence of 1,8-cineole), verbenoniferum (with verbenone > 18%) and camphoripherum (> 20% camphor) (Pintore et al. 2002; Napoli et al. 2010). Many other chemotypes were further defined, but a large part of this variability appeared to be related to harvest season, geographic origin, and climatic pattern (Salido et al. 2003; Zaouali et al. 2005; Varela et al. 2009; Napoli et al. 2010; Jordán et al. 2011). By combining chemical and agro-morphological data from a wild rosemary collection from southern Italy, three biotypes were also classified (De Mastro et al. 2004): (1) long shoots, high number of axillary shoots, small-sized leaves and a high yield of camphor-rich (> 40%) essential oils; (2) medium-sized shoots and leaves, low number of small-sized axillary shoots, low essential oil yield with the predominance of α-pinene/verbenone; and (3) low number of large-sized leaves, a fair number of axillary shoots and quite small shoots, intermediate essential oil yield, with a predominance of α-pinene (> 20%), verbenone, and 1,8-cineole. However, due to the polygenic fashion and the environment effects on many agro-morphological and chemical traits, they cannot be easily used to distinguish closely related samples (Zaouali et al. 2012). Therefore, a more robust and stable characterization of rosemary germplasm might include more reliable plant descriptors and markers, such as floral morphology, genome size and molecular profiles.
Nuclear DNA content showed a key role in systematics and a useful tool in biodiversity estimation (Kellogg 1998; Leitch et al. 2005). Flow cytometry is an effective and fast approach to assess the amount of nuclear DNA and relative genome size in all biological species (Dolezel and Bartos 2005; Dolezel et al. 2007). Genome variation could be an indicator of genetic divergence and speciation process (Murray 2005; Garnatje et al. 2007), highlighting possible molecular mechanisms involved in these processes (Petrov et al. 2000; Bennetzen et al. 2005; Harkess et al. 2016).
Among molecular markers, microsatellites (SSRs—Simple Sequence Repeats) are co-dominant and highly informative markers, abundant and uniformly distributed throughout plant genomes, and broadly used to genotype a wide range of plant species (Carimi et al. 2011; Jiao et al. 2012; Mercati et al. 2015; Fu et al. 2017). Until now, studies on R. officinalis genetic diversity are limited, both for wild germplasm and cultivated varieties. Currently, only few works report the characterization of limited collection using different types of molecular markers, such as Random Amplified Polymorphic DNA (RAPD) (Angioni et al. 2004; Zaouali et al. 2012), nuclear ribosomal sequences (ITS) (Rosselló et al. 2006), allozymes (Zaouali and Boussaid 2008; Zaouali et al. 2012), nuclear (nSSR) and plastidial (cpSSR) Simple Sequence Repeat (Segarra-Moragues and Gleiser 2009; Mateu-Andrés et al. 2013). Preliminary information available about the genetic variability of rosemary in western Mediterranean basin support the hypothesis that this area could be a diversification center of R. officinalis (Mateu-Andrés et al. 2013). More recently, High Resolution Melting (HRM) approach was also proposed as a cost- and time-effective system to characterize rosemary populations (Nunziata et al. 2018, 2019). The system is an alternative method to capillary electrophoresis, providing percentage of HRM curves confidence for each locus, named GCP (genotype confidence percentage), as a direct measure of the genetic similarities, but HRM method is not able to furnish “true” genetic profiles. Indeed, HRM approach assumes that melting curves should be as different as fragments are diverse. As well known, the system shows many sources of error, and GCP, based on a Euclidean and non-genetic distance, is not linearly proportional to similarity of sequences (Hewson et al. 2009; Chagné 2015). As a consequence, many common statistical analyses adopted in population genetics, based on allele frequency, cannot be developed (e.g., expected and observed heterozygosity, fixation index, genetic differentiation, structure analysis etc.). Finally, unlike more common capillary electrophoresis approach and the widespread PCR instruments, easily available in all molecular biology laboratories, the HRM system requires specific qPCR equipment and software.
To our knowledge, a comprehensive characterization of rosemary, including morphological, chemical and genetic analyses is missing. In the present work, a R. officinalis collection, counting wild and cultivated genotypes, representing the whole Sicilian genetic background for this species, has been characterized by means of a multidisciplinary approach. With this purpose, morphological traits and VOCs patterns were evaluated, flow cytofluorimetric analysis was performed, and the entire collection was genotyped by a panel of nuclear SSRs. These are still the most accessible, fast and low-cost system (being able to work in multiplex) currently available. This technique is able to furnish unique and repeatable profiles for each genotype and population, useful also to build a reference dataset in rosemary.
Methods
Arrangement of plants collection and sampling for morphological observations
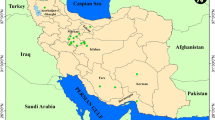
With the aim to cover the lack of knowledge about wild and cultivated rosemary from Sicily, a collection activity started in the 2013 winter season. Vegetative parts of both wild and cultivated plants were collected, mostly growing in the Northern coastal area of Sicily (Fig. 1; Table 1). Since the surveyed area covered most of the basiphilous rocky substrates where native R. officinalis populations may be retrieved (Rosmarinetea officinalis class), the collected samples may be considered representative of the genetic background of R. officinalis from Sicily. To sample a representative collection, according to plant density, almost 3–15 plants for each population were collected. As suggested by Zaouali et al. (2005), since R. officinalis propagates vegetatively, plants were considered different when growing at a distance > 20 m; from each mother plant, 5–10 stem cuttings were picked up and soon inserted into 104-cells polystyrene trays filled with a mixed soil:peat (70:30 v:v) substrate. The trays were constantly surveyed to evaluate the survival and establishment of plants. After plant rooting, they were transplanted into a collection field in the experimental farm “Sparacia” (Department of Agricultural, Food and Forest Sciences, University of Palermo, Cammarata, Agrigento, Italy, 37°38°06″ N; 13°45′47″ E), with the aim to preserve the genetic collection of rosemary. In the field site, both climatic pattern and soil conditions are typical of the Mediterranean dry environments, with 350–600 mm average annual rainfall, mainly distributed throughout the fall-winter period, dry and hot summers, and typically clayey soils. Prior to transplant, 1 t ha−1 organic pelletized fertilizer was spread and buried by soil work; transplant was done arranging plants at a 1 × 1 m distance.
Growth and development of established plants were periodically surveyed. In December 2017, representative samples for each population (1–9 plants each) were harvested (Table 1). Fresh young herbaceous twigs were used for genome size, flow cytometry and morphological traits evaluation, using the most important traits: number of nodes within 10 cm, mean internode length (cm), number of leaves for whorl, average dimensions (length and width in mm) of leaves (Table 2). The same leaf samples were furthermore collected for molecular analysis, directly frozen in liquid nitrogen and then stored at − 80° C until use.
Analysis of VOCs
In late spring 2017, when plants were at a vegetative stasis after blooming, samples from young herbaceous twigs (2–3 for each individual, amounting about 20 g of fresh material) were collected to perform VOCs (volatile organic compounds) analyses. They were identified through the HS-SPME (Head Space-Solid Phase MicroExtraction) coupled with GC–MS. This technique, already successfully used to analyze volatiles in many medicinal and aromatic plants (Carrillo and Tena 2006; Carrubba et al. 2009, 2011; D’Auria and Racioppi 2015; Sgorbini et al. 2015), may allow a quick and effective qualitative screening among individuals based on major VOCs emitted by plants. Since no solvent is required, this procedure may allow reducing the size of sample and its manipulation. The fiber was the 2 cm, 50 μm DVB/CAR/PDMS (divinylbenzene/carboxen/polydimethylsiloxane from Supelco). Before its use, the SPME fiber was conditioned for 2 h at 250 °C in the inlet of a gas-chromatograph.
With this purpose, leaves were separated from the collected twigs and put (approx. 0.5 g for each sample) in a 5 mL vial, immediately sealed with a silicon septum and left for at least 24 h at 25 °C for stabilization and achievement of equilibrium conditions. Thereafter, the SPME fiber was inserted, with the help of a manual holder system, in the silicon septum of the vial. After 30 min at 25 °C, the SPME fiber was recovered and immediately inserted into the injector port of the gas chromatograph allowing for 2 min desorption at 250 °C. Three replicates of each sample were made.
A GC–MS Thermo with autosampler was used for the chromatographic analyses. A capillary column SLB-5MS from Supelco (30 m × 250 μm × 0.25 μm film thickness) was used as stationary phase under the following experimental chromatographic conditions: the injector was in splitless mode with a temperature of 250 °C, helium carrier gas at 1 mL min−1; oven temperature program: 5 min isotherm at 40 °C followed by a linear temperature increase of 4 °C min−1 up to 200 °C held for 2 min. MS scan conditions: source temperature 230 °C, interface temperature 280 °C, EI energy 70 eV, mass scan range 33–350 amu.
The Retention Indexes (R.I.) were experimentally determined relatively to the retention time of a series of n-alkanes (C10–C24) with linear interpolation and they were compared with retention index NIST database on-line (https://webbook.nist.gov/chemistry/name-ser/).
Identification of the individual components was based on comparison of both the retention time and the mass spectrum with those of authentic compounds. Tentatively identification of other components was based on a matching with a score over 90% with mass spectra reported in Wiley7 and NIST05 library. Standards, required to confirm some assignments, were obtained from Merck (Milano, Italy) and used without further purification.
Genome size and flow cytometry evaluation
One hundred mg of fresh leaf tissue was used to determine the ploidy level, while 150 mg of the same tissue were collected to determine DNA content per nucleus, using 50 mg of fresh pea (Pisum sativum L.) leaf tissue as internal standard (2C = 9.07 pg DNA). The legume was chosen from a list of recommended plants as excellent standard for DNA content evaluation (Johnston et al. 1999; Dolezel et al. 2007). To separate nuclei from rosemary cells, leaf tissues were chopped and dispersed into the nuclei extraction buffer (Partec solution CyStain® UV Precise P, 250 tests) added with one drop of Tween 20 and 1% w/v PVP, which was subsequently filtered (30-μm Cell-Trics filter). To reduce mechanical damage, the scalpel blades used for chopping were replaced every three samples. The nuclei were stained in 4,6-diamidino-2-phenylindole (DAPI) staining buffer (Partec Cystain UV precise P). Routinely, 3000–4000 nuclei were measured per sample and histograms of DNA content were generated using Partec software package (Partec-FlowMax®). The 2C DNA content was calculated based on the fluorescence intensity of the G1 peaks of both the internal standard and rosemary samples. The same operator on the same machine, adopting three biological replicates for each sample, performed the analyses.
DNA extraction and microsatellite analysis
Genomic DNA was extracted and purified from leaves (100 mg) using DNeasy Plant Mini Kit (Qiagen, Milan, Italy). Stock solutions of DNA were resuspended in 70 µL Nuclease-free water (Merk Millipore Corporation). DNA quantity and quality were measured using Biophotometer® D30 (Eppendorf, Hamburg, Germany) and stored at − 20 °C. Molecular investigations were carried out by amplifying seven nuclear microsatellites (nSSR) Roff101, Roff135, Roff246, Roff424, Roff438, Roff515 and Roff850, from Segarra-Moragues and Gleiser (2009). PCRs were performed in 20 µl reaction mixture starting from 50 ng DNA as described in Mercati et al. (2013a), using different annealing temperatures (Ta), depending on primer pairs used. The fragments were analyzed on an ABI PRISM 3500 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Data analysis
All quantitative data, including morphological traits and VOCs, were submitted to statistical analysis by means of the statistical package Minitab® v 17.1.0. A preliminary univariate ANOVA by location was carried out, and whenever the ANOVA showed a significant result, mean differences were validated through Tukey’s test. The differences between wild and cultivated populations were detected by calculating a single DF contrast within the factor “locations” (Gomez and Gomez 1984).
The alleles were sized by Gene Mapper v. 4.1 software (Table S1). The main genetic parameters, including the number of alleles per locus (N), number of effective alleles (Ne), major allele frequency (M), observed (Ho) and expected heterozygosity (He), Inbreeding coefficient (F), Polymorphism Information Content (PIC), were evaluated for each SSR used using GenAlEx6 (Peakall and Smouse 2006) and PowerMarker (Liu and Muse 2005) software.
Principal Component Analysis (PCA) of both morphological traits and VOCs was carried out using R/FactoMiner (Le et al. 2008).
A Pearson’s correlation analysis (p < 0.05) was also carried out by Hmisc R/package (https://cran.r-project.org/web/packages/Hmisc/index.html) to confirm PCA results. A scatter plot showing correlation coefficients between traits and their significance was developed by R/Performance Analytic (https://cran.r-project.org/web/packages/PerformanceAnalytics/index.html).
To study the genetic relationships among rosemary populations, cluster analysis based on UPGMA (Unweighted Pair Group Method with Arithmetic Mean) algorithm was performed. The phylogenetic tree was developed by R/poppr (Kamvar et al. 2014) with Bruvo’s distance (Bruvo et al. 2004). The bootstrap analysis was performed based on 1000 re-samplings.
A model-based (Bayesian) clustering was performed to estimate genetic relationship among samples and the population structure by STRUCTURE software (Pritchard and Wen 2003). The program was set as previous reported in Mercati et al. (2013b) and twenty independent runs for K ranging from 1 to 10 were carried out. An ad hoc statistic, proposed by Evanno et al. (2005), was used to determine the most probable K value, to compensate for overestimation of subgroup number by STRUCTURE. Samples with membership probabilities ≥ 0.8 were assigned to the corresponding subgroups and lines with membership < 0.8 were assigned to a mixed subgroup.
Finally, a Discriminant Analysis of Principal Components (DAPC), implemented in the R/adegenet (Jombart and Ahmed 2011), was also carried out to validate and confirm cluster and STRUCTURE results. The number of PCs (principal components) retained was evaluated using the cross-validation approach. To verify the assignment of individuals to clusters, the K-means algorithm, ‘find.clusters’, was used.
Availability of germplasm specimens
The rosemary genotypes used for the trial are available at the germplasm ex situ collection maintained in the experimental farm “Sparacia” (Cammarata, Agrigento, Italy, 37°38°06″ N; 13°45′47″ E). The collection is cured by the Department of Agricultural, Food and Forest Sciences, University of Palermo in compliance with the Regional Sicilian Government Project “Biodiversity preservation—Public Conservation Centers—Safeguard and exploitation of Sicilian herbaceous crop populations and varieties”. PSR Sicilia 2007–2013: Misura 214/2, Azione A. (https://bancagermoplasma.it/psr-misura-2142a/). Specimens are available upon request to the authors.
Results
Morphological traits and volatile organic compounds analysis
Three years after transplanting, many plants showed an erect growth habitus (Table 2). All exhibited a pale violet corolla ground color (except MAR population, whose corolla was mainly light blue). Analysis of variance (ANOVA) highlighted significant differences among populations for only two morphological traits (length of leaves—LL, and number of leaves per whorl—NL), while no significant difference was observed between wild and cultivated plants. The cultivated population named CAS showed the longest leaves, with a mean leaf length of 18.5 mm, whereas the cultivated population PA exhibited the shortest (11.8 mm) leaves arranged in dense whorls (Table 2). The means for each VOC detected by HS-SPME and the related univariate ANOVA are reported in Table 3. Seven volatiles out of twelve showed significant differences among populations; α-pinene showed the largest differences, averaging 20.4% and 40.2% in wild and cultivated populations, respectively. Many compounds that were showing significant differences among populations, also highlighted significant differences between groups (“W vs. C”). By contrast, 1,8-cineole did not show significant differences among populations at univariate ANOVA, but a significant differentiation between wild and cultivated plants was detected by single DF contrast (Table 3). In detail, rather all wild populations exhibited a 1,8-cineole content higher than 40% (on average 46.2%) with an outstanding higher value in plants from L7 population, whereas cultivated plants showed a 30.5% average content of the same compound (Table 3).
PCA on morphological traits did not allow us to define distinct clusters for wild and cultivated populations, although about 70% variability was explained (Figure S1). As a whole, the first axis seemed to be more related with leaves width, whereas the second PC with their length. As expected, mean length of internodes and number of nodes per 10 cm, being inversely correlated, were located on opposite quadrants of the PCA score plot; number of leaves per whorl followed the same trend of number of nodes (Figure S1).
By contrast, although the multivariate analysis on VOCs explained a lower value of total variability (49%), PCA results allowed us to distinguish wild from cultivated rosemary populations (Fig. 2a). Indeed, six out of seven samples, belonging to the cultivated populations, were clearly separated by PCA first component (Dim1). In addition, 62% of samples collected in Torrenova (ME) (all TOR samples, and one plant each belonging to L1 and L2 populations), were separated by the second component (Dim2) from the others. Limonene, α-pinene, and γ-terpinene were most weighing for Dim1 able to separate wild and cultivated populations. Sabinene, camphene, 1,8-cineole and linalool mainly contributed to the variability explained by Dim2 (Fig. 2a). These evidences were confirmed by Pearson’s correlation analysis (Fig. 2b), showing positive and negative significant correlations (p < 0.05). Among these, 1,8-cineole vs. α-pinene and limonene showed the higher (negative) correlation coefficients (Fig. 2b).
a Principal Component Analysis (PCA) referred to main VOCs detected on wild (blue triangles) and cultivated (red circles) populations of R. officinalis. VOCs associated to samples separation were indicated (green arrows) in the plot, underlining their significance values (0.2 < cos2 < 0.8). b Pearson’s correlation matrix of selected VOCs. Positive and negative correlations are displayed in blue and red color, respectively. Size and color intensity are proportional to the correlation coefficients. The significant correlations (p < 0.05) were highlighted
Flow cytometry and genome size evaluation
To evaluate the genome size and ploidy level/genetic stability among accessions, belonging to Sicilian R. officinalis germplasm, flow cytometry approach was used. No significant differences in the ploidy level estimation were detected in our collection. In all plants studied, the genome size recorded was 2C values ± 2.50 pg (1227 Mbp/C) (Figure S2).
Genetic diversity of rosemary Sicilian germplasm
Variation at seven nuclear SSR loci was evaluated on rosemary collection from Sicily. All the loci were polymorphic scoring a high mean PIC value (0.701) with an allele number ranging from 5 to 14 alleles per locus (Table 4) and a mean of major allele frequency of 0.427. Overall, genetic diversity, measured as expected heterozygosity, appeared high (He = 0.731) with an observed heterozygosity (Ho) ranging from 0.511 to 0.956 (Table 4). The inbreeding coefficient (F = − 0.070) was negative, but could be considered in equilibrium.
A phylogenetic tree was defined based on genetic distances, cluster analysis and UPGMA algorithm (Fig. 3). Five main clusters were defined (I, II, III, IV and V), and the accessions were clustered based on their geographic origins (Fig. 3). Interestingly, all cultivated samples were grouped in cluster I, assembled in two private sub-clusters. The remaining four plants, belonging to cluster I, were from AL population. In cluster II, three private sub-clusters were found including all samples from Levanzo (LEV), Cefalù (L7) and two accessions from Castel di Tusa (ME) (L6). Clusters III and IV grouped plants from L3 and L4 populations, respectively. Finally, the largest numbers of samples (42%) were grouped in cluster V, divided into two smaller sub-clusters: the first one included all samples (9) from L5 population, while the second included the samples belonging to L5 population and all the samples from Torrenova (ME) (L1, L2 and TOR populations) and S. Stefano di Camastra (ME).
To infer population structure by determining the number of groups in the germplasm collection, STRUCTURE analysis was performed. Following the Evanno et al. (2005) statistic, K = 7 was identified as the optimum number of genetic groups (K). Using the admixture coefficient (Q) ≥ 0.8 as cutoff of probability to assign each sample to a group identified, 33 out of 45 samples (73%) were assigned to a specific group (Table S2). In detail, all plants collected in Levanzo (LEV population) were assigned to group 1 (pink); L5 and L6 populations belonged to group 4 (orange) and group 5 (light red), respectively; four out of 5 plants from L4 population were assigned to group 6 (dark red); and finally, seven out of 8 plants collected in Torrenova (ME) and STEF population from S. Stefano di Camastra (ME) belonged to group 7 (light blue) (Table S2; Fig. 4). The other samples showed an admixture genetic structure. Although samples from cultivated plants have an admixture profile (blue and green groups), they showed a typical shape, that is very similar to samples belonging to AL population, in agreement to cluster analysis.
Admixture proportions of wild and cultivated plants belonging to Sicilian R. officinalis germplasm. Each vertical bar represents a sample and the color proportion for each bar represents the posterior probability of assignment of each individual to one of seven groups identified. The range of assignment probability varies from 0 to 100%
In the DAPC analysis, cross-validation indicated that seven PCs and five DAs were useful to describe the genetic diversity of rosemary collection. These results agreed with both phylogenetic and STRUCTURE analysis. The samples were clustered based on their origin. In particular samples showing the admixture profiles K2/K3 (all cultivated genotypes and AL wild population; Fig. 4; Table S1), belonging to cluster I (Fig. 3), were separated from the other groups (Fig. 5). Similarly, LEV, L6, and L7 populations, belonging to K1, K5 and K1/K5 (Fig. 4; Table S1), respectively, and grouped in the cluster II (Fig. 3), were more genetically different than the other wild populations (Fig. 5). Finally, although the samples belonging to L1, L2, L4, L5, STEF and TOR showed different genetic pools (Fig. 4; Table S1), they were very closely related (Fig. 5). DAPC analysis allowed us to split the Sicilian germplasm in three main groups, separated in the different quadrants (Fig. 5): group I, represented by cultivated genotypes and AL wild population; group II, contained LEV, L6, and L7 population; and group III with samples belonging to L1, L2, L3, L4, L5, STEF and TOR populations. Interestingly, based on Fst and Nei genetic distance (Nei 1978), the differences between group I and group II were similar to the values obtained comparing group II and III, both represented by wild populations. In addition, group I was closer to group III (Nei = 0.383) than II to III (Nei = 0.628) (Table 5).
Discussion
A significant number of papers were addressed to explore many aspects of morphological, phytochemical and genetic variability of R. officinalis. To our knowledge, few efforts were devoted as far to characterize this species through a multidisciplinary approach.
In Sicily, rosemary is used since ancient times, for both medicinal and food purposes (Lentini and Venza 2007). The main sources for local supply are the collection from wild populations and cultivated individuals. However, most of the traditional rosemary cultivations are represented by single individuals, mostly grown in gardens and orchards in the close surroundings of human settlements, whereas specialized and intensive cultivations are only limited to a few hectares (Migliore and Saggio Scaffidi 2007).
Our results allowed arguing that most of cultivated plants/populations derived from native wild mother plants. Since most of the wild biotypes are widespread in hardly accessible mountainous and steeply sloping areas, it is possible that a number of valuable individuals were brought to cultivation with the purpose to have more easy-to-use available plant material (Burkhart and Jacobson 2009). It seems likely that the choice was concerned mainly with leaves size (the major source of aromatic stuff), and this hypothesis may probably explain the larger size of the leaves in the cultivated individuals, and the extensive homogeneity for this trait of the cultivated populations. Otherwise, since limited interest was paid to other aspects, the other morphological traits, such as the colour of corolla, showed homogeneity across all samples. At the same time, it would be not surprising that some individuals, classified among the “wild” biotypes, would otherwise belong to formerly cultivated (“escaped to cultivation” and naturalized) plants.
Although some distinction could be made at population level based on plant leaves size, morphological traits were not able to achieve a satisfactory discrimination among groups. This lack of discrimination among populations suggests that, once brought to cultivation in homogeneous conditions (hence, once minimized the variability due to the environment), the remaining fluctuations among the major morphological traits are not high enough to discriminate genotypes. Most variations in such traits seem to be due to the environment (as expected), rather than under genetic control. Thus, the perplexity expressed by Zaouali et al. (2012) as concerns the utility of morphological traits for assessing differences among populations sounds reasonable.
The VOC content seems more able to discriminate among populations. Of course, the available data did not allow us to distinguish among chemotypes, whose proper determination in rosemary requires a different experimental procedure (Napoli et al. 2010). Notwithstanding, VOCs obtained by HS-SPME showed a sharp separation among groups of populations, mainly noticeable in the relative content in α-pinene (on average, 40.7% in cultivated biotypes and 20.4 in wild ones) and 1,8-cineole (46.2 in wild biotypes and 30.5 in cultivated ones). Therefore, they can be classified as cineoliferum (or A) chemotype, as reported in previous studies (Li et al. 2016; Nunziata et al. 2019).
Flow cytometry revealed stable genome size in our collection, both in wild and cultivated populations. The genome size recorded (± 2.50 pg) was in agreement to the values available in the literature for the species (Pellicer et al. 2010). However, the procedure adopted in this study could be used as a reference for all species experiencing separation difficulties, including many medicinal plants (Greilhuber et al. 2007). Indeed, this procedure allowed to isolate the nuclei coping with the complexity of the substances contained in rosemary cells.
Microsatellite analysis underlined a suitable and significant biodiversity among Sicilian germplasm. Comparing the genetic variability of our collection to that reported by Segarra-Moragues and Gleiser (2009), the unique available report utilizing nSSR in rosemary, number of alleles per locus, observed and expected heterozygosity agreed. A more recent study based on cpSSR markers identified ten haplotypes among a widespread germplasm collection belonging to whole Mediterranean basin (Mateu-Andrés et al. 2013), but biased towards populations from Spain (23 out of 47). Samples collected from different Italian regions, including plants from Agrigento and Messina (Sicily), belonged to the two most common haplotypes (H2 and H4) and clustered in two main branches, together with Algerian, French, Moroccan and Spanish genotypes (Mateu-Andrés et al. 2013), highlighting a close genetic background. These results were confirmed by Nunziata et al. (2019) using HRM technique. However, due to the limits of this last approach, the genetic background of Sicilian populations included in that study could be partially misclassified. Indeed, genotypes from Torrenova (TOR) and S. Stefano di Camastra (STEF), two very close locations, showed high genetic diversity able to classify these genotypes in different clusters, while STEF population appeared very close to samples belonging to AL population from Vittoria (RG), a location on the other side of Sicily (Nunziata et al. 2019). Our molecular analysis, through “standard” genotyping by SSRs, supported for the first time the evidences of well distinguished genetic profiles belonging, respectively, to wild and cultivated populations. In addition, clustering and the identification of genetic pools (K = 7) are correlated to geographic origins of populations. Therefore, they seem somehow dependent upon the anthropization (disturbance level) of the original collection site. Hence, the AL population, although belonging to the wild collection, lies close to the cultivated groups, probably due to the high level of disturbance of the original AL grown area. DAPC analysis confirmed previous results, highlighting a clear genetic diversity that allowed us to distinguish three main groups in the collection. In particular, group I represented by cultivated genotypes and AL wild population, with K2/K3 admixture profile, showed a major similarity to group II (K1, K5, and the admixture K1/K5) than what emerged from the comparison between the two wild population groups (II and III). To note, within group III (K4, K6, K7, admixture K4/K6 and K5/K7) L3 individuals, collected from a high and hardly accessible calcareous rock, were distinguished from all the other populations. In summary, the genetic analysis underlined an interesting richness of biodiversity among Sicilian germplasm, so far never highlighted, that can be useful to plan future breeding programs to exploit this important resource.
Conclusions
The multidisciplinary approach applied in this work has been able to fully characterize the Sicilian germplasm collection, covering the lack of knowledge about its genome size and stable SSR genetic profiles. Morphological, chemical and genetic observations, offered distinct points of view of rosemary’s diversity; however, taking into account all data together allowed us to depict the relationships among populations that would have not been possible otherwise. The Sicilian rosemary has been confirmed as an important component of plant biodiversity in the Tyrrhenian region, whose conservation has been possible due to the limited and—by far—sustainable use by local populations. The new inputs from R&D sector have, however, opened an impressive series of new opportunities for rosemary utilization, and it is easy to foresee that, as soon as requirements become higher, this equilibrium condition will soon show its weakness. Until now, the local germplasm did not seem to be mixed with genetic material from outside. However, further studies through nSSR genotyping of a wider rosemary germplasm collection will support the preservation that will probably become necessary in a near future.
Author contribution statement
AC designed the project and experiments. AC and MS collected plants, managed collection field, collected and analyzed morphological and chemical data. LA performed and discussed flow cytometry. FS, FM, AM and AL performed DNA extraction and SSR analyses. FS and FM interpreted and discussed genetic analyses. AC, FS and FM performed and discussed statistical analyses, and FM performed multivariate analysis. AC and FM wrote the first draft of the manuscript. All Authors edited and approved the final version of the manuscript.
References
Alipour M, Saharkhiz MJ (2016) Phytotoxic activity and variation in essential oil content and composition of rosemary (Rosmarinus officinalis L.) during different phenological growth stages. Biocatal Agric Biotechnol 7:271–278
Andrade JM, Faustino C, Garcia C, Ladeiras D, Reis CP, Rijo P (2018) Rosmarinus officinalis L: an update review of its phytochemistry and biological activity. Future Sci OA 4:FSO283
Angioni A, Barra A, Cereti E, Barile D, Coisson JD, Arlorio M, Dessi S, Coroneo V, Cabras P (2004) Chemical composition, plant genetic differences, antimicrobial and antifungal activity investigation of the essential oil of Rosmarinus officinalis L. J Agric Food Chem 52:3530–3535
Araniti F, Lupini A, Mercati F, Statti GA, Abenavoli MR (2013) Calamintha nepeta L. (Savi) as source of phytotoxic compounds: bio-guided fractionation in identifying biological active molecules. Acta Physiol Plant 35:1979–1988. https://doi.org/10.1007/s11738-013-1236-7
Araniti F, Marrelli M, Lupini A, Mercati F, Statti GA, Abenavoli MR (2014) Phytotoxic activity of Cachrys pungens Jan, a Mediterranean species: separation, identification and quantification of potential allelochemicals. Acta Physiol Plant 36:1071–1083. https://doi.org/10.1007/s11738-013-1482-8
Atak M, Mavi K, Uremis I (2016) Bio-herbicidal effects of oregano and rosemary essential oils on germination and seedling growth of bread wheat cultivars and weeds. Rom Biotechnol Lett 21:11149–11159
Bennetzen JL, Ma J, Devos KM (2005) Mechanisms of recent genome size variation in flowering plants. Ann Bot 95:127–132
Burkhart EP, Jacobson MG (2009) Transitioning from wild collection to forest cultivation of indigenous medicinal forest plants in eastern North America is constrained by lack of profitability. Agrofor Syst 76:437–453
Bruvo R, Michiels NK, D'Souza TG, Schulenburg H (2004) A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol Ecol 13:2101–2106
Carimi F, Mercati F, De Michele R, Fiore MC, Riccardi P, Sunseri F (2011) Intra-varietal genetic diversity of the grapevine (Vitis vinifera L.) cultivar ‘Nero d'Avola’ as revealed by microsatellite markers. Genet Res Crop Evol 58(7):967–975. https://doi.org/10.1007/s10722-011-9731-4
Carrillo JD, Tena MT (2006) Determination of volatile compounds in antioxidant rosemary extracts by multiple headspace solid-phase microextraction and gas chromatography. Flavour Fragr J 21:626–633. https://doi.org/10.1002/ffj.1630
Carrubba A, Ascolillo V, Pagan Domenech AT, Saiano F, Aiello P (2009) Modifications over time of volatile compounds in Coriander (Coriandrum sativum L). Acta Hort 826:43–49
Carrubba A, Militello M, Saiano F, Pagan Domenech AT (2011) Comparison between different techniques for volatiles analyses in Coriander (Coriandrum sativum L.). Acta Hort 925:151–154
Chagné D (2015) Application of the high-resolution melting technique for gene mapping and SNP detection in plants. Methods Mol Biol 1245:151–159. https://doi.org/10.1007/978-1-4939-1966-6_11
CREA (2013) PlantA-Res. Rete Nazionale delle Risorse Genetiche Vegetali per l'Alimentazione e l'Agricoltura. Piante Aromatiche e Medicinali. Rosmarino. https://planta-res.politicheagricole.it/schede_descr/ROSMARINO.pdf. Accessed 10 Sep 2019
D’Auria M, Racioppi R (2015) The effect of drying on the composition of volatile organic compounds in Rosmarinus officinalis, Laurus nobilis, Salvia officinalis and Thymus serpyllum. A HS-SPME-GC-MS study. J Essent Oil Bear Pl 5:1209–1223. https://doi.org/10.1080/0972060X.2014.895213
De Mastro G, Ruta C, Mincione A, Poiana M (2004) Bio-morphological and chemical characterization of rosemary (Rosmarinus officinalis L.) biotypes. Acta Hort 629:471–482
Dolezel J, Bartos J (2005) Plant DNA flow cytometry and estimation of nuclear genome size. Ann Bot 95:99–110
Dolezel J, Greilhuber J, Suda J (2007) Flow cytometry with plant cells. Analysis of genes, chromosomes and genomes. Wiley, Weinheim
Drew BT, González‐Gallegos JG, Xiang C-L, Kriebel R, Drummond CP, Walker JB, Sytsma KJ (2017) Salvia united: the greatest good for the greatest number. Taxon 66(1):133–145. https://doi.org/10.12705/661.7
Euro+Med (2018) Rosmarinus. https://ww2.bgbm.org/EuroPlusMed/. Accessed 10 Sep 2019
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Ferrer-Gallego P, Ferrer-Gallego R, Roselló R, Peris JB, Guillén A, Gómez J, Laguna E (2014) A new subspecies of Rosmarinus officinalis (Lamiaceae) from the eastern sector of the Iberian Peninsula. Phytotaxa 172:61–70
Fu Y, Yang M, Horbach C, Kessler D, Diederichsen A, You FM, Wang H (2017) Patterns of SSR variation in bread wheat (Triticum aestivum L.) seeds under ex situ gene-bank storage and accelerated ageing. Genet Res Crop Evol 64:277–290. https://doi.org/10.1007/s10722-015-0349-9
Garnatje T, Garcia S, Canela MA (2007) Genome size variation from a phylogenetic perspective in the genus Cheirolophus Cass. (Asteraceae): biogeographic implications. Plant Syst Evol 264:117–134
Gianguzzi L, Papini F, Cusimano D (2015) Phytosociological survey vegetation map of Sicily (Mediterranean region). J Maps. https://doi.org/10.1080/17445647.2015.1094969
Gomez KA, Gomez AA (1984) Statistical procedures for agricultural research. John Wiley and sons Inc, New York
Greilhuber J, Temsch EM, Loureiro JCM (2007) Nuclear DNA content measurement. In: Doležel J, Greilhuber J, Suda J (eds) Flow cytometry with plant cells. Analysis of genes, chromosomes and genomes. Wiley-VCH Verlag, Weinheim, pp 67–101
Harkess A, Mercati F, Abbate L, McKain M, Pires JC, Sala T, Sunseri F, Falavigna A, Leebens-Mack J (2016) Retrotransposon proliferation coincident with the evolution of dioecy in asparagus. G3 Genes Genomes Genet 6(9):2679–2685
Hatch LC (2013) Cultivars of woody plants, 2.0th edn, vol 3. TCR Press, WI, p 114
Hewson K, Noormohammadi AH, Devlin JM, Mardani K, Ignjatovic J (2009) Rapid detection and non-subjective characterisation of infectious bronchitis virus isolates using high-resolution melt curve analysis and a mathematical model. Arch Virol 154(4):649–660. https://doi.org/10.1007/s00705-009-0357-1
Homma AKO (1992) The dynamics of extraction in Amazônia: a historical perspective. Adv Econ Bot 9:23–31
Homma AKO (1996) Utilization of forest products for amazonian development: potential and limitations. In: Lieberei R, Reisdorff C, Machado AD (eds) Interdisciplinary research on the conservation and sustainable use of the amazonian rain forest and its information requirements. Report on the workshop held in Brasilia, Brazil, November 20–22, 1995. Hamburg, Germany
Jiao Y, Jia H, Li X, Chai M, Jia H, Chen Z, Wang G, Chai C, Van de Weg E, Gao Z (2012) Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra). BMC Genom 13:201. https://doi.org/10.1186/1471-2164-13-201
Johnston JS, Bennett MD, Rayburn AL, Galbraith DW, Price HJ (1999) Reference standards for determination of DNA content of plant nuclei. Am J Bot 86:609–613
Jombart T, Ahmed I (2011) Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071
Jordán MJ, Lax V, Martínez C, Aouissat M, Sotomayor JA (2011) Chemical intraspecific variability and chemotype determination of Rosmarinus officinalis L. in the region of Murcia. Acta Hort 925:109–114
Kamvar ZN, Tabima JF, Grünwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. Peer J 2:e281
Kellogg EA (1998) Relationships of cereal crops and other grasses. Proc Natl Acad Sci USA 95:2005–2010
Lamrani Alaoui M, Hassikou R (2018) Rapid risk assessment to harvesting of wild medicinal and aromatic plant species in Morocco for conservation and sustainable management purposes. Biodivers Conserv 27:2729–2745. https://doi.org/10.1007/s10531-018-1565-3
Le S, Josse J, Husson F (2008) FactoMineR: an R package for multivariate analysis. J Stat Softw 25:1–18. https://doi.org/10.18637/jss.v025.i01
Leitch IJ, Soltis DE, Soltis PS, Bennett MD (2005) Evolution of DNA amounts across land plants (Embryophytaz). Ann Bot 95:207–217
Lentini F, Venza F (2007) Wild food plants of popular use in Sicily. J Ethnobiol Ethnomed 3:15. https://doi.org/10.1186/1746-4269-3-15
Li G, Cervelli C, Ruffoni B, Shachter A, Dudai N (2016) Volatile diversity in wild populations of rosemary (Rosmarinus officinalis L.) from the Tyrrhenian Sea vicinity cultivated under homogeneous environmental conditions. Ind Crops Prod 84:381–390. https://doi.org/10.1016/j.indcrop.2016.02.029
Liu K, Muse SV (2005) Powermarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Mamoci E, Cavoski I, Simeone V, Mondelli D, Al-Bitar L, Caboni P (2011) Chemical composition and in vitro activity of plant extracts from Ferula communis and Dittrichia viscosa against postharvest fungi. Molecules 16:2609–2625. https://doi.org/10.3390/molecules16032609
Mateu-Andrés I, Aguilella A, Boisset F, Currás R, Guara M, Laguna E, Marzo A, Puche MF, Pedrola J (2013) Geographical patterns of genetic variation in rosemary (Rosmarinus officinalis) in the Mediterranean basin. Bot J Linn Soc 171:700–712. https://doi.org/10.1111/boj.12017
Médail F, Diadema K (2009) Glacial refugia influence plant diversity patterns in the Mediterranean Basin. J Biogeogr 36:1333–1345. https://doi.org/10.1111/j.1365-2699.2008.02051.x
Médail F, Quézel P (1999) Biodiversity hotspots in the Mediterranean Basin: setting global conservation priorities. Conserv Biol 6:1510–1513
Mercati F, Leone M, Lupini A, Sorgonà A, Bacchi M, Abenavoli MR, Sunseri F (2013a) Genetic diversity and population structure of a common bean (Phaseolus vulgaris L.) collection from Calabria (Italy). Genet Resour Crop Evol 3:839–852. https://doi.org/10.1007/s10722-012-9879-6
Mercati F, Riccardi P, Leebens-Mack J, Abenavoli MR, Falavigna A, Sunseri F (2013b) Single nucleotide polymorphism isolated from a novel EST dataset in garden asparagus (Asparagus officinalis L.). Plant Sci 203–204:115–123
Mercati F, Longo C, Poma D, Araniti F, Lupini A, Mammano MM, Fiore MC, Abenavoli MR, Sunseri F (2015) Genetic variation of an Italian long shelf-life tomato (Solanum lycopersicon L.) collection by using SSR and morphological fruit traits. Genet Resour Crop Evol 62:721–732. https://doi.org/10.1007/s10722-014-0191-5
Mercati F, Fontana I, Gristina AS, Martorana A, El Nagar M, De Michele R, Fici S, Carimi F (2019) Transcriptome analysis and codominant markers development in caper, a drought tolerant orphan crop with medicinal value. Sci Rep 9:2045–2322. https://doi.org/10.1038/s41598-019-46613-x
Migliore G, Saggio Scaffidi C (2007) La filiera delle piante officinali in Sicilia. In: Crescimanno M (ed) Le piante officinali in Sicilia Potenzialità di sviluppo della coltivazione con metodo biologico. Università degli Studi di Palermo Dip. ESAF, Palermo, pp 75–116 (in Italian)
Morales R (2010) Género Rosmarinus L. In: Morales R et al (eds) Flora iberica, Real Jardín Botánico, CSIC, Madrid, pp 327 331
Mulas M, Mulas G (2005) Cultivar selection from rosemary (Rosmarinus officinalis L.) spontaneous populations in the Mediterranean area. Acta Hort 676:127–133
Murray BG (2005) When does intraspecific C-value variation become taxonomically significant? Ann Bot 95:119–125
Napoli EM, Curcuruto G, Ruberto G (2010) Screening of the essential oil composition of wild Sicilian rosemary. Biochem Syst Ecol 4:659–670
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nunziata A, Cervelli C, De Benedetti L (2018) Genotype confidence percentage of SSR HRM profiles as a measure of genetic similarity in Rosmarinus officinalis. Plant Gene 14:64–68
Nunziata A, De Benedetti L, Marchioni I, Cervelli C (2019) High resolution melting profiles of 364 genotypes of Salvia rosmarinus in 16 microsatellite loci. Ecol Evol 9:3728–3739
Peakall R, Smouse PE (2006) GenAlEx6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pellicer J, Estiarte M, Garcia S, Garnatje T, Peñuelas J, Sardans J, Vallès J (2010) Genome size unaffected by moderate changes in climate and phosphorus availability in Mediterranean plants. Afr J Biotech 9(37):6070–6077
Petrov DA, Sangster TA, Johnston JS, Hartl DL, Shaw KL (2000) Evidence for DNA loss as a determinant of genome size. Science 287:1060–1062
Pignatti S (1982) Flora d'Italia, vol II. Edagricole:500, Bologna
Pintore G, Usai M, Bradesi P, Juliano C, Boatto G, Tomi F, Chessa M, Cerri R, Casanova J (2002) Chemical composition and antimicrobial activity of Rosmarinus officinalis L. oils from Sardinia and Corsica. Flavour Fragr J 17:15–19
Pritchard JK, Wen W (2003) Documentation for STRUCTURE software version 2. https://web.stanford.edu/group/pritchardlab/software/readme/readme.html. Accessed 20 Dec 2019
Rosselló JA, Cosín R, Boscaiu M, Vicente O, Martínez I, Soriano P (2006) Intragenomic diversity and phylogenetic systematics of wild rosemaries (Rosmarinus officinalis L. s.l., Lamiaceae) assessed by nuclear ribosomal DNA sequences (ITS). Plant Syst Evol 262:1–12
Rosúa JL (1981) El complejo Rosmarinus eriocalyx-tomentosus en la península ibérica. Anales Jard Bot Madrid 2:587–595
Salido S, Altarejos J, Nogueras M, Sanchez A, Luque P (2003) Chemical composition and seasonal variations of rosemary oil from Southern Spain. J Essent Oil Res 15:10–14
Sánchez-Camargo AdP, Herrero M (2017) Rosemary (Rosmarinus officinalis) as a functional ingredient: recent scientific evidence. Curr Opin Food Sci 14:13–19
Schippmann U, Leaman DJ, Cunningham AB (2002) Impact of cultivation and gathering of Medicinal Plants on biodiversity: global trends and issues. In: FAO, 2002, “Biodiversity and the ecosystem approach in Agriculture, Forestry and Fisheries”, Inter-Departmental Working Group on Biological Diversity for Food and Agriculture, Rome, p 21
Segarra-Moragues JG, Gleiser G (2009) Isolation and characterisation of di and tri nucleotide microsatellite loci in Rosmarinus officinalis (Lamiaceae), using enriched genomic libraries. Conserv Genet 3:571–575
Sgorbini B, Bicchi C, Cagliero C, Cordero C, Liberto E, Rubiolo P (2015) Herbs and spices: characterization and quantitation of biologically-active markers for routine quality control by multiple head space solid-phase microextraction combined with separative or non-separative analysis. J Chromatogr A 1376:9–17
UPOV (2000) Working paper on test guidelines for Rosemary (Rosmarinus officinalis L.). Technical working party for vegetables, thirty-fourth session, Angers, France, September 11–15, 2000. https://www.upov.int/edocs/mdocs/upov/en/twv/34/twv_34_14.pdf Accessed 10 Sep 2019
Varela F, Navarrete P, al R, Fanlo M, Melero R, Sotomayor JA, Jordán MJ, Cabot P, Sánchez de Ron D, Calvo R, Cases A (2009) Variability in the chemical composition of wild Rosmarinus officinalis L. Acta Hort 826:167–174
Zaouali Y, Boussaid M (2008) Isozyme markers and volatiles in Tunisian Rosmarinus officinalis L. (Lamiaceae): a comparative analysis of population structure. Biochem Syst Ecol 36:11–21
Zaouali Y, Chograni H, Trimech R, Boussaid M (2012) Genetic diversity and population structure among Rosmarinus officinalis L. (Lamiaceae) varieties: var. typicus Batt. and var. troglodytorum Maire. based on multiple traits. Ind Crops Prod 38:166–176
Zaouali Y, Messaoud C, Ben Salah A, Boussaïd M (2005) Oil composition variability among populations in relationship with their ecological areas in Tunisian Rosmarinus officinalis L. Flavour Fragr J 20:512–520
Acknowledgements
The collection field was carried out within the Project “Biodiversity preservation—Public Conservation Centers—Safeguard and exploitation of Sicilian herbaceous crops populations and varieties”. PSR Sicilia 2007–2013: Misura 214/2, Azione A.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Carrubba, A., Abbate, L., Sarno, M. et al. Characterization of Sicilian rosemary (Rosmarinus officinalis L.) germplasm through a multidisciplinary approach. Planta 251, 37 (2020). https://doi.org/10.1007/s00425-019-03327-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-019-03327-8