Abstract
Main conclusion
Two sunflower hydroxyacyl-[acyl carrier protein] dehydratases evolved into two different isoenzymes showing distinctive expression levels and kinetics’ efficiencies.
β-Hydroxyacyl-[acyl carrier protein (ACP)]-dehydratase (HAD) is a component of the type II fatty acid synthase complex involved in ‘de novo’ fatty acid biosynthesis in plants. This complex, formed by four intraplastidial proteins, is responsible for the sequential condensation of two-carbon units, leading to 16- and 18-C acyl-ACP. HAD dehydrates 3-hydroxyacyl-ACP generating trans-2-enoyl-ACP. With the aim of a further understanding of fatty acid biosynthesis in sunflower (Helianthus annuus) seeds, two β-hydroxyacyl-[ACP] dehydratase genes have been cloned from developing seeds, HaHAD1 (GenBank HM044767) and HaHAD2 (GenBank GU595454). Genomic DNA gel blot analyses suggest that both are single copy genes. Differences in their expression patterns across plant tissues were detected. Higher levels of HaHAD2 in the initial stages of seed development inferred its key role in seed storage fatty acid synthesis. That HaHAD1 expression levels remained constant across most tissues suggest a housekeeping function. Heterologous expression of these genes in E. coli confirmed both proteins were functional and able to interact with the bacterial complex ‘in vivo’. The large increase of saturated fatty acids in cells expressing HaHAD1 and HaHAD2 supports the idea that these HAD genes are closely related to the E. coli FabZ gene. The proposed three-dimensional models of HaHAD1 and HaHAD2 revealed differences at the entrance to the catalytic tunnel attributable to Phe166/Val1159, respectively. HaHAD1 F166V was generated to study the function of this residue. The ‘in vitro’ enzymatic characterization of the three HAD proteins demonstrated all were active, with the mutant having intermediate K m and V max values to the wild-type proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The first steps of ‘de novo’ fatty acid biosynthesis in plants are localized in plastids and mitochondria, where the multienzymatic complexes: acetyl-CoA carboxylase (ACCase) and fatty acid synthase (FAS) work co-ordinately (Yasuno et al. 2004; Harwood 2005). Plant and bacteria FAS complexes differ from the Type I complex in mammals which is coded for by a single gene. A heterodimer of the latter’s resulting polypeptide synthesizes fatty acids (Smith et al. 2003). In plant plastids and bacteria, the type II FAS complex, generating mostly up to 16- or 18-carbon fatty acids, is an easily dissociable, multisubunit complex consisting of monofunctional enzymes each encoded by a discrete gene (Høj and Mikkelsen 1982; Shimakata and Stumpf 1982a). This complex is formed by four enzymes a β-ketoacyl-[ACP]-synthase (KAS I, II or III), a β-ketoacyl-[ACP]-reductase (KAR), a β-hydroxyacyl-[ACP]-dehydratase (HAD) and an enoyl-[ACP]-reductase (ENR). The enzyme carrying out the third step of each cycle of fatty acid synthesis, HAD, is responsible for converting β-hydroxyacyl-ACP to trans-2-enoyl-ACP, by releasing a water molecule.
Two genes coding for HAD have been identified in Arabidopsis, At2g22230 and At5g10160. Microarray assays showed both genes are highly expressed during lipid biosynthesis in seed development (Schmid et al. 2005). Plant HAD enzymes have been previously purified from other plant tissues such as spinach leaves and safflower seeds (Shimakata and Stumpf 1982a, b), and several putative genes have been identified in public databases but, up to now, only one of the Arabidopsis HAD genes (At2g22230) has been heterologous expressed in Escherichia coli, purified and its activity demonstrated by mass spectrometry (Brown et al. 2009).
Some bacteria such as Escherichia coli, on the other hand, contain two β-hydroxyacyl-[ACP]-dehydratase enzymes: FabA is required for unsaturated fatty acid synthesis and has a dual dehydratase/isomerase function acting primarily on C10 substrates (Heath and Rock 1996), and FabZ with only a dehydratase function that is the principal enzyme involved in fatty acid elongation from C4 to C18 (Mohan et al. 1994). The sequences of FabA and FabZ are remarkably similar, but can be distinguished by an active site Asp in FabA versus a Glu in FabZ (White et al. 2005). Li et al. (2009) have cloned and analysed different Arachis hypogea FAS genes, among them the β-hydroxyacyl-[ACP]-dehydratase. The deduced protein sequence showed similarities with the bacteria E. coli β-hydroxyacyl-[ACP] dehydratase, EcFabZ, maintaining a high level of conservation in the catalytic residues. Such comparisons suggest that A. hypogea HAD has an analogous function to EcFabZ. Similar analyses and conclusions were reached for the Arabidopsis HAD proteins (Brown et al. 2009).
The Pseudomonas aeruginosa, Plasmodium falciparum and Helicobacter pylori FabZ dehydratase structures have been intensely studied (Kimber et al. 2004; Zhang et al. 2008; Maity et al. 2011). Contrariwise, a plant HAD protein remains to be crystallized. The FabZ enzyme belongs to the hot-dog protein family, whose monomer structure consists of six β-pleated sheets surrounding a long α-helix, resulting in an L shape. Initially, an antiparallel dimeric structure is required that is formed by the interaction of the β3-sheet of one monomer with the last two turns of the α3-helix of a second monomer. The dimers then form hexamers. The catalytic residues, histidine and glutamic acid are located at the dimeric interface (White et al. 2005), specifically in the long, narrow hydrophobic tunnel formed upon dimerization. In P. aeruginosa FabZ a highly mobile tyrosine residue, near to the ACP binding domain, blocks the entrance of the catalytic tunnel. When this residue rotates 120º the tunnel opens allowing entrance of the substrate, which is then susceptible to dehydration (Kimber et al. 2004).
Mass spectrophotometric analyses of AtHAD demonstrated the ability of the enzyme to use either crotonyl-ACP or 3-hydroxybutyryl-ACP as substrates (Brown et al. 2009). Two other plant HADs, from spinach and safflower, have been biochemically characterized in plant extracts. Their enzymatic activities, monitored spectrophotometrically using crotonyl-ACP as substrate, showed very similar K m values of 9.7 and 9.0 μM, respectively. The reversible reaction was favored in the ‘in vitro’ assays (Shimakata and Stumpf 1982a, b) as in bacteria, such as P. falciparum. Only when the next enzyme in the FAS cycle, ENR, is included in the experiment, did the direction of the reaction change to that observed ‘in vivo’ (Sharma et al. 2003). In some bacteria and protozoa species, including P. falciparum, H. pylori and Toxoplasma gondii, the readily available substrate crotonyl-CoA has been used instead of crotonyl-ACP for characterization of β-hydroxyacyl-[ACP]-dehydratase activity (Sharma et al. 2003; Liu et al. 2005; Dautu et al. 2008).
In the present study we have characterized two different Helianthus annuus (sunflower) HAD genes and their products with the goal of optimizing the total lipid content in developing seeds. The predicted structural models of these proteins showed some interesting differences between both isoforms. A mutant affecting the opening of the active site pocket in HAD1 was generated to study the effect of the residue occluding the entrance to the FabZ active site tunnel as the homologous residue in the sunflower HAD genes differ. The three proteins, heterologously expressed in E. coli were functional and their kinetic parameters, determined in vitro, question whether this residue has an analogous function in the sunflower enzymes.
Materials and methods
Plant material
Sunflower (Helianthus annuus L.) wild-type line CAS-6 (Sunflower Collection of Instituto de la Grasa, CSIC, Seville, Spain) was grown and materials collected as described by González-Thuillier et al. (2015).
Cloning of cDNAs encoding sunflower HAD
Approximately 0.1 g of each tissue was ground in liquid N2 with precooled sterile mortar and pestle. Total RNA was extracted using a Spectrum Plant Total RNA kit (Sigma-Aldrich, St. Louis, MO, USA). To eliminate residual DNA from total RNA, a DNA-free kit (Ambion, Austin, TX, USA) was used, and mRNA was isolated from total RNA (1 µg) using the GenElute mRNA Miniprep kit (Sigma-Aldrich). The corresponding cDNA was synthesized using a Ready-To-Go T-Primed First Strand Kit (GE Healthcare Life Science, Little Chalfont, UK) from DNA-free mRNA.
The A. thaliana HAD protein sequence encoded by the At2g22230 gene was used to search sunflower expressed sequence tags (ESTs) publicly available in the NCBI database (http://www.ncbi.nlm.nih.gov/) to find putative mRNAs encoding HAD homologs using the TBLASTN algorithm (Altschul et al. 1997). ESTs corresponding to internal sequences of two genes were identified. These ESTs were aligned using the ClustalX v.2.0.10 program (Larkin et al. 2007) to group those with similar or identical sequences. Polymerase chain reaction (PCR) fragments were amplified with two pairs of primers designed from these groups, F-HAD1/R-HAD1 and F-HAD2/R-HAD2 (Table 1; all the primers were synthesized by Eurofins MWG Operon, Ebersberg, Germany). The 5′-ends were obtained using the Smart™-RACE cDNA amplification kit (Clontech, Mountain View, CA USA) and specific reverse internal oligonucleotides pairs for each cloned sequence: R-HAD1.2, R-HAD1.3 and R-HAD1.4; and R-HAD2.2, R-HAD2.3 and R-HAD2.4 (Table 1). The 3′-end of the cDNAs was obtained by PCR using the external oligo FA2Z (Table 1), complementary to the sequences incorporated during the initial cDNA synthesis, and specific internal oligos for each cloned sequence: F-HAD1.2 and F-HAD2.2 (Table 1). Once the ends of the two genes were identified the complete sequences were amplified with the specific pair of primers F-atg_HAD1/R-stop_HAD1 and F-atg_HAD2/R-stop_HAD2 for HaHAD1 and HaHAD2, respectively. The PCR fragments were cloned into the pMBL-T vector (Dominon, North Kingstown, RI, USA), transformed into XL1-Blue, and several clones were sequenced on both strands by Secugen SL (Madrid, SP). The identity of the clones was confirmed using the BLASTX algorithm (Altschul et al. 1997), distinguishing two different gene types coding for HAD isoforms, HaHAD1 and HaHAD2 (684 base pairs (bp) and 666 bp, respectively). These cDNA sequences were deposited in GENBANK under Accession numbers HM044767 |gi:302634223| and GU595454 |gi:291480628|, respectively.
cDNA and protein sequence analyses
Protein sequences homologous to the predicted sequences of HaHAD1 and HaHAD2 were identified using the BLASTP algorithm (Altschul et al. 1997). Alignment of the amino acid sequences, including the transit peptides, for plastid HAD proteins publicly available at GENBANK was performed using the ClustalX v.2.0.10 program with the default settings (Larkin et al. 2007). These entire alignments were used to generate a phylogenetic tree based on the neighbour-joining algorithm (Saitou and Nei 1987), and the resulting ‘phenogram’ was drawn using the MEGA program 4.0.2 (Tamura et al. 2007). Transit peptides were identified through alignment with known plastid HAD sequences and using the network-based program TargetP V1.1 (Emanuelsson et al. 2007).
Modelling of the three-dimensional structures of HaHAD1 and HaHAD2
Homology modelling studies were performed as detailed in González-Mellado et al. (2010). The structure of the P. falciparum β-hydroxyacyl-ACP dehydratase, FabZ, sequence with accession number UniProtKB Q965D7 (Kostrewa et al. 2005) was used as a model for both sunflower sequences, being the most homologous HAD for which X-ray structure information was available in the database RCSB PDB (Berman et al. 2000) (PDB Entry: 1Z6B).
‘In vitro’ site-directed mutagenesis
To induce a transversional substitution, T→G, at position 496 in the HaHAD1 gene, the QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA) was used together with (a) the primers named t496 g_antisense and t496g (Table 1), designed via the Agilent Technologies web page (www.agilent.com/genomics/qcpd), (b) the Kit’s High Fidelity DNA polymerase and (c) the HaHAD1 gene cloned into pQE80L. Following the manufacturer’s protocol, PCR products were transformed into XL1-Blue supercompetent cells and several of the resulting clones subjected to sequencing to confirm the desired mutation, that is, the mutant gene, HaHAD1 F166V.
Genomic DNA Southern blot analysis
Genomic DNA from 2.5 g of sunflower leaves was isolated as described previously (González-Mellado et al. 2010). DNA samples were digested with different restriction enzymes (all from New England Biolabs, Hitchin, UK) and electrophoresed in a 0.8 % agarose gel. The gel was soaked in 250 mM HCl for 30 min, then washed three times in distilled water and finally blotted onto a Hybond-N + transfer membrane (GE Healthcare). The filter was probed with [α-32P]dCTP-labelled HaHAD1 and HaHAD2 gene-specific DNA probes, 684 bp and 666 bp, respectively, obtained by PCR amplification with the following pairs of primers: F-atg_HAD1/R-stop_HAD1 for HaHAD1 and F-atg_HAD2/R-stop_HAD2 for HaHAD2 (Table 1). Hybridization was performed as detailed previously (González-Thuillier et al. 2015). Images of radioactive filters were obtained as described by González-Mellado et al. (2010).
Real time quantitative PCR (RT-qPCR)
The cDNAs obtained from developing seeds and different vegetative tissues as described in González-Thuillier et al. (2015) were subjected to RT-qPCR with the following gene-specific pair of primers: F-HAD1qpcr/R-HAD1qpcr for HaHAD1 and F-HAD2qpcr/R-HAD2qpcr for HaHAD2 (Table 1) using SYBR Green (QuantiteTect™ SYBR® Green PCR Kit, Qiagen, Hilden, DE) in an MiniOpticon system (Bio-Rad, Hercules, CA, USA). The reaction was run, a calibration curve constructed and the sunflower actin gene HaACT1 (GenBank Accession number FJ487620) used as the internal reference as detailed previously (González-Thuillier et al. 2015). Three biological and two technical replicates were carried out on each sample.
Expression of sunflower recombinant HAD proteins in E. coli
The expression vector pQE-80L contains an N-terminal 6× histidine affinity tag to facilitate protein purification. The genes HaHAD1 and HaHAD2 were amplified using the primer pairs F-HAD1SacI/R-HAD1XmaI and F-HAD2BamHI/R-HAD2PstI (Table 1), respectively. The PCR product, corresponding to each gene missing the signal peptide contained the desired restriction sites at each end, SacI/XmaI (HaHAD1) and BamHI/PstI (HaHAD2) for directional cloning into pQE80L after restriction with such enzymes. Constructions pQE-80L::HaHAD1 and pQE-80L::HaHAD2 were transformed into competent E. coli XL1-Blue strain (Stratagene, La Jolla, CA, USA), respectively.
E. coli cells harboring recombinant plasmids were grown under continuous shaking at 37 °C in SB medium (2.5 % tryptone, 1.5 % yeast extract, 0.5 % NaCl, pH 7.5) containing ampicillin (100 µg ml−1). Heterologous expression of these two sunflower genes, as well as the mutant HaHAD1 F166V, was induced at OD 0.8 with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and grown for an additional 2 h. The cells were collected by centrifugation at 4000g for 15 min. The cell pellets were resuspended in 10 ml of 50 mM Tris (pH 8.0), 1 mg ml−1 lysozyme, 0.1 mg ml−1 DNAse, 5 mM DTT and lysed by sonication at 0 °C using a Branson sonicator (Model SLPe; Thomas Scientific, Swedesboro, NJ, USA), applying 10 pulses of 20 s. The lysed bacterial suspension was centrifuged for 40 min at 20,000g. The protein was purified from the resulting supernatant by immobilized metal affinity chromatography (IMAC) and submitted to kinetic characterization as previously described (Moreno-Perez et al. 2011). Protein concentration was measured using a BCA Protein Assay Kit Pierce from Thermo Scientific (Waltham, MA, USA).
Protein gel electrophoresis
Recombinant proteins were submitted to SDS–polyacrylamide gel electrophoresis (PAGE) and the gels stained with Coomassie blue as detailed previously (González-Thuillier et al. 2015).
Growth-rate and fatty acids analysis in E. coli
50 ml cultures of E. coli carrying either pQE80L::HaHAD1 or pQE80L::HaHAD2 were grown for the sunflower proteins’ expression. The OD of the bacterial culture (three replicates per construct) was measured in a spectrophotometer Ultrospec 3300pro (GE Healthcare) at 30 min intervals, as an index of increasing cellular content. Based on this information, the specific growth rates were calculated from the plots generated from the increase in log OD600 with time using the mean of three independent experiments to calculate each value in the growth curves.
Culture cells were harvested by centrifugation (15 min at 3200g), washed and resuspended in water (1/10 from the initial volume) for fatty acid methyl ester analysis. 150 µg of heptadecanoic acid was added to the samples to serve as an internal standard for quantification. Extraction of the lipids, preparation of the methyl esters and their analysis by gas chromatography is described in González-Mellado et al. (2010). Statistical significance of the results was estimated using Student’s t test (SigmaPlot 8.0, SPSS, Chicago, IL, USA), with P < 0.05 considered significant.
In vitro characterization of HaHAD1, HaHAD2 and HaHAD1 F166V
To study their activity, the reverse reaction of these enzymes, which is favored in vitro, dehydration was measured using crotonyl-CoA (Sigma-Aldrich) as substrate and monitoring the decrease in absorbance at 260 nm, similar to the procedure described in Shimakata and Stumpf (1982b) and Dautu et al. (2008) with some modifications. The purified proteins HaHAD1, HaHAD2 or HaHAD1 F166V were added at 5.0 μg ml−1 to reaction buffer (17.9 mM potassium phosphate, 1.6 mM DTT pH 6.8) to give a final volume of 600 μl. The reaction was started by adding the crotonyl-CoA substrate at different concentrations between 20 and 100 μM. The background at 260 nm, decreasing due either to substrate instability or to the E. coli extract’s endogenous activity was also monitored using purified protein extract from induced cultures containing empty expression vector. This background activity was subtracted from sample values. The reaction was carried out at 25 °C and monitored each minute using the spectrophotometer Ultrospec 3300pro (GE Healthcare) and the software Datrys Life Science (2.2.0.0 Version). The extinction coefficient of crotonyl-CoA ε 260 = 22.6 × 103 M−1 cm−1 (Dautu et al. 2008; Shimakata and Stumpf 1982b) was used to convert absorbance measurements to concentration of β-hydroxyacyl-CoA produced from crotonyl-CoA. For each enzymatic assay three independent experiments were done with each of the three purified proteins. Kinetic parameter analysis was carried out as described in González-Thuillier et al. (2015).
Results
Cloning and sequence analysis of two sunflower β-hydroxyacyl-[ACP]-dehydratase cDNAs
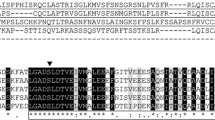
Two groups of ESTs for plastid HAD enzymes in sunflower were identified on the basis of their homology to the Arabidopsis HAD gene (At2g22230). Two cDNAs, consisting of 684 bp for HaHAD1 and 666 bp for HaHAD2, were amplified from 15 days after flowering (DAF) developing seed mRNA (see Materials and methods). The amino acid sequences, deduced from the cDNA of HaHAD1 and HaHAD2 genes using bioinformatics tools, consisted of 227 and 221 residues, respectively. Through alignment with known HAD sequences and using the network-based method TargetP V1.1 to identify chloroplast transit peptides (Emanuelsson et al. 2007), Asn56 and Ala51 of the sunflower HAD1 and HAD2 sequences, respectively, were the best candidates for the N-termini of the mature proteins (Fig. 1). Both transit peptides contained phosphorylation motifs specific for binding the 14-3-3 protein that together with HSP70 can form a cytosolic guidance complex towards the chloroplast (May and Soll 2000) (Fig. 1). These motives were predicted between the Pro35 and Ser42 in HaHAD1 and between the Pro33 and Ser39 in HaHAD2. In the case of HaHAD1 proteolytic processing of the transit peptide would produce a 171 residues protein with a putative molecular mass of 18.9 kDa and a pI of 5.74, while HaHAD2 would have 170 residues with a mass of 18.6 kDa and a pI of 5.67. These two sunflower amino acid sequences with 75 % identity were compared with homologous proteins from different phylogenetic groups such as A. thaliana, Oryza sativa, A. hypogea or P. aeruginosa (Fig. 1).
Alignment of the deduced amino acid sequences of H. annuus β-hydroxyacyl-[ACP]-dehydratase HaHAD1 and HaHAD2, with the closely related sequences from A. hypogea, AhHAD (gi|268527763|), A. thaliana, AtHAD1 (gi|18399910|), Oryza sativa, OsHAD (gi| 115464091|) and Pseudomonas aeruginosa, PaFabZ (gi|674745580|). Identical residues are highlighted as black boxes, highly conserved residues as dark grey boxes and weakly conserved residues as light grey boxes. The active sites are indicated with black triangles and those residues participating in the ACP binding site are indicated with white triangles. Bold red letters indicate the residue involved in tunnel opening regulation, Tyr88 in P. aeruginosa. Arrows show residues located at the catalytic tunnel, from the first (solid) and second (dash) monomer of the homodimer. The putative N-terminal plastid transit peptides of HaHAD1 and HaHAD2, are underlined, 56 and 51 amino acids, respectively. The putative phosphorylation motives are highlighted in green and enclosed in a rectangle
This alignment shows, excluding the transit peptide, which is the less conserved region, identical or highly conserved domains between the HADs. The prediction of the structures and the location of catalytic residues in both sunflower proteins was performed using the SWISS-MODEL server (http://swissmodel.expasy.org/) and the well-studied P. aeruginosa FabZ structure (Kimber et al. 2004). The comparison suggests that the residues involved in recognition and binding of ACP are Arg174, Arg176 and Gln195 in HaHAD1 and Arg167, Arg169 and Gln188 in HaHAD2. The predicted catalytic residues of HaHAD1 are His129 and Glu143 and in HaHAD2 are His122 and Glu136. While the arginine is highly conserved in different kingdoms such as plant, bacteria, and protozoa, the glutamine residue is plant exclusive. Kimber and collaborators (2004) propose that the catalytic residues localize to the hydrophobic tunnel spanning both monomers which has been identified in the FabZ crystal structure from microbes. For HaHAD1 these residues for the first monomer are Leu92, His94, Ala146, Gln147, Gly150, Asp163, Asn164, Phe165, Phe166, Phe167, Ala168 and Met221, and for the second monomer Phe130, Ile135, Met136, Pro137, Gly138, Val139, Met141, Phe175, Lys177, Pro178 and Val179. In the case of HaHAD2, the equivalents are Leu85, His87, Ala139, Gln140, Gly143, Glu156, Asn157, Phe158, Val159, Phe160, Ala161 plus Leu214, and Phe123, Ile128, Met129, Pro130, Gly131, Val132, Met134, Phe168, Lys170, Pro171 plus Val172. The tunnel residues in sunflower HaHAD1 and HaHAD2 are identical except for Asp163 to Glu156, Phe166 to Val159 and Met221 to Leu214. The homologues of Tyr88 in P. aeruginosa FabZ are Phe166 in HaHAD1 and Val159 in HaHAD2.
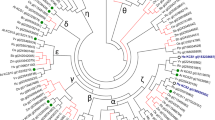
Whole sequenced genome availability of different organisms from many phylogenetic groups together with bioinformatic tools permits identification of groups potentially sharing a common ancestor with HAD from plants (Fig. S1). As the dendrogram shows both sunflower HADs emerge relatively close in the phylogeny. These protein sequences have ≥81 % identity with their plant homologues, such as Vitis vinifera, A. thaliana or A. hypogea. The dendrogram (Fig. S1) also reveals that plant HADs are relatively close to their Chlorophyta and Cyanobacteria homologues.
Tertiary structure prediction of HaHAD proteins
The amino acid sequences of HaHAD1 and HaHAD2 were compared to the known structure of the protozooan protein from P. falciparum FabZ (UniProtKB Q965D7) (Berman et al. 2000). The 47 % sequence identity, of the sunflower proteins, to that of PfFabZ (Fig. 2) infers that the structure of the latter could be used as model for the sunflower proteins. The distribution of the deduced α helices and β sheets of HaHAD1 and HaHAD2 are very similar to that of the model (Fig. 2). This suggests that the sunflower enzymes will also display an “L” shape formed by three α helices (α1-α3) surrounded by six β sheets (β1–β6) that has previously been described in other fatty acid synthesis related proteins, such as acyl-ACP thioesterases (Mayer and Shanklin 2005; Serrano-Vega et al. 2005). The deduced secondary structures of sunflower HADs differ slightly from the PfFabZ structure, as β3 and β6 are longer in the former proteins.
Comparison between the deduced amino acid sequences of H. annuus β-hydroxyacyl-[ACP]-dehydratase, HaHAD1 and HaHAD2, and P. falciparum, PfFabZ (Kostrewa et al. 2005). Identical residues are highlighted as black boxes, highly conserved residues as dark grey boxes and weakly conserved residues as light grey boxes. Residues in the active sites (black triangles), ACP binding residues (white triangles) and residues involved in the tunnel opening (black dotted triangle) are also shown. PfFabZ structural elements are named as in Kimber et al. (2004); h α-helix, s β-sheet
The sunflower HAD 3D structures generated by the program Swiss-PdbViewer show very similar assemblies. A minor difference is shown when the monomers of the sunflower proteins are represented; an extra residue in the loop between α3 and β3 in HaHAD1 compared to HaHAD2 (Fig. 3). When the molecular surfaces of the homodimers were represented, however, more differences between the two deduced protein structures occur (Fig. 3c, d, e, f). In HaHAD1, as in most of the available plant HAD sequences, the tunnel is completely closed by phenylalanine 166 whereas in HaHAD2 with valine 159 in the analogous position the tunnel is always open. Valine is smaller and lacks the aromatic ring present in phenylalanine acting as a “lid” (Fig. 3c d, e, f).
Proposed structural models for sunflower β-hydroxyacyl-[ACP]-dehydratase homodimers, HaHAD1 (a, c, e) and HaHAD2 (b, d, f), modelled from that known for PfFabZ (1z6b) (Kostrewa et al. 2005). a and b Ribbon diagrams. c and d Views of the molecular surfaces. e and f Internal views of the active sites and catalytic tunnel. Residues in α-helices are red, in β-sheets yellow, in the tunnel opening green, and in the catalytic site blue. For HaHAD1/HaHAD2: in α3, Glu143/Glu136; in loop following α2, His129/His122; in β3, Phe166/Val159
Southern blot analysis of HaHAD1 and HaHAD2
Two DNA gel blot analyses were performed on genomic DNA extracted from sunflower leaves to evaluate the number of copies of HaHAD1 and HaHAD2 in the genome. An aliquot of the gDNA was digested with restriction enzymes having one (AleI) or no (HindIII) cutting sites within the cDNA of HaHAD1. Figure 4a shows a single band obtained with HindIII, indicating that HaHAD1 is a single copy gene. With AleI three bands (circa 1.5, 8 and 10 kb) are seen instead of the expected two. To understand these results a new search in the TAIR database (http://www.arabidopsis.org/index.jsp) was carried out revealing that AtHAD (At2g22230) has three introns within the gene sequence. Nevertheless, given the length of the gene (smaller than 1.5 kb) and the band sizes obtained, an AleI restriction site was not expected in the introns. Incomplete digestions of the gDNA could explain the release of three fragments containing sequences of the gene, however. Therefore, assuming an AleI restriction site localized upstream or downstream of the gene, complete digestion would generate, 1.5 and 8 kb bands, but an incomplete digestion a circa 10 kb fragment. This hypothesis supports the idea of a single copy of HaHAD1gene in the sunflower genome. Similarly, the number of copies of HaHAD2 gene in the genome was studied. An aliquot of genomic DNA was digested with AleI, having one target site within the HaHAD2 cDNA, and others lacking targets sites in the cDNA (SphI, XbaI, SacI and NcoI,). Only one band was detected when a target site was lacking and two bands when a target site was present, inferring that HaHAD2 is also a single copy gene (Fig. 4b). This concurs with the observation that a blast search of available H. annuus ESTs performed with the HaHAD1 and HaHAD2 sequences retrieved only nucleotide fragments corresponding to the latter.
Tissue expression profiles of sunflower β-hydroxyacyl-[ACP]-dehydratases
The expression of the HaHAD1 and HaHAD2 genes was analysed by RT-qPCR in roots, stems, 1–7 days after germination (DAG) cotyledons, leaves of 20-day-old seedlings and 12–30 DAF seeds in different developmental stages (Fig. 5). Both sunflower genes showed the highest levels of expression in seeds, particularly HaHAD2, as has been described previously for AtHAD [At2g22230; Schmid et al. 2005)]. HaHAD1 and HaHAD2 presented different expression patterns. HaHAD2 showed the highest level of expression in 12DAF seeds, decreasing gradually as the seed developed similar to the profile found for AtHAD (Schmid et al. 2005). HaHAD1, by contrast, showed a constant level of expression during seed development. HaHAD2 was much more highly expressed than HaHAD1 in 12 and 20 DAF seeds, whereas HaHAD1 exhibited slightly more expression in the rest of tissues compared to HaHAD2, especially in the roots where only traces of expression were detected.
Expression levels of β-hydroxyacyl-[ACP]-dehydratase in developing seeds and vegetative tissues from H. annuus, HaHAD1 and HaHAD2, determined by RT-qPCR from sunflower line CAS-6. Stages in cotyledon development are designated days after germination (DAG) and in seed development days after flowering (DAF). Data are mean values ± SD of three independent biological samples
Fatty acid modifications in E. coli cells expressing recombinant HaHAD1 and HaHAD2
The constructions pQE80L::HaHAD1 and pQE80L::HaHAD2, missing the deduced plastid targeting sequences, respectively, were transformed into XL1-Blue cells. The growth rate of the cells expressing HaHAD1, HaHAD2 or the empty vector as background control was monitored at 37 °C as explained in “Materials and methods”. Lower growth rates were observed when expression of either HaHAD1 or HaHAD2 was induced compared to the control. The cells expressing the first gene showed half of the growth rate compared to the cells without induction or the control (0.33/0.74/0.76 h−1, respectively). A similar decrease, albeit of a lower magnitude (0.57/0.8/0.76 h−1, respectively) was found when the second gene was induced. Induced expression of both genes resulted in cessation of growth after 2–3 h.
A small increase, but non-statistically significant, in the bacteria fatty acid content occured when the HaHAD1 or HaHAD2 gene was present, but was not induced. Induction resulted in further increases, especially significant for HaHAD1. This accumulation was accompanied by modification of the fatty acid profiles (Table 2). Namely, expression resulted in a circa 2-fold increase of the saturated:unsaturated fatty acids which was accompanied by a circa 33–50 % reduction in the C16:0/C18:0 ratio. The C16:1/C18:1 ratios by comparison showed little change.
Purification of the recombinant HaHAD1, HaHAD2 and HaHAD1 F166V proteins and their biochemical characterization
Given that the deduced tertiary structures revealed differences at the catalytic tunnel between the two sunflower proteins; the differing amino acid, phenylalanine 166, was mutagenized to a valine in HaHAD1. The two wild-type constructs as well as the pQE80L::HaHAD1 F166V gene were expressed in XL1-Blue cells and the resulting proteins purified (Fig. S2).
‘In vivo’ β-hydroxyacyl-(ACP]-dehydratase eliminates a H2O molecule from β-hydroxyacyl-ACP to generate an enoyl-ACP. This reversible reaction, hydration, is favored under ‘in vitro’ conditions; hence crotonyl-ACP or crotonyl-CoA are used as substrates (Shimakata and Stumpf 1982a, b; Sharma et al. 2003; Liu et al. 2005; Dautu et al. 2008). Crotonyl-CoA was used in the present experiments. All three heterologously expressed proteins, HaHAD1, HaHAD2 and HaHAD1 F166V were active (Fig. S3) with the production of β-hydroxyacyl-CoA dependent on concentration and time of the reaction. The slopes generated from the graphs and the Hill equations were used to calculate K m and V max values. This resulted in a K m for crotonyl-CoA of 68.49 ± 6.15 µM and V max of 13.04 ± 1.22 µmol min−1 mg−1 for HaHAD1 protein and a K m of 97.73 ± 8.67 µM and V max of 29.16 ± 2.07 µmol min−1 mg−1 for HaHAD2 protein. These results showed that HaHAD2 presented higher activity and lower substrate affinity than HaHAD1 for crotonyl-CoA. The HaHAD1 F166V protein had a K m for crotonyl-CoA of 82.30 ± 6.01 µM and the V max 21.61 ± 1.80 µmol min−1 mg−1. These results indicate that the mutant protein was more active with crotonyl-CoA than HaHAD1 but less so than HaHAD2.
Discussion
Sunflower seeds are one of the most important sources of oil. To provide a basic groundwork for improving and modifying the quality of its oil we are characterizing the structural genes and their encoded proteins taking part in its synthesis. Below we discuss the peculiarities of the β-hydroxyacyl-[ACP]-dehydratases, the third of four components of the FAS enzyme complex. Previously studied are the enoyl-[ACP]-reductase and β-ketoacyl-[ACP]-synthase III (González-Mellado et al. 2010; González-Thuillier et al. 2015). Remaining are the β-ketoacyl-[ACP]-reductase and the other two β-ketoacyl-[ACP]-synthases, I and II.
Two β-hydroxyacyl-[ACP]-dehydratase genes, HaHAD1 and HaHAD2, were isolated and cloned from sunflower developing seed cDNA. The deduced proteins had transit peptides targeting the mature protein to the plastid or chloroplast. Both transit peptides showed a phosphorylation motif, as was the case for most of the previously studied sunflower proteins involved in fatty acid biosynthesis, such as FatA-type thioesterases (Serrano-Vega et al. 2005), the ω3-desaturase HaFAD7 (Venegas-Calerón et al. 2006) and the condensing enzyme HaKASIII (González-Mellado et al. 2010). Southern blot analysis of HaHAD1 and HaHAD2 indicated that both are most likely single copy genes as was observed for H. annuus enoyl-[ACP]-reductases (González-Thuillier et al. 2015).
The phylogenetic analysis of the sunflower HADs with related proteins from different taxonomic groups pointed to a common origin for these two proteins. According to the location of both proteins in the phylogenetic tree the duplication of HAD occurred recently. The existence of a common ancestor with cyanobacterial hydroxyacyl-[ACP]-dehydratase concurs with the cyanobacterial origin of the chloroplast via endosymbiosis (Goksøyr 1967), as was observed for the KASIII and ENR proteins from the sunflower FAS complex (González-Mellado et al. 2010; González-Thuillier et al. 2015). During evolution the plastid/chloroplast HAD gene sequences have acquired transit peptides, the major difference to the cyanobacterial HAD genes.
According to the RT-qPCR analysis both sunflower β-hydroxyacyl-[ACP]- dehydratases had higher levels of expression in seeds than other tissues. The profiles were not identical, however. HaHAD2, expressed throughout seed development, had markedly higher levels of mRNA in 12 and 20 DAF seeds. HaHAD1 was expressed at a relatively low, constant level throughout seed development, and remarkably, also in all tested plant tissues. Such results imply that HaHAD1 functions as a housekeeping gene, ensuring maintenance of FAS activity for polar membrane lipid synthesis and repair. HaHAD2, by comparison, is the gene primarily responsible for synthesis of storage lipids. This concurs with the fact that the maximum rate of lipid accumulation in developing sunflower seeds takes place 18–19 DAF (Martínez-Force et al. 2000). The expression changes of HaHAD2 during seed development mimic that of the previously analyzed Arabidopsis At2g22230 gene (Schmid et al. 2005) and of the sunflower HaENR2 gene (González-Thuillier et al. 2015). The low level of mRNA for both HaHAD genes in young cotyledons was unexpected given the elevated levels of mRNA found for both sunflower HaENR genes in this tissue. During the change from dark/reserve to photosynthetically active cotyledons and the phase of rapid expansion of young leaves higher quantities of polar membrane lipids are required.
Expression of the sunflower β-hydroxyacyl-[ACP]-dehydratses in E. coli resulted in a decreased growth rate, especially for HaHAD1 containing cells. Concomitant with these changes an increase of total fatty acids occurred. Presumably this results from an increase in fatty acid synthesis that the bacteria initiate in an attempt to produce enough unsaturated fatty acids to maintain membrane fluidity. This will be unsuccessful due to the massive expression of the plant enzyme and its incapability to isomerize and as a consequence the bacteria will die (see below). The reduction by 9 and 5 % of the C18 fatty acids in the bacteria heterologously overexpressing either HaHAD1 or HaHAD2, respectively, is puzzling as sunflower enzymes presumably have a higher affinity for a C16 substrate than the bacterial enzyme.
In bacteria two β-hydroxyacyl-[ACP]-dehydratase enzymes, FabA and FabZ, take part in fatty acid biosynthesis. FabA can isomerize as well as dehydrate. The β-hydroxyacyl-[ACP] dehydratases from plants are more similar to FabZ. In E. coli the separation between unsaturated and saturated fatty acid biosynthesis occurs during chain elongation at 10 carbons atoms when isomerization is carried out by FabA. The production of unsaturated versus saturated fatty acids relies, therefore at least in part, on the FabA/FabZ ratio (Fig. S4) (Brock et al. 1967). The normal result is circa 50 % each of saturated and monoenoic fatty acids (Garwin et al. 1980) which is close to the 0.8 ratio found for the vector control cells (Fig. S4). Heterologously expressing the sunflower genes lacking isomerase activity increased the amount of saturated fatty acids to 76 and 70 % for HaHAD1 and HaHAD2, respectively (Fig. S4). Early work has shown that in E. coli below 20 % unsaturated fatty acids is insufficient to maintain membrane fluidity at 35 °C, and hence leads to cell death (Cronan and Gelmann 1973).
Using the P. aeruginosa FabZ crystal structure as a model, monomer structures were predicted for HaHAD1 and HaHAD2 with α helices and β strands forming the “hot dog” structure analogous to those described previously (Kimber et al. 2004; Kostrewa et al. 2005). The functional residues, catalytic site as well as recognition and ACP binding sites were localized in conserved regions of these polypeptides. A minor difference between the sunflower structures was a one residue longer loop between α3 and β3 in HaHAD1 than HaHAD2. As the dehydratases function as dimers (Sharma et al. 2003; Kimber et al. 2004; White et al. 2005; Swarnamukhi et al. 2006) dimeric structures were modelled for HaHAD1 and HaHAD2. This revealed an interesting structural difference resulting from the presence of a phenylalanine in position 166 in HaHAD1 versus a valine in position 159 in HaHAD2. Only in the Nostoc genera of the Cyanobacteria is a valine also found. The specified residue is located at the entrance of the catalytic tunnel formed between the monomers upon dimerization, thereby potentially regulating substrate access to the catalytic site. These two residues present very different structures at the tunnel entrance. In HaHAD1 Phe166 totally blocks the entrance while in HaHAD2 Val159 does not. To probe whether this residue is important for enzyme activity, its mutation to a valine in HaHAD1 giving HaHAD1 F166V was accomplished. All three proteins were heterologously expressed and purified.
The activity of the proteins to carry out the reverse reaction, hydration of crotonyl-CoA to β-hydroxyacyl-CoA was tested in vitro. All three were active. The K m for crotonyl-CoA of the HaHAD1 enzyme was lower than for HaHAD2 (68.49 and 97.73 µM, respectively), indicating a higher substrate affinity of the first enzyme for crotonyl-CoA. K m values between 82.6 and 86 µM, within the range of the ones observed for the sunflower proteins, were previously described for these proteins in different microorganisms, such as H. pylori, T. gondii and P. falciparum (Sharma et al. 2003; Liu et al. 2005; Dautu et al. 2008). The V max values were contrary, however, as HaHAD2 exhibited a greater reaction velocity than HaHAD1, 29 and 13 µmol min−1 mg−1, respectively. Together these results suggest that although HaHAD2 needs a higher concentration of crotonyl-CoA than HaHAD1, the former is more efficient in generating the product than HaHAD1. For the HaHAD1 F166V protein the V max and K m were 82.30 µM and 21.61 µmol min−1 mg−1, respectively, that is, intermediate to the values obtained for the two wild-type enzymes. Combined these observations intimate that the residue at the tunnel opening may have little influence of the entrance of the substrate to the active site. Additional subtle structural differences must exist that contribute to the different K m and V max values found for these enzymes.
An interesting correlation occurs between the enzyme kinetic and RT-qPCR results. During early seed development high HaHAD2 gene expression levels occur when ‘de novo’ fatty acid synthesis is highly active, and therefore larger amounts of substrate are available for dehydration. This concurs with the enzymatic analyses revealing that the HaHAD2 enzyme requires higher levels of substrate than HaHAD1. This characteristic makes HaHAD2 the more suitable enzyme to carry out the production of large amounts of fatty acid during, for instance, lipid accumulation in the seed even though it carries out the dehydration more slowly than HaHAD1 does. The appearance of this isoform, HaHAD2, with the valine substitution and the accompanying mutations resulting in a higher substrate concentration requirement and lower reaction rate, should be a recent evolutionary event due to the predominance of Phe166 in the plant kingdom HAD sequences. In any case, the real relevance in oil biosynthesis of this isoform and its changes respect to the most phylogenetically conserved during sunflower seed development must be stated taking in account that these proteins form part of the FAS complex interacting with other subunits for which other isoforms and temporal expression windows have been described (González-Mellado et al. 2010; González-Thuillier et al. 2015), in such a way that coevolution cannot be discarded.
Author contribution statement
IGT, MVC and EMF conceived and designed research. IGT conducted experiments and contributed to data elaboration. RS contributed to the RT-qPCR and biochemical analyses. IGT, MVC, PWK and EMF analyzed data. IGT, MVC, RG, PWK and EMF wrote the manuscript. All authors read and approved the manuscript.
Abbreviations
- ACP:
-
Acyl carrier protein
- DAF:
-
Days after flowering
- DAG:
-
Days after germination
- ENR:
-
Enoyl-ACP reductase
- FAS:
-
Fatty acid synthase
- HAD:
-
β-Hydroxyacyl-ACP dehydratase
References
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242
Brock DJH, Kass LR, Bloch K (1967) Beta-hydroxydecanoyl thioester dehydrase.II, Mode of action. J Biol Chem 242:4432–4440
Brown A, Affleck V, Kroon J, Slabas A (2009) Proof of function of a putative 3-hydroxyacyl-acyl carrier protein dehydratase from higher plants by mass spectrometry of product formation. FEBS Lett 583:363–368
Cronan JE, Gelmann EP (1973) Estimate of minimum amount of unsaturated fatty-acid required for growth of Escherichia coli. J Biol Chem 248:1188–1195
Dautu G, Ueno A, Munyaka B, Carmen G, Makino S, Kobayashi Y, Igarashi M (2008) Molecular and biochemical characterization of Toxoplasma gondii β-hydroxyacyl-acyl carrier protein dehydratase (FABZ). Parasitol Res 102:1301–1309
Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953–971. doi:10.1038/nprot.2007.131
Garwin JL, Klages AL, Cronan JE (1980) Beta-ketoacyl-acyl carrier protein synthase-II of Escherichia coli. Evidence for function in the thermal regulation of fatty acid synthesis. J Biol Chem 255:3263–3265
Goksøyr J (1967) Evolution of eucaryotic cells. Nature 214:1161. doi:10.1038/2141161a0
González-Mellado D, von Wettstein-Knowles P, Garcés R, Martínez-Force E (2010) The role of beta-ketoacyl-acyl carrier protein synthase III in the condensation steps of fatty acid biosynthesis in sunflower. Planta 231:1277–1289. doi:10.1007/s00425-010-1131-z
González-Thuillier I, Venegas-Calerón M, Garcés R, von Wettstein-Knowles P, Martínez-Force E (2015) Sunflower (Helianthus annuus) fatty acid synthase complex: enoyl-[acyl carrier protein]-reductase genes. Planta 241:43–56. doi:10.1007/s00425-014-2162-7
Harwood J (2005) Fatty acid biosynthesis. In: Murphy DJ (ed) Plant lipids: Biology, utilisation and manipulation. Blackwell Publishing, Oxford, pp 27–66
Heath RJ, Rock CO (1996) Roles of the FabA and FabZ beta-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J Biol Chem 271:27795–27801
Høj PB, Mikkelsen JD (1982) Partial separation of individual enzyme-activities of an ACP-dependent fatty-acid synthetase from barley chloroplasts. Carlsberg Res Commun 47:119–141. doi:10.1007/Bf02914031
Kimber MS, Martin F, Lu YJ, Houston S, Vedadi M, Dharamsi A, Fiebig KM, Schmid M, Rock CO (2004) The structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa. J Biol Chem 279:52593–52602. doi:10.1074/jbc.M408105200
Kostrewa D, Winkler FK, Folkers G, Scapozza L, Perozzo R (2005) The crystal structure of PfFabZ, the unique beta-hydroxyacyl-ACP dehydratase involved in fatty acid biosynthesis of Plasmodium falciparum. Protein Sci 14:1570–1580. doi:10.1110/Ps.051373005
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Li MJ, Li AQ, Xia H, Zhao CZ, Li CS, Wan SB, Bi YP, Wang XJ (2009) Cloning and sequence analysis of putative type II fatty acid synthase genes from Arachis hypogaea L. J Biosciences 34:227–238
Liu WZ, Luo C, Han C, Peng SY, Yang YM, Yue JM, Shen X, Jiang HL (2005) A new beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Helicobacter pylori: molecular cloning, enzymatic characterization, and structural modeling. Biochem Bioph Res Comm 333:1078–1086
Maity K, Venkata BS, Kapoor N, Surolia N, Surolia A, Suguna K (2011) Structural basis for the functional and inhibitory mechanisms of β-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Plasmodium falciparum. J Struct Biol 176:238–249. doi:10.1016/j.jsb.2011.07.018
Martínez-Force E, Cantisan S, Serrano-Vega MJ, Garcés R (2000) Acyl-acyl carrier protein thioesterase activity from sunflower (Helianthus annuus L.) seeds. Planta 211:673–678
May T, Soll J (2000) 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell 12:53–63. doi:10.1105/Tpc.12.1.53
Mayer KM, Shanklin J (2005) A structural model of the plant acyl-acyl carrier protein thioesterase FatB comprises two helix/4-stranded sheet domains, the N-terminal domain containing residues that affect specificity and the C-terminal domain containing catalytic residues. J Biol Chem 280:3621–3627. doi:10.1074/jbc.M411351200
Mohan S, Kelly TM, Eveland SS, Raetz CR, Anderson MS (1994) An Escherichia coli gene (FabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. Relation to FabA and suppression of mutations in lipid A biosynthesis. J Biol Chem 269:32896–32903
Moreno-Perez A, Sánchez-García A, Salas JJ, Garcés R, Martínez-Force E (2011) Acyl-ACP thioesterases from macadamia (Macadamia tetraphylla) nuts: cloning, characterization and their impact on oil composition. Plant Physiol Biochem 49:82–87
Saitou N, Nei M (1987) The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37:501–506. doi:10.1038/Ng1543
Serrano-Vega MJ, Garcés R, Martínez-Force E (2005) Cloning, characterization and structural model of a FatA-type thioesterase from sunflower seeds (Helianthus annuus L.). Planta 221:868–880. doi:10.1007/s00425-005-1502-z
Sharma SK, Kapoor M, Ramya TNC, Kumar S, Kumar G, Modak R, Sharma S, Surolia N, Surolia A (2003) Identification, characterization, and inhibition of Plasmodium falciparum beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ). J Biol Chem 278:45661–45671. doi:10.1074/jbc.M304283200
Shimakata T, Stumpf PK (1982a) The procaryotic nature of the fatty-acid synthetase of developing Carthamus tinctorius L (safflower) seeds. Arch Biochem Biophys 217:144–154. doi:10.1016/0003-9861(82)90488-X
Shimakata T, Stumpf PK (1982b) Purification and characterizations of β-ketoacyl-[acyl-carrier-protein] reductase, β-hydroxyacyl-[acyl-carrier-protein] dehydrase, and enoyl-[acyl-carrier-protein] reductase from Spinacia oleracea leaves. Arch Biochem Biophys 218:77–91. doi:10.1016/0003-9861(82)90323-X
Smith S, Witkowski A, Joshi AK (2003) Structural and functional organization of the animal fatty acid synthase. Prog Lipid Res 42:289–317. doi:10.1016/S0163-7827(02)00067-X
Swarnamukhi PL, Sharma SK, Bajaj P, Surolia N, Surolia A, Suguna K (2006) Crystal structure of dimeric FabZ of Plasmodium falciparum reveals conformational switching to active hexamers by peptide flips. FEBS Lett 580:2653–2660. doi:10.1016/j.febslet.2006.04.014
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi:10.1093/molbev/msm092
Venegas-Calerón M, Muro-Pastor AM, Garcés R, Martínez-Force E (2006) Functional characterization of a plastidial omega-3 desaturase from sunflower (Helianthus annuus) in cyanobacteria. Plant Physiol Biochem 44:517–525. doi:10.1016/j.plaphy.2006.09.005
White SW, Zheng J, Zhang YM, Rock CO (2005) The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem 74:791–831. doi:10.1146/annurev.biochem.74.082803.133524
Yasuno R, von Wettstein-Knowles P, Wada H (2004) Identification and molecular characterization of the β-ketoacyl-[acyl carrier protein] synthase component of the Arabidopsis mitochondrial fatty acid synthase. J Biol Chem 279:8242–8251. doi:10.1074/jbc.M308894200
Zhang L, Liu WZ, Hu TC, Du L, Luo C, Chen KX, Shen X, Jiang HL (2008) Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ). J Biol Chem 283:5370–5379
Acknowledgments
We thank Dr. Alicia M. Muro-Pastor for help with Southern blot analysis and A. González-Callejas and B. Lopez-Cordero for skilful technical assistance. This work was supported by the “Ministerio de Economia y Competitividad” and FEDER project, AGL2011-23187. IGT was supported by a JAE-CSIC contract, in part financed by the European Social Fund.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2015_2410_MOESM1_ESM.tif
Fig. S1 Phylogenetic comparison of plant and algae β-hydroxyacyl-[ACP]- dehydratase homologues of the E. coli FabZ protein. H. annuus sequences are shown in bold (TIFF 59094 kb)
425_2015_2410_MOESM2_ESM.tif
Fig. S2 Purification of H. annuus recombinant HAD proteins. Coomassie blue-stained SDS-PAGE of empty pQE80L vector, recombinant HaHAD1, HaHAD2 and HaHAD F166V from XL1-Bl grown at 37 °C. S, soluble fraction; AF, affinity purified using one passage through a Ni2+-charged HisTrap FF column; kDa, size markers (TIFF 781 kb)
425_2015_2410_MOESM3_ESM.tif
Fig. S3 Activity of H. annuus β-hydroxyacyl-[ACP]-dehydratase proteins, HaHAD1, HaHAD2 and HaHAD1. The in vitro reverse reactions were performed in 17.9 mM potassium phosphate buffer, 1.6 mM DTT at pH 6.8, 25 °C with HaHAD1 (5 μg ml−1), HaHAD2 (5 μg ml−1) and HaHAD1 F166V (5 μg ml−1). The reaction was started by adding the substrate (crotonyl-CoA) at different concentrations (20-100 μM) and following the decrease in absorbance at 260 nm as crotonyl-CoA was hydrated. Results correspond to the average of three independent determinations (TIFF 31640 kb)
425_2015_2410_MOESM4_ESM.tif
Fig. S4 Schematic representation of E. coli fatty acid biosynthesis comparing the effect on the ratio of saturated to unsaturated fatty acids expressing H. annuus β-hydroxyacyl-[ACP]-dehydratases, HaHAD1 or HaHAD2 with functional EcFabZ. Proteins involved in fatty acid biosynthesis in E. coli are represented. FabZ, β-hydroxyacyl-[ACP]-dehydratase Z; FabA, β-hydroxydecanoyl-[ACP]-dehydratase A; FabB, β-ketoacyl-[ACP]-synthase I; FabI, enoyl-[ACP]-reductase. Symbols in grey define the bacterial pathway; in black represent the effect of expression of H. annuus β-hydroxyacyl-[ACP]-dehydratases in the E. coli cells (TIFF 1684 kb)
Rights and permissions
About this article
Cite this article
González-Thuillier, I., Venegas-Calerón, M., Sánchez, R. et al. Sunflower (Helianthus annuus) fatty acid synthase complex: β-hydroxyacyl-[acyl carrier protein] dehydratase genes. Planta 243, 397–410 (2016). https://doi.org/10.1007/s00425-015-2410-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2410-5