Abstract
The pathophysiology of hypertrophic scar (HS) shares similarities with cancer. HOXC10, a gene significantly involved in cancer development, exhibits higher expression levels in HS than in normal skin (NS), suggesting its potential role in HS regulation. And the precise functions and mechanisms by which HOXC10 influences HS require further clarification. Gene and protein expressions were analyzed using raeal-time quantitative polymerase chain reaction (RT–qPCR) and western blot techniques. Cell proliferation and migration were evaluated using EdU proliferation assays, CCK-8 assays, scratch assays, and Transwell assays. Chromatin immunoprecipitation (ChIP) and dual-luciferase reporter assays were conducted to investigate the interactions between HOXC10 and STMN2. HOXC10 and STMN2 expression levels were significantly higher in HS tissues compared with NS tissues. Silencing HOXC10 led to decreased activation, proliferation, migration, and fibrosis in hypertrophic scar fibroblasts (HSFs). Our findings also indicate that HOXC10 directly targets STMN2. The promotional effects of HOXC10 knockdown on HSF activation, proliferation, migration, and fibrosis were reversed by STMN2 overexpression. We further demonstrated that HOXC10 regulates HSF activity through the TGF-β/Smad signaling pathway. HOXC10 induces the activation and fibrosis of HSFs by promoting the transcriptional activation of STMN2 and engaging the TGF-β/Smad signaling pathway. This study suggests that HOXC10 could be a promising target for developing treatments for HS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hypertrophic scars (HS) commonly develop following severe burns or skin injuries, manifesting as a fibroproliferative condition during the skin’s healing process (Chiang et al. 2016). This condition is characterized by increased activity of fibroblasts and deposition of collagen (Čoma et al. 2021). A crucial aspect of HS’s pathogenesis involves the transformation of fibroblasts into myofibroblasts, which are identified by their expression of α-smooth muscle actin (α-SMA) (Lingzhi et al. 2020). These myofibroblasts play a critical role in promoting the synthesis of collagen types I and III (Tai et al. 2021). The presence of HS can result in significant cosmetic and functional deficits, leading to both physical and psychological distress (Rabello et al. 2014). Despite the prevalence and impact of HS, effective prevention and treatment strategies remain elusive, underscoring the urgent need for the development of novel clinical interventions.
The homeobox (HOX) gene family plays a pivotal role in developmental biology, significantly influencing skin development (Scott and Goldsmith 1993). HOX genes are implicated in orchestrating skin cell proliferation and are essential for the regeneration of skin wounds (Stelnicki et al. 1998). Analysis of high-throughput sequencing data from the GEO dataset GSE210434 revealed 65 genes markedly upregulated in fibroblasts derived from HS tissues compared to normal skin (NS) tissues, with HOXC10 emerging as a gene of interest for further study. Previous investigations have primarily explored HOXC10 in the context of its involvement in tumor growth, cell death, and the processes of cell migration and invasion (Yu et al. 2022; He et al. 2023). Additionally, another study reported elevated expression levels of HOXC10 in HS tissues compared to non-affected tissues, suggesting that HOXC10 may play a role in regulating HS (Kang et al. 2023). Given this background, we speculate that HOXC10 may significantly impact the development and progression of HS, warranting further exploration of its function in this specific pathological condition.
Bioinformatics analysis JASPAR suggested that HOXC10 could potentially target STMN2, a gene implicated in the regulation of fibrosis according to previous research (Asselah et al. 2005). For instance, STMN2 overexpression has been shown to elevate levels of osteopontin, α-SMA, and vimentin in vascular smooth muscle cells (VSMCs) (Ke et al. 2022). Furthermore, STMN2 has been identified as a critical player in fibrotic processes (Hu and Zhou 2023). However, the specific mechanism by which HOXC10 and STMN2 interact to influence fibrosis in HS remains unclear.
Previous studies have reported that the TGF-β/Smad signaling pathway plays a role in regulating HS formation, offering the potential for mitigating scar keloids and HS formation (Zhang et al. 2020b). Liang et al. also indicated in a recent study that CLC-3 suppressed fibrosis formation in HS by mediating the TGF-β/Smad signaling pathway (Liang et al. 2024). Interestingly, evidence suggests STMN2 may facilitate the nuclear translocation of Smad2/3, thereby intensifying TGFβ signal transduction (Zhong et al. 2021).
Therefore, based on the above data, we hypothesize that HOXC10 contributes to the fibrosis of HS by facilitating the transcriptional activation of STMN2, which in turn activates the TGF-β/Smad signaling pathway.
Materials and methods
Ethics approval and human tissue samples
Skin samples, including 20 normal and 20 HS tissues, were obtained during plastic surgery procedures at the hospital, following the ethical guidelines approved by the Human Research Ethics Committee of the Hospital. These guidelines are in alignment with the principles outlined in the Declaration of Helsinki. Prior to tissue acquisition, written informed consent was obtained from all participating patients. Using these HS tissues, primary human hypertrophic scar fibroblasts (HSFs) were subsequently isolated for research purposes.
Isolation and culture of HSFs and NSFs
The process of isolating and culturing human HSFs and normal skin fibroblasts (NSFs) involved the digestion of the dermal layers from both HS and normal skin tissues. This was accomplished by finely chopping the tissues and incubating them in 0.1 mg/ml collagenase type I at 37 °C for 3 h. Following isolation, the fibroblasts derived from HSFs and NSFs were cultured in Dulbecco’s modified Eagle medium (DMEM) with low glucose (31,600,034, Gibco, Grand Island, NY, USA) enriched with 10% fetal bovine serum (FBS, 35-081-CV, Corning, NY, USA), 100 U/ml penicillin (Thermo Fisher Scientific, San Jose, CA, USA), and 100 μg/ml streptomycin (Thermo Fisher Scientific). Fibroblasts from the third to fifth passages were selected for further experimental investigations.
RNA extraction and quantitative real-time polymerase chain reaction (RT–qPCR)
Total RNA from cells was extracted using the Trizol reagent (Invitrogen, Gaithersburg, MD, USA). For reverse transcription, 1 µg of RNA was used with the PrimeScript cDNA Synthesis Kit (6110A, Takara, Osaka, Japan), according to the instructions provided by the manufacturer. Quantitative PCR analyses were conducted utilizing the TaqMan® Universal PCR Master Mix (4,305,719, Thermo Fisher Scientific). Primers for the assays were obtained from Origene Biotech (Wuxi, Jiangsu, China), with the following specific sequences:
HOXC10 forward: 5′-GAGCGAAAAGGAGAGGGCCAAA-3′, reverse: 5′-TCCGCTCTTTGCTGTCAGCCAA-3′;
STMN2 forward: 5′-CCAGAAGAAACTGGAGGCTGCA-3′, reverse: 5′-GCTTTTCCTCCGCCATCTTGCT-3′;
GAPDH forward: 5′-GTCTCCTCTGACTTCAACAGCG-3′, reverse 5′-ACCACCCTGTTGCTGTAGCCAA-3′.
Relative mRNA expression levels were quantified employing the 2−ΔΔCt method and were normalized against GAPDH expression.
Western blot
Protein lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors and incubated at 4 °C for 30 min to ensure complete lysis (Beyotime Inc., Hiamen, Jiangsu, China). Protein concentrations were determined with the BCA Protein Assay Kit (10,741,395, Thermo Fisher Scientific). Subsequently, 30 μg of protein from each lysate was resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE; 4–20% Tris–Glycine gels, Thermo Fisher Scientific) and transferred onto polyvinylidene fluoride (PVDF) membranes (Merck Millipore, MA, USA). Membranes were blocked for 1 h in Odyssey Blocking Buffer (Li-Cor, Lincoln, NB, USA) containing 0.1% Tween-20 and then incubated with primary antibodies specific for Collagen I (ab138492, 1:1000, Abcam, Cambridge, UK), Collagen III (ab184993, 1:1000, Abcam), α-SMA (ab5694, 1:1000, Abcam), HOXC10 (ab153904, 1:1000, Abcam), STMN2 (ab185956, 1:10,000, Abcam), TGF-β (ab215715, 1:1000, Abcam), phosphorylated Smad-2 (p-Smad-2, ab280888, 1:1000, Abcam), Smad-2 (ab40855, 1:2000, Abcam), phosphorylated Smad-3 (p-Smad-3, ab51451, 1:2000, Abcam), Smad-3 (ab40854, 1:1000, Abcam), and GAPHD (ab8245, 1:1000, Abcam) as a loading control. After washing with phosphate-buffered saline (PBS), the membranes were exposed to secondary antibodies (ab7090, 1:500, Abcam or ab150165, 1:500, Abcam) for 60 min at room temperature. Detection of the protein bands was achieved using an enhanced chemiluminescence (ECL) system (WBULS0100, Merck Millipore).
Cell transfection and treatment
small-interfering RNA sequences designed to silence HOXC10 expression (si-HOXC10) and a non-targeting control sequence (si-NC) were acquired from Origene (Shanghai, China). For the overexpression of HOXC10 (oe-HOXC10) and STMN2 (oe-STMN2), their respective full-length sequences were amplified via PCR and cloned into pcDNA3.1 vectors provided by System Biosciences (SBI, Mountain View, CA, USA). Vectors lacking any insert acted as negative controls (oe-NC). These constructs were then introduced into HSFs using Lipofectamine 3000 (Invitrogen) and incubated for 48 h, with the addition of a leukocyte activation cocktail (BD Biosciences GmbH, Heidelberg, Germany) during the final 5 h to stimulate the cells.
To explore the influence of the TGF-β and Smad signaling pathways on HSF behavior, cells transfected with either oe-NC or oe-HOXC10 were grown to approximately 80% confluence. They were then seeded at a density of 1 × 10^5 cells/mL in culture plates. The cells underwent treatment with a TGF-β signaling pathway inhibitor, LY2109761 (S2704 Selleckchem, Houston, TX, USA), at a concentration of 10 μmol/L for 2 h.
Cell proliferation assay
The Cell Counting Kit-8 (CCK-8, TransGen, Beijing, China) and the EDU assay kit (RIBOBIO, Guangzhou, China) were employed to assess cell proliferation. HSFs were seeded at a density of 5 × 103 cells per well in 96-well plates. At time intervals of 0, 24, 48, and 72 h, 10 μL of CCK-8 solution and 90 μL of medium were incubated at 37℃ for 1 h. Subsequently, the absorbance at 450 nm was measured using a microplate reader (Bio-Rad, Hercules, CA, USA). In addition, to directly observe proliferating cells, an experiment incorporating 5-ethynyl-2-deoxyuridine (EdU) was conducted following the manufacturer’s instructions. In brief, HSFs were seeded in 96-well plates at a density of 5 × 103 cells per well and cultured for 24 h. The cells were then incubated with 50 μmol/mL EdU in the medium for 3 h. Following incubation, the cells were fixed with 4% paraformaldehyde and then incubated with 50 μL of 2 mg/mL glycine at room temperature for 5 min. After a 10 min wash with 100 μL of 0.01 M PBS containing 0.5% Triton X-100 (PBS-T), the cells were incubated with 100 μL of 1× Apollo staining reaction solution in the dark for 30 min. Subsequently, 30 μL of DAPI was added and incubated in the dark for 5 min. The number of EdU-positive cells was then counted under a BX51 fluorescence microscope, 40×/0.75 ph2 correction (Olympus, Tokyo, Japan). The mean value for each condition represents the average of these biological replicates.
Scratch assay
HSFs were cultured in six-well plates at a density of 5 × 104 cells/cm2 in DMEM medium for 24 h to form a confluent monolayer. After removing the DMEM medium, a scratch was created using a sterile 200 μL pipette tip. The cells were washed twice with PBS to remove debris and then incubated with a serum-free medium. The scratch width was captured using a DMi8 optical microscope (Leica, Wetzlar, Germany) to monitor the healing process. The microscope’s “Mark-and-Find” feature was utilized to record specific positions within each well, capturing multiple areas along the denuded surface. Time-lapse phase contrast imaging was performed, recording a frame every 15 min over a 24 h incubation period using an AxioCam single-channel camera and AxioVision software (Carl Zeiss, AG, Jena, Germany). Images taken at 0 and 24 h were presented to show the scratch healing results.
Transwell assay
HSFs were prepared in serum-free DMEM at a concentration of 1 × 106 cells per 100 μL and placed into the upper compartment of the Transwell chamber. To stimulate cell migration, 600 μL of DMEM supplemented with 10% FBS was added to the lower compartment. Following a 48 h incubation period, non-migrating cells were removed from the upper surface of the membrane. Cells that had migrated through the membrane were fixed and stained with crystal violet for 5 min. The stained cells, indicative of successful migration, were visualized and documented using a DMi8 optical microscope, 20×/0.4, corr 0–2 mm DIC.
Chromatin immunoprecipitation (ChIP) assay
HSFs were harvested and lysed using the RIPA lysis buffer provided by Merck Millipore. After lysis, the cells were subjected to cross-linking and sonication. The resulting lysates were combined with 900 μL of ChIP Dilution Buffer, 20 μL of 50× Protease Inhibitor Cocktail, and 60 μL of Protein A Agarose/Salmon Sperm DNA beads, followed by an incubation period of overnight at 4 °C. The lysates were then centrifuged to collect the supernatant, which was carefully transferred into a fresh tube. To this, 1 μL of either a HOXC10 antibody (NBP1-71,933, 1:1000, Novus Biologicals, CO, USA) or an IgG control antibody (ab205718, 1:1000, Abcam) was added, and the mixture was incubated overnight at 4 °C to allow for immunoprecipitation. After the immunoprecipitation and subsequent washes, 1 μL of RNase A was added to each sample for RNA removal, followed by an hour long incubation at 37 °C. Then, the samples were treated with 10 μL of 0.5 M EDTA, 20 μL of 1 M Tris–HCl, and 2 μL of 10 mg/mL proteinase K, and incubated at 45 °C for 2 h to digest proteins. The DNA was subsequently extracted, and its purity and concentration were assessed using agarose gel electrophoresis.
Dual-luciferase reporter assay
To investigate the effect of HOXC10 on STMN2 promoter activity, both the wild-type and mutant variants of HOXC10 (HOXC10-wt, HOXC10-mut) were cloned into the psiCHECK2 vector (Promega, Madison, WI, USA). The 293T cells were then transfected with either the HOXC10-wt or HOXC10-mut or the empty constructs using Lipofectamine 3000 (Invitrogen). Following 48 h of incubation, luciferase activity was assessed using the Dual-Luciferase Reporter Assay System (Promega).
Bioinformatic database analysis
GEO dataset GSE210434 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE210434) was used to assess the differential gene expression between fibroblasts derived from HS tissues compared to NS tissues. The online bioinformatic database JASPAR (https://jaspar.elixir.no/) was utilized to predict the binding relationship between HOXC10 and the promoter sequence of STMN2.
Statistical analysis
The statistical evaluation of the data was conducted using SPSS version 20.0 (IBM, Armonk, NY, USA). Results are presented as the mean ± standard deviation, derived from a minimum of three independent experiments. Differences between two groups were assessed using unpaired Student’s t-tests, while comparisons among more than two groups were performed through one-way analysis of variance (ANOVA), followed by Tukey’s post hoc test for multiple comparisons. A p value < 0.05 was deemed indicative of statistical significance.
Results
HOXC10 and STMN2 were upregulated in skin tissues from HS patients
The analysis of high-throughput sequencing expression profiles from the GEO dataset GSE210434 revealed that 65 genes were significantly upregulated in the HS group compared with the NS group (Fig. 1a). Among these genes, HOXC10, highlighted as the prominent round red dot, exhibited the highest expression levels and was therefore selected for further experiments. Both gene and protein expression levels of HOXC10 were increased in human tissues from HS patients compared with those from the control group (Fig. 1b, c). As shown by the high-throughput sequencing expression profiles from the GEO dataset GSE210434, STMN2 expression, highlighted as the prominent round red dot, was found to be upregulated in the skin tissue of HS patients (Fig. 1d). Furthermore, qRT–PCR and western blot data confirmed that STMN2 was elevated in the skin tissues derived from HS patients in relation to those from the NS group (Fig. 1e). These data suggest that HOXC10 and STMN2 may play critical roles in the regulation of HS formation.
HOXC10 and STMN2 were upregulated in skin tissues from HS patients. a High-throughput sequencing expression profile analysis of the GEO dataset GSE210434 revealed transcriptomic differences between fibroblasts from normal scar (NS) tissue and hypertrophic scar (HS) tissue. b The relative mRNA expression of HOXC10 in NS and HS tissues was assessed via RT–qPCR, n = 20. c The protein expression of HOXC10 in NS and HS tissues was assessed via western blot, n = 3. d The same high-throughput sequencing analysis indicated that STMN2 was upregulated in HS tissues compared to NS tissues, n = 3. e The relative mRNA expressions of STMN2 in NS and HS tissues were examined via RT–qPCR, n = 20. f The protein expressions of STMN2 in NS and HS tissues were examined via western blot, n = 3. *p < 0.05, **p < 0.01, ***p < 0.001
Knockdown of HOXC10 inhibited HSF viability, proliferation, migration, and fibrosis.
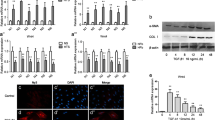
To confirm the regulatory effect of HOXC10 on HSF behavior, HOXC10 was knocked down or overexpressed in HSFs (Fig. 3d, e). Overexpression of HOXC10 significantly enhanced the HSF viability at 24, 48, and 72 h, increased proliferation at 24 h, and promoted migration at 24 h (Figs. 2a–d). In contrast, HOXC10 silencing yielded opposite effects. Furthermore, HOXC10 overexpression markedly increased the expression of collagen I, collagen III, and α-SMA in HSFs (Fig. 2e), while HOXC10 depletion reduced the expression levels of these proteins. Collectively, these findings indicate that viability, proliferation, migration, and fibrotic responses in HSFs were inhibited following HOXC10 knockdown, whereas they were augmented upon HOXC10 overexpression.
Knockdown of HOXC10 inhibited HSF viability, proliferation, migration, and fibrosis. HOXC10 was knocked down or overexpressed in human skin fibroblasts (HSFs) by transfecting them with sh-HOXC10 or shNC for knockdown, and oe-HOXC10 or oe-NC for overexpression. a Cell viability was assessed using the CCK-8 assay after 24, 48, and 72 h. b Cell proliferation was investigated using the EdU assay after 24 h (scale bar, 100 µm). c–d The migration of the HSFs was examined using both the scratch assay after 24 h and the Transwell assay after 48 h (scale bar, 100 µm). e The expression levels of collagen I, collagen III, and α-SMA were detected by western blot, n = 3. The length of the scale bar is 100 µm. *p < 0.05, **p < 0.01, ***p < 0.001
HOXC10 targeted STMN2 and regulated its expression
To further elucidate the downstream mechanisms underlying HOXC10-mediated modulation of HSF behavior, we utilized the JASPAR database to predict the presence of a potential HOXC10 binding consensus sequence (GTAATTAAA) within the STMN2 promoter, identified between positions 766 and 774 (Fig. 3a). Support for this hypothesis was provided by dual-luciferase reporter assays and ChIP experiments, which verified HOXC10’s direct interaction with the STMN2 promoter, thus implicating it in the transcriptional regulation of STMN2 (Figs. 3b, c). Notably, HOXC10 overexpression resulted in elevated levels of both HOXC10 and STMN2 (Fig. 3d, e), while silencing HOXC10 led to a decrease in their expressions. Collectively, these findings demonstrate that HOXC10 directly binds to the STMN2 promoter, serving as a positive regulator of its expression.
HOXC10 targeted STMN2 and regulated its expression. a The JASPAR database predicted the binding relationship between HOXC10 and the promoter sequence of STMN2 (https://jaspar.elixir.no/). b The luciferase reporter gene assay was used to determine the activity of the STMN2 promoter in HEK293T cells transfected with either wild-type (WT) or mutated HOXC10. c The ChIP assay was utilized to detect the binding of HOXC10 to the STMN2 promoter in HSFs. d,e) PCR and western blot were used to detect the expression levels of HOXC10 and STMN2 in HSFs, n = 3. *p < 0.05, **p < 0.01, ***p < 0.001
The stimulatory effect of STMN2 overexpression on HSF viability, proliferation, migration, and fibrosis was reversed by HOXC10 knockdown
To delve deeper into the contributions of STMN2 and HOXC10 to HSF behavior, we conducted experiments involving the overexpression of STMN2 combined with the knockdown of HOXC10 in HSFs. Overexpression of STMN2 led to enhanced HSF viability at 24, 48, and 72 h, increased proliferation at 24 h, and promoted migration at 24 h (Figs. 4a–d). However, these effects were diminished upon HOXC10 knockdown. The levels of collagen I, collagen III, α-SMA, TGF-β, phosphorylated Smad2 (p-Smad-2), and phosphorylated Smad3 (p-Smad-3) were elevated with STMN2 overexpression (Fig. 4e). However, the stimulatory impact of STMN2 on the expression of these proteins was negated following HOXC10 depletion. In summary, the increased HSF viability, proliferation, migration, fibrosis, and activation of the TGF-β/Smad signaling pathway induced by STMN2 overexpression were mitigated by HOXC10 knockdown.
The stimulatory effect of STMN2 overexpression on HSF viability, proliferation, migration, and fibrosis was reversed by HOXC10 knockdown. STMN2 was overexpressed in human skin fibroblasts (HSFs) by transfection with oe-STMN2, with oe-NC serving as the overexpression control. Additionally, HOXC10 knockdown was achieved using sh-HOXC10, with shNC as the knockdown control. a Cell viability was assessed using the CCK-8 assay after 24, 48, and 72 h. b Cell proliferation was investigated using the EdU assay after 24 h (scale bar, 100 µm). c,d The migration of the HSFs was examined using both the scratch assay after 24 h and the Transwell assay after 48 h (scale bar, 100 µm). e The expression levels of collagen I, collagen III, α-SMA, β-SMA, p-Smad-2, and p-Smad-3 were detected by western blot, n = 3. The length of the scale bar is 100 µm. *p < 0.05, **p < 0.01, ***p < 0.001
HOXC10 enhanced HSF viability, proliferation, migration, and fibrosis by activating the TGF-β/Smad signaling pathway
To verify the involvement of the TGF-β/Smad signaling pathway in the HOXC10-regulated behavior of HSFs, the pathway inhibitor LY2109761 was applied to HSFs overexpressing HOXC10. The addition of LY2109761 notably counteracted the enhancing effects of HOXC10 overexpression on HSF viability at 24, 48, and 72 h, as well as on proliferation and migration at 24 h (Figs. 5a–d). Moreover, the increase in protein expression induced by HOXC10 overexpression, including collagen I, collagen III, α-SMA, TGF-β, phosphorylated Smad2 (p-Smad-2), and phosphorylated Smad3 (p-Smad-3), was diminished following LY2109761 treatment (Fig. 5e). Taken together, the activation of the TGF-β/Smad signaling pathway is critical in mediating the effects of HOXC10 on HSF viability, proliferation, migration, and fibrosis.
HOXC10 enhanced HSF viability, proliferation, migration, and fibrosis by activating the TGF-β/Smad signaling pathway. HOXC10 was overexpressed in human skin fibroblasts (HSFs) by transfecting them with oe-HOXC10 or oe-NC. Afterward, the HSFs were treated with LY2109761. a Cell viability was assessed using the CCK-8 assay after 24, 48, and 72 h. b Cell proliferation was investigated using the EdU assay after 24 h (scale bar, 100 µm). c–d The migration of the HSFs was examined using both the scratch assay after 24 h and the Transwell assay after 48 h (scale bar, 100 µm). e The expression levels of collagen I, collagen III, α-SMA, β-SMA, p-Smad-2, and p-Smad-3 were detected by western blot. n = 3. The length of the scale bar is 100 µm. *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
HS, characterized by an overgrowth of fibrous tissue, results from the overactive proliferation of fibroblasts and increased collagen accumulation (Zhou et al. 2023). The pathophysiology of HS shares similarities with cancer, including unregulated cell growth, a hypoxic environment within cells, and an imbalance between epithelial and mesenchymal tissues (Chen et al. 2023). Research has shown that HOXC10 is significantly implicated in oncogenesis (Fang et al. 2021). The expression of HOXC10 was abnormally elevated in HS compared to NS (Kang et al. 2023), suggesting a potential role of HOXC10 in regulating HS. Despite these findings, the specific mechanisms by which HOXC10 contributes to these processes, particularly in the context of HS, remain to be clarified. In our research, we found that both HOXC10 and STMN2 exhibited higher expression levels in HS tissues in comparison to NS tissues. Silencing HOXC10 led to a decrease in HSF activation, proliferation, migration, and fibrosis. Additionally, our findings indicate that HOXC10 directly targeted STMN2. HOXC10 knockdown counteracted the promotional effects of STMN2 overexpression on HSF activation, proliferation, migration, and fibrosis. We further established that HOXC10 influenced HSF activities through the TGF-β/Smad signaling pathway. This investigation lays the groundwork for potentially utilizing HOXC10 as a therapeutic target in the management of HS.
HOXC10, a member of the HOX gene family, was documented to facilitate various types of tumor tumorigenesis and progression. For example, HOXC10 led to an increased proliferation, migration, and invasion of glioma cell lines (Li et al. 2018). Another study indicated that silencing HOXC10 inhibited cell proliferation and induced apoptosis in osteosarcoma cells by modulating caspase 3 activity. Additionally, as a highly conserved transcription factor, HOXC10 has been identified to collaborate with multiple target molecules, contributing to the onset and progression of cancer (Kang et al. 2023). Nevertheless, the role of HOXC10 in the regulation of HS has been infrequently documented. Recent studies have discovered that HOXC10 expression is upregulated in HS tissues compared to NS tissues. Consistent with these findings, we also observed abnormally high levels of HOXC10 expression in HS tissues. Moreover, silencing HOXC10 significantly reduced the proliferation, migration, and fibrosis of HSF, confirming the role of HOXC10 in mediating HS.
Our research further revealed that HOXC10 targets the promoter region of STMN2, leading to an upregulation of STMN2 expression in HS. The knockdown of HOXC10 mitigated the increase in HSF proliferation, migration, and fibrosis caused by overexpression of STMN2. In line with these findings, STMN2 has been identified as a key factor in fibrosis regulation (Paradis et al. 2010; Asselah et al. 2005). Previous studies have shown an increase in STMN2 levels in hepatic fibrosis, linking it to the severity of the fibrosis stage (Paradis et al. 2010; Asselah et al. 2005). STMN2 is known to promote fibrogenesis through the activation of sympathetic neurotransmitters (Paradis et al. 2003). However, our study is the first to report the interaction between HOXC10 and STMN2, highlighting their collective role in the fibrosis of HSF in HS.
The TGF-β/Smad signaling pathway stands as the central regulator of collagen synthesis in fibroblasts and myofibroblasts (Finnson et al. 2020). Its prolonged activation causes the sustained overactivation of these cells, crucial for the abnormal collagen deposition seen in scar formations, including HS (Zhang et al. 2020a). Consequently, therapeutic approaches have been developed to inhibit the TGF-β/Smad signaling pathway (Zhang et al. 2020a). For instance, silencing eIF3a was shown to mitigate TGF‑β1‑induced α-SMA and collagen I protein expression in keloid fibroblasts (Li and Zhao 2018). Additionally, the knockdown of microRNA-21 decreased the expression of fibrosis markers such as collagen I, collagen III, fibronectin, and α-SMA by inhibiting the TGF-β/Smad pathway (Li et al. 2016). In our research, we further elucidated the pivotal role of the TGF-β/Smad signaling pathway in HOXC10-mediated HS. We demonstrated that HOXC10 promotes HSF activation, proliferation, migration, and fibrosis through this signaling pathway.
In conclusion, our study has elucidated that HOXC10 facilitates HSF activation, proliferation, migration, and fibrosis by upregulating the expression of its downstream target, STMN2, through the activation of the TGF-β/Smad signaling pathway. Our findings suggest that targeting HOXC10 could represent a promising strategy for developing novel therapeutic interventions for HS.
While the use of human HSFs provides valuable insights in this study, a significant limitation is the potential impact of fibroblast origin on HOXC10 expression and function. Fibroblasts from different embryonic origins exhibit distinct HOX expression profiles, leading to heterogeneity in their behavior. This variability could influence how HOXC10 operates in the context of HS and potentially affect the generalizability of our findings. Future research should include fibroblasts from multiple body sites to ensure broader applicability of the results. Furthermore, these in vitro models may not fully capture the complex in vivo environment of HS. Future studies should employ more comprehensive in vivo models to validate our findings and provide a more accurate representation of the disease’s pathological processes.
Data availability
The data that support the fndings of this study are not openly available and are available from the corresponding author upon reasonable request.
Abbreviations
- α-SMA:
-
α-Smooth muscle actin
- ANOVA:
-
One-way analysis of variance
- CCK-8:
-
Cell counting kit-8
- ChIP:
-
Chromatin immunoprecipitation
- ECL:
-
Enhanced chemiluminescence
- FBS:
-
Fetal bovine serum
- HOX:
-
Homeobox
- HS:
-
Hypertrophic scars
- HSFs:
-
Hypertrophic scar fibroblasts
- NS:
-
Normal skin
- NSFs:
-
Normal skin fibroblasts
- PBS:
-
Phosphate-buffered saline
- PVDF:
-
Polyvinylidene fluoride
- RIPA:
-
Radio-immunoprecipitation assay
- shRNAs:
-
Short hairpin RNAs
- VSMCs:
-
Vascular smooth muscle cells
References
Asselah T, Bièche I, Laurendeau I et al (2005) Liver gene expression signature of mild fibrosis in patients with chronic hepatitis C. Gastroenterology 129(6):2064–2075. https://doi.org/10.1053/j.gastro.2005.09.010
Chen Z, Han F, Du Y et al (2023) Hypoxic microenvironment in cancer: molecular mechanisms and therapeutic interventions. Signal Transduct Target Ther 8(1):70. https://doi.org/10.1038/s41392-023-01332-8
Chiang RS, Borovikova AA, King K et al (2016) Current concepts related to hypertrophic scarring in burn injuries. Wound Repair Regen 24(3):466–477. https://doi.org/10.1111/wrr.12432
Čoma M, Fröhlichová L, Urban L et al (2021) Molecular changes underlying hypertrophic scarring following burns involve specific deregulations at all wound healing stages (inflammation, proliferation and maturation). Int J Mol Sci. https://doi.org/10.3390/ijms22020897
Fang J, Wang J, Yu L et al (2021) Role of HOXC10 in cancer. Front Oncol 11:684021. https://doi.org/10.3389/fonc.2021.684021
Finnson KW, Almadani Y, Philip A (2020) Non-canonical (non-SMAD2/3) TGF-β signaling in fibrosis: mechanisms and targets. Semin Cell Dev Biol 101:115–122. https://doi.org/10.1016/j.semcdb.2019.11.013
He X, Wang H, Wang R et al (2023) HOXC10 promotes esophageal squamous cell carcinoma progression by targeting FOXA3 and indicates poor survival outcome. Heliyon 9(10):e21056. https://doi.org/10.1016/j.heliyon.2023.e21056
Hu Y, Zhou J (2023) Identification of key genes and functional enrichment analysis of liver fibrosis in nonalcoholic fatty liver disease through weighted gene co-expression network analysis. Genomics Inform 21(4):e45. https://doi.org/10.5808/gi.23051
Kang M, UHK, E-JO et al. (2023) Differential expression of tension-sensitive HOX genes in fibroblasts is associated with different scar types. Life Science Weekly, 2023/07/18/, p 1063
Ke X, Guo W, Peng Y et al (2022) Investigation into the role of Stmn2 in vascular smooth muscle phenotype transformation during vascular injury via RNA sequencing and experimental validation. Environ Sci Pollut Res Int 29(3):3498–3509. https://doi.org/10.1007/s11356-021-15846-7
Li T, Zhao J (2018) Knockdown of elF3a inhibits TGF-β1-induced extracellular matrix protein expression in keloid fibroblasts. Mol Med Rep 17(3):4057–4061. https://doi.org/10.3892/mmr.2017.8365
Li G, Zhou R, Zhang Q et al (2016) Fibroproliferative effect of microRNA-21 in hypertrophic scar derived fibroblasts. Exp Cell Res 345(1):93–99. https://doi.org/10.1016/j.yexcr.2016.05.013
Li S, Zhang W, Wu C et al (2018) HOXC10 promotes proliferation and invasion and induces immunosuppressive gene expression in glioma. FEBS J 285(12):2278–2291. https://doi.org/10.1111/febs.14476
Liang Q, Pan F, Qiu H et al (2024) CLC-3 regulates TGF-β/Smad signaling pathway to inhibit the process of fibrosis in hypertrophic scar. Heliyon 10(3):e24984. https://doi.org/10.1016/j.heliyon.2024.e24984
Lingzhi Z, Meirong L, Xiaobing F (2020) Biological approaches for hypertrophic scars. Int Wound J 17(2):405–418. https://doi.org/10.1111/iwj.13286
Paradis V, Bièche I, Dargère D et al (2003) Molecular profiling of hepatocellular carcinomas (HCC) using a large-scale real-time RT-PCR approach: determination of a molecular diagnostic index. Am J Pathol 163(2):733–741. https://doi.org/10.1016/s0002-9440(10)63700-5
Paradis V, Dargere D, Bieche Y et al (2010) SCG10 expression on activation of hepatic stellate cells promotes cell motility through interference with microtubules. Am J Pathol 177(4):1791–1797. https://doi.org/10.2353/ajpath.2010.100166
Rabello FB, Souza CD, Farina Júnior JA (2014) Update on hypertrophic scar treatment. Clinics (sao Paulo) 69(8):565–573. https://doi.org/10.6061/clinics/2014(08)11
Scott GA, Goldsmith LA (1993) Homeobox genes and skin development: a review. J Invest Dermatol 101(1):3–8. https://doi.org/10.1111/1523-1747.ep12358258
Stelnicki EJ, Kömüves LG, Kwong AO et al (1998) HOX homeobox genes exhibit spatial and temporal changes in expression during human skin development. J Invest Dermatol 110(2):110–115. https://doi.org/10.1046/j.1523-1747.1998.00092.x
Tai Y, Woods EL, Dally J et al (2021) Myofibroblasts: function, formation, and scope of molecular therapies for skin fibrosis. Biomolecules. https://doi.org/10.3390/biom11081095
Yu J, Chen X, Zhao S et al (2022) HOXC10 promotes metastasis in colorectal cancer by recruiting myeloid-derived suppressor cells. J Cancer 13(12):3308–3317. https://doi.org/10.7150/jca.76945
Zhang T, Wang X-F, Wang Z-C et al (2020a) Current potential therapeutic strategies targeting the TGF-β/Smad signaling pathway to attenuate keloid and hypertrophic scar formation. Biomed Pharmacother 129:110287. https://doi.org/10.1016/j.biopha.2020.110287
Zhong FJ, Sun B, Cao MM et al (2021) STMN2 mediates nuclear translocation of Smad2/3 and enhances TGFβ signaling by destabilizing microtubules to promote epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett 506:128–141. https://doi.org/10.1016/j.canlet.2021.03.001
Zhou S, Xie M, Su J et al (2023) New insights into balancing wound healing and scarless skin repair. J Tissue Eng 14:20417314231185850. https://doi.org/10.1177/20417314231185848
Funding
None.
Author information
Authors and Affiliations
Contributions
Xin Zhou carried out the conceptualization, investigation, resources, software, validation, visualization, writing—original draft, and writing—review and editing. Song Lin carried out the investigation, data curation, formal analysis, methodology, project administration, supervision, and writing—review and editing. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, X., Lin, S. HOXC10 promotes hypertrophic scar fibroblast fibrosis through the regulation of STMN2 and the TGF-β/Smad signaling pathway. Histochem Cell Biol 162, 403–413 (2024). https://doi.org/10.1007/s00418-024-02317-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-024-02317-6