Abstract
Following skeletal muscle injury, both myogenic and immune cells interact closely during the regenerative process. Although icing is still a common acute treatment for sports-related skeletal muscle injuries, icing after muscle injury has been shown to disrupt macrophage accumulation and impair muscle regeneration in animal models. However, it remains unknown whether icing shortly after injury affects macrophage-related phenomena during the early stages of muscle regeneration. Therefore, we focused on the distribution of M1/M2 macrophages and cytokines expressed predominantly by macrophages during the early stages of muscle regeneration after muscle crush injury. Icing resulted in a decrease, not retardation, in the accumulation of M1 macrophages, but not M2 macrophages, in injured muscles. Consistent with the decrease in M1 macrophage accumulation, icing led to a reduction, instead of delay, in the level of tumor necrosis factor-α (TNF-α) expression. Additionally, at subsequent timepoints, icing decreased the number of myogenic precursor cells in the regenerating area and the size of centrally nucleated regenerating myofibers. Together, our findings suggest that icing after acute muscle damage by crushing disturbs muscle regeneration through hindering tM1 macrophage-related phenomena.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Skeletal muscle regeneration is a tightly coordinated process that requires the interaction between skeletal muscle and the immune system (Tidball 2011). Following muscle injury, satellite cells, i.e., muscle stem cells, activate and differentiate into myoblasts, which then proliferate (Huard et al. 2002; Sciorati et al. 2016). Following cell proliferation, myoblasts terminally differentiate and fuse with each other, forming centrally nucleated regenerating myofibers that restore preinjury conditions (Huard et al. 2002; Sciorati et al. 2016). During the muscle regeneration process, macrophages, which are the most abundant immune cell population, exert their influence as two phenotypically distinct subsets, M1 and M2 macrophages (also known as proinflammatory and antiinflammatory macrophages, respectively) (Mantovani et al. 2004; Sciorati et al. 2016; Tidball 2017). To facilitate the restoration process, therapeutic interventions are applied to damaged skeletal muscles (Freitas et al. 2007; Jarvinen et al. 2007; Li et al. 2010; Oyaizu et al. 2018; Kawashima et al. 2021a, b). Among these interventions, icing has traditionally been used as one of the acute treatments to inhibit the inflammatory response, including pain and swelling, following soft-tissue injury (Swenson et al. 1996; Bleakley et al. 2004). However, several studies using animal models have reported that icing after skeletal muscle injury impairs muscle regeneration (Takagi et al. 2011; Shibaguchi et al. 2016; Kawashima et al. 2021a, b). Notably, previous studies showed that icing after muscle injury delays the accumulation of macrophages and impairs muscle regeneration (Takagi et al. 2011; Kawashima et al. 2021a, b). However, the macrophage-related phenomena that are responsible for the impairment of muscle regeneration by icing remain unclear.
Following skeletal muscle injury, monocytes are recruited from blood vessels and differentiate into M1 macrophages (Sciorati et al. 2016). Then M1 macrophages infiltrate into the injured sites, release inflammatory cytokines, and phagocytose necrotic muscle debris (Arnold et al. 2007; Smith et al. 2008). M1 macrophages then undergo a phenotypic transition to M2 macrophages during muscle regeneration, where they perform a reparative function during the later stages of the regenerative process (Vannella and Wynn 2016; Tidball 2017; Santa et al. 2019). Although inflammation has been considered harmful (Swenson et al. 1996), the inflammatory response involving M1 macrophages is indispensable to complete skeletal muscle regeneration (Tidball 1995; Wang et al. 2014; Chazaud 2020). M1 macrophages express multiple inflammatory cytokines involved in muscle regeneration, one of which is represented by tumor necrosis factor-α (TNF-α), which promotes the proliferation of myoblasts (Li 2003; Tidball et al. 2014; Santa et al. 2019). While previous studies have shown that icing delays macrophage recruitment during the early stages of muscle regeneration (Takagi et al. 2011; Kawashima et al. 2021a, b), whether icing affects important phenomena such as M1 macrophage accumulation, proinflammatory cytokine expression, and myoblast proliferation, occurring in the early stages of muscle regeneration, is still poorly understood.
The aim of the present study was to examine the chronological changes in M1/M2 macrophage accumulation and the expression of macrophage-related cytokines during the early stages of the icing-induced muscle regenerative process. In this study, icing after muscle injury decreased the accumulation of M1 macrophages, which was accompanied by decreased TNF-α expression, decreased Pax7+ cell accumulation, and eventually delayed muscle regeneration. Thus, our results demonstrated that disruption of M1 macrophage-related events occurs during the early stages of icing-treated muscle regeneration.
Material and methods
Animals
One hundred and twenty-four 8 week-old male Wistar rats weighing 180–200 g (Japan SLC Inc., Shizuoka, Japan) were used in the study and allowed free access to food and water throughout the experiments. All animals were housed in a controlled environment maintained at 22 ± 2 °C with a 12-h light and dark cycle. Additionally, this study was approved by the Institutional Animal Care and Use Committee and was conducted in accordance with the Kobe University Animal Experimentation Regulations (approval numbers P180706 and P200906).
Experimental protocol
Muscle injury was induced by crushing the extensor digitorum longus (EDL) muscle according to previous reports (Takagi et al. 2011; Hatade et al. 2014; Takeuchi et al. 2014; Miyakawa et al. 2020; Kawashima et al. 2020). The animals were anesthetized by isoflurane inhalation, the anterolateral surface of their left hindlimb was shaved, and a longitudinal incision approximately 20 mm long was made to expose the EDL muscle. The middle section of the muscle belly was crushed for 30 s with forceps attached to a 500 g weight. Immediately after the injury, the skin was closed with a 4–0 suture. The animals were then randomly divided into two groups: non-icing and icing. Five minutes after the injury, the animals in the icing group were given ice packs made by enclosing crushed ice in a polyethylene bag. The surface temperature of the ice pack ranged from 0.3 °C to 1.3 °C. For 20 min, an ice pack was applied transcutaneously to the injured EDL muscle with minimum compression controlled to merely touching the skin. In our previous investigation using the same subjects and icing method (Takagi et al. 2011), icing could decrease muscle surface temperature by more than 10 °C, but caused little decrease in rectal temperature. At 6 and 12 h after injury, as well as 1, 2, 3, 4, 5, 6, and 7 days after injury, the animals were euthanized, and EDL muscles were harvested. These samples were immediately frozen in dry-ice-cooled acetone and stored at a constant temperature of −80 °C until analysis. The samples were used for morphological and histochemical analyses at all timepoints following injury (n = 5 per group at each timepoint). Additionally, the samples were used for western blot analysis 1, 2, and 3 days following injury (n = 4 per group at each timepoint).
Morphological analysis
Transverse cryosections (10 μm) of frozen EDL muscles were cut and mounted on glass slides using a cryostat (CM-3050S or CM-1850, Leica Microsystems, Wetzlar, Germany). As described below, some sections were stained with hematoxylin and eosin (H&E), while others were used for immunohistochemistry. By staining with H&E and comparing with morphological characteristics in figures of previous reports (Goetsch et al. 2003; Srikuea et al. 2010; Takagi et al. 2011; Takeuchi et al. 2014), every section made in this study exhibited the features of an injury site. All H&E-stained sections were viewed using an Olympus BX50 microscope (Olympus, Tokyo, Japan) equipped with Olympus UplanApo 20×/0.70 NA or UplanApo 40×/0.85 NA objectives. The images were recorded with an EOS Kiss X8i camera (maximum resolution 6000 × 4000 pixels, Canon Inc., Tokyo, Japan) using the EOS utility software (Canon Inc.).
Immunohistochemistry
Cryosections were air-dried for 15 min at room temperature, fixed with 4% paraformaldehyde, washed in phosphate-buffered saline (PBS), blocked and permeabilized for 1 h at room temperature with PBS containing 10% normal goat serum and 1% Triton X 100, and incubated overnight at 4 °C or for 1 h at room temperature with primary antibodies. A mouse monoclonal anti-CD68 antibody (1:200, MCA341GA, Batch# 0515, Bio-Rad, Hercules, CA, USA, RRID:AB_566872), which is a glycosylated type I membrane protein predominantly expressed in late endosomes and lysosomes of macrophages, was used to detect macrophages in this study. Because the datasheet for the anti-F4/80 antibody (ab6640, Lot#GR3327073-2; Abcam, Cambridge, UK, RRID:AB_1140040) used in the previous study (Kawashima et al. 2021a, b) contains information that this antibody recognizes the antigen of mouse F4/80 that is expressed by murine macrophages, it was impossible to use the same antibody in this study using rats. According to the datasheet of mouse monoclonal anti-CD68 antibody (MCA341GA, Batch# 0515, Bio-Rad, Hercules, CA, USA, RRID:AB_566872), this antibody recognizes rat ED1 antigen (known as CD68), and the CD68 expression has been observed in both M1 and M2 macrophages in rats (Fujita et al. 2014; Fan et al. 2021). In addition to the anti-CD68 antibody, rabbit polyclonal anti-mannose receptor (CD206) antibody (1:2000, ab64693, lot# GR3256060-1, Abcam, Cambridge, UK, RRID:AB_1523910), which is a C-type lectin primarily present on the surface of M2 macrophages, rabbit polyclonal anti-iNOS antibody (1:100, ab3523, lot# GR3230433-8, Abcam, RRID:AB_303872), which is one of the isoenzymes of nitric oxide synthase and is highly expressed by lipopolysaccharide and other stimuli induced by M1 macrophages, mouse monoclonal anti-Pax7 antibody (1:50, sc-81648, lot# G1916, Santa Cruz Biotechnology, CA, USA, RRID:AB_2159836), rabbit polyclonal anti-TNF-α antibody (1:100, ab6671, lot# GR3214221-23, Abcam, RRID:AB_305641), and rabbit polyclonal anti-dystrophin antibody (1:200, sc-15376, lot# F0910, Santa Cruz Biotechnology, RRID:AB_2091230) were used as primary antibody and were diluted with PBS containing 5% normal goat serum. After washing the sections three times in PBS, the samples were visualized for 2 h at room temperature with appropriate species-specific Alexa Fluor 488 and/or 568 or 594 fluorescence-conjugated secondary antibodies (1:1000, Thermo Fisher Scientific, Waltham, MA, USA) diluted in PBS containing 5% normal goat serum. Following a PBS wash, the sections were mounted in a Vectashield mounting medium (Vector Laboratories, Burlington, ON, Canada) containing 4′, 6-diamino-2-phenylindole (DAPI), to visualize the nuclei. The samples were then viewed on a ZEISS Axio Vert. A1 with an objective (LD A-Plan 40×/0.55 NA, Zeiss, Oberkochen, Germany) and photographed with an AxioCam MRm (maximum resolution 1388 × 1040 pixels) using the AxioVision Rel. 4.8 software (Zeiss).
Control experiments were conducted with the undamaged muscle, using double staining for CD68 and iNOS or CD206, and single staining for TNF-α and Pax7. These experiments were performed as positive or negative control of these antibodies in rat muscles. In these control experiments, animals of the same strain and age with those in this study were used. In the undamaged muscle sample, no CD68+iNOS+ cells and TNF-α could be detected (Supplementary Fig. 1a–d, i–k). Meanwhile, several CD68+CD206+ cells were observed in the undamaged muscle (Supplementary Fig. 1e–h), which may indicate tissue-resident macrophages (Watanabe et al. 2019). Pax7+ cell was observed at the periphery of skeletal muscle myofiber (Supplementary Fig. 1l–n), where quiescent satellite cells are known to exist (Yin et al. 2013).
Quantification of muscle regeneration
Muscle regeneration was quantified using the previously described method of measuring the cross-sectional area (CSA) of centrally nucleated myofibers, which indicated regeneration of myofibers at 7 days after injury (Takagi et al. 2011). In the sections used for H&E staining, the CSA of 100 muscle fibers with a central nucleus at 7 days after injury was determined using ImageJ software (http://rsbweb.nih.gov/ij).
Quantitative analyses of macrophages and Pax7+ cells
The identification of M1 and M2 macrophages was conducted in accordance with a report published previously (Kawashima et al. 2021a, b). CD68 (a pan-macrophage marker expressed in M1 and M2 macrophages; Taylor et al. 2005), iNOS (a representative marker of M1 macrophages), and CD206 (a representative marker of M2 macrophages) were used for immunolabeling. M1 macrophages were identified as CD68+iNOS+ cells using double immunolabeling for CD68 and iNOS (e.g., Bencze et al. 2012; Le Moal et al. 2018; Kawashima et al. 2021a, b). According to previous reports, CD68+CD206− cells were also quantified as M1 macrophages (Tidball and Villalta 2010; Villalta et al. 2011; Kawashima et al. 2021a, b). M2 macrophages were identified as CD68+CD206+ cells using double immunolabeling for both (Villalta et al. 2011; Rayagiri et al. 2018). Additionally, the following method was used to quantify the number of macrophages and Pax7+ cells. To summarize, five non-overlapping injured fields per animal were captured at ×40 magnification using an objective lens. Furthermore, macrophages and Pax7+ cells were counted and expressed as number of cells per field. A total of five fields were averaged, and a single value was recorded per animal. All quantitative analyses were performed using the ImageJ software described above, and positive cells were detected by one observer (A.M.).
Western blot analysis
Portions (approximately 25 mg) of frozen injured muscle samples were homogenized in PRO-PREP (iNtRON Biotechnology Inc., Gyeonggi-do, Korea). The homogenates were centrifuged at 15,000g for 30 min at 4 °C, and supernatants were collected. Proteins (50 μg per lane) were separated by 7.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (GE Healthcare, Chalfont St Giles, UK). After blocking for 1 h in Tris-buffered saline with Tween 20 (TBS-T) containing 20 g/L skim milk or 20 g/L albumin bovine, F–V (Nacalai Tesque, Inc., Kyoto, Japan), the membranes were incubated overnight at 4 °C with the primary antibodies, rabbit polyclonal anti-TNF-α antibody (1:500, ab6671, lot# GR3214221-23, Abcam, RRID: AB_305641) and rabbit polyclonal anti-insulin-like growth factor (IGF)-1A antibody (1:500, 20,214–1-AP, lot# 00,039,622, Proteintech, Chicago, IL, RRID: AB_10666736). The membranes were then incubated for 1 h at room temperature with horseradish peroxidase (HRP)-conjugated species-specific secondary antibodies. Blots were developed using the Luminata Forte Western HRP Substrate (WBLUF0100; Merck Millipore) and recorded with OptimaShot CL-420α (Fujifilm Wako Pure Chemical Co., Osaka, Japan). The concentrations of the corresponding protein bands were analyzed by using ImageJ software. The expression levels of the samples were normalized to those of the standard samples and expressed as fold changes relative to the values in the non-icing group at 1 day after injury.
Statistical analysis
Data are expressed as mean ± standard error. Comparisons were made using Welch’s t-test or a two-way analysis of variance (ANOVA) (group × time, group × CSA distribution), followed by Tukey’s post hoc test. The statistical differences were considered significant at P < 0.05. Additionally, the statistical analyses were performed using the R statistical package (version 4.0.5, R Foundation for Statistical Computing Platform, Vienna, Austria).
Results
Morphological features of the muscle regenerative process in the non-icing and icing groups
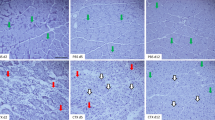
One day after skeletal muscle injury, necrotic myofibers were observed as swollen and round cells with faintly stained sarcoplasm, and several mononuclear cells infiltrated the necrotic myofibers in both the non-icing and icing groups (Fig. 1a, e). At 2 days, the contours of necrotic myofibers in the non-icing group became unrecognizable with the robust mononuclear cell infiltration (Fig. 1b), whereas the infiltration of mononuclear cells was mild in the icing group (Fig. 1f). At 3 days, some myotubes, which were formed by the fusion of the myoblasts, were observed in the non-icing group (Fig. 1c, d). In contrast, the icing group retained a high number of mononuclear cells while myotubes were barely detectable at the same timepoint (Fig. 1g). These morphological results were consistent with previous research demonstrating that icing delayed muscle regeneration following muscle injury (Takagi et al. 2011; Miyakawa et al. 2020).
Morphological characteristics of the regenerating process after skeletal muscle injury by hematoxylin and eosin staining. a–g Transverse sections of the extensor digitorum longus muscle in the non-icing (a–d) and icing (e–g) groups at 1 day (a, e), 2 days (b, f), and 3 days (c, d, g) after injury. Asterisks indicate the necrotic myofibers. Arrowheads indicate myotubes. d A magnified image of boxed area in c. Scale bars, 30 µm
To evaluate muscle regeneration, the CSA of the regenerating muscle fibers characterized by their central nucleus was determined at 7 days after injury (Fig. 2). Quantification of the CSAs revealed those in the icing group (652.91 ± 17.86 µm2) to be significantly less than those in the non-icing group (798.33 ± 50.97 µm2) (Fig. 2c). At 7 days post-injury, the distribution of CSA revealed that the icing group had a higher proportion of centrally nucleated myofibers smaller than 500 µm2 than the non-icing group (Fig. 2d), suggesting that icing weakens muscle regeneration.
Comparison of regenerating myofiber size at 7 days after skeletal muscle injury. a, b Transverse sections of the extensor digitorum longus muscle stained with hematoxylin and eosin in non-icing (a) and icing (b) groups. Arrowheads indicate regenerating muscle fibers with a central nucleus. Scale bars, 60 µm. c Mean cross-sectional area (CSA) of regenerating muscle fibers. d CSA distribution of regenerating muscle fibers. Data pooled from 100 fibers per animal. Values are expressed as mean ± standard error (n = 5), and statistical significance was assessed with Welch’s t-test (c) or two-way analysis of variance followed by Tukey’s post hoc test (d). The interaction between group and time is significant in d.*P < 0.05, **P < 0.01; statistical difference between groups
Icing after skeletal muscle injury diminishes M1 macrophage accumulation
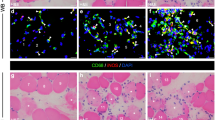
To identify the mononuclear cells observed in injured myofibers, double immunolabeling with CD68 (a pan-macrophage marker; Taylor et al. 2005) and iNOS (a representative M1 macrophage marker; e.g., Bencze et al. 2012; Le Moal et al. 2018; Kawashima et al. 2021a, b) was performed (Fig. 3). M1 macrophages were identified as cells that were double-positive for both markers. At 6 h after injury, several M1 macrophages were detected at the injured sites for both the groups (Fig. 3a, e). Subsequently, until 1 day after injury, the number of M1 macrophages increased in both the groups (Fig. 3b, f, i, j). Correspondingly, a remarkable accumulation of M1 macrophages was noted at 2 days after injury in the non-icing group, and the number of these cells drastically decreased the following day (Fig. 3c, d, i, j). However, there were no significant changes in the number of M1 macrophages between 1 and 3 days after injury in the icing group (Fig. 3f–i). Additionally, at 2 days after injury, the icing group had significantly fewer M1 macrophages than the non-icing group (Fig. 3i). Owing to their close proximity to regenerating muscle cells expressing iNOS from 4 days after injury (Kaliman et al. 1999), iNOS+ macrophages became indistinguishable from regenerating muscle cells expressing iNOS (Supplementary Fig. 2). As a result, CD68+CD206− cells were quantified as M1 macrophages at 4 days after injury, in accordance with previous reports (Tidball and Villalta 2010; Villalta et al. 2011; Kawashima et al. 2021a, b). Moreover, CD68+iNOS+ cells and CD68+CD206− cells had similar spatial distributions at 1 day after injury in the non-icing group (Supplementary Fig. 3), and thus were considered to be the same cells in this study. The number of M1 macrophages decreased similarly in the non-icing and icing groups from 4 days after injury (Fig. 4). Together, these results suggest that icing after skeletal muscle injury reduces the accumulation of M1 macrophages.
Chronological changes in M1 macrophage accumulation after skeletal muscle injury. a–h Representative images of M1 macrophages at the injured sites in non-icing (a–d) and icing (e–h) groups at 6 h (a, e), 1 day (b, f), 2 days (c, g), and 3 days (d, h) after injury. Transverse sections of the extensor digitorum longus muscle were analyzed by immunofluorescence for CD68 (green) and iNOS (red). Nuclei were stained with 4′, 6-diamino-2-phenylindole (DAPI, blue). Scale bars, 20 µm. i Quantification of the number of M1 macrophages after the injury. j Percentages of CD68+iNOS+ cells in total CD68+ cells after injury. Arrowheads indicate M1 macrophages identified as the double-positive color (yellow) of CD68 and iNOS surrounding a nucleus (blue). Five non-overlapping fields per animal were counted (n = 4, per group per timepoint). Values are expressed as the mean ± standard error, and statistical significance was assessed with two-way analysis of variance followed by Tukey’s post hoc test. The interaction between group and time is significant in i. **P < 0.01; statistical difference between groups. §P < 0.05; statistical differences from the previous timepoint in the same group. The main effect is significant for time, not for group, in j
Quantification of CD68+CD206− cell (M1 macrophage) number after skeletal muscle injury. a, b Transverse sections of the extensor digitorum longus muscle in non-icing (a) and icing (b) groups were analyzed by immunofluorescence for CD68 (green) and CD206 (red) at 4 days after injury. Nuclei were stained with 4′, 6-diamino-2-phenylindole (DAPI, blue). Arrowheads indicate M1 macrophages identified as the green color surrounding a nucleus (blue). Scale bars, 20 µm. c Quantification of the number of M1 macrophages. Five non-overlapping fields per animal were counted (n = 5, per group per timepoint). Values are expressed as mean ± standard error, and statistical significance was assessed with two-way analysis of variance followed by Tukey’s post hoc test. The interaction between group and time is significant. **P < 0.01; statistical difference between groups. §P < 0.05; statistical differences from the previous timepoint in the same group
Icing after skeletal muscle injury decreases TNF-α protein expression in the injured area
Quantification of TNF-α expression levels in injured muscles using western blot analysis revealed a constant level of expression from day 1 to 3 after injury in the non-icing group (Fig. 5a, b). Interestingly, the TNF-α expression level was significantly lower in the icing group than in the non-icing group at 3 days after injury (Fig. 5a, b).
Chronological changes in the expression levels of tumor necrosis factor-α (TNF-α) and its distribution in the injured sites after skeletal muscle injury. a, b Representative blots (a) and quantification (b) of TNF-α at 1, 2, and 3 days after injury in non-icing and icing groups. The expression levels of protein were normalized to the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels and expressed as the fold change (arbitrary unit, a.u.) from the values in the non-icing group at 1 day after injury. Values are expressed as mean ± standard error (n = 4, per group per timepoint), and statistical significance was assessed with two-way analysis of variance followed by Tukey’s post hoc test. The interaction between group and time is significant in b. *P < 0.05; statistical difference between groups. c, d Representative images of TNF-α-positive area in the injured sites in non-icing group at 3 days after injury. Transverse sections of the extensor digitorum longus muscle were analyzed by immunofluorescence for TNF-α (red) and CD68 (green) (c) or Pax7 (green) (d). Nuclei were stained with 4′, 6-diamino-2-phenylindole (DAPI, blue). Arrowheads in c indicate CD68+ cells, and those in d show Pax7+ cells. Scale bars, 20 µm
To investigate the distribution of TNF-α in the injured area at 3 days after injury, double immunostaining for TNF-α (red) and CD68 (green) or Pax7 (green), a transcription factor expressed in quiescent and activated satellite cells and proliferating myoblasts (Yin et al. 2013), was performed (Fig. 5c, d). TNF-α was expressed in a subset of CD68+ macrophages (Fig. 5c), in areas adjacent to Pax7+ cells (Fig. 5d), and in the interstitial spaces (Fig. 5c, d).
Icing after skeletal muscle injury reduces Pax7+ cell accumulation in injured sites
Activated and migrating satellite cells or proliferating myoblasts are detected in injured sites (Yin et al. 2013; Tian et al. 2016). To observe Pax7+cells in injured sites, dystrophin (red) was used. Dystrophin is a cytoskeletal protein that disappears in response to myofiber damage (Lovering and De Deyne 2004). No Pax7+ cells were observed in the injured area at 2 days after the injury in either group (data not shown). However, at 3 days after injury, several Pax7+ cells were detected at the injured sites of both the groups (Fig. 6a, d, g). Furthermore, at days 4 and 5 after injury, the number of Pax7+ cell in the injured sites increased in both groups, but appeared to be less abundant in the icing group (Fig. 6b, c, e, f). Moreover, two-way ANOVA demonstrated the effects of time and group to be significant (Fig. 6g), further indicating that the icing group consistently had fewer Pax7+ cells in the injured area than the non-icing group. These findings suggest that icing after muscle injury reduces Pax7+ cell accumulation.
Pax7+ cell distribution in the regenerating area after skeletal muscle injury. a–f Representative images of Pax7+ cells in non-icing (a–c) and icing (d–f) groups at 3 days (a, d), 4 days (b, e), and 5 days (c, f) after injury. Transverse sections of the extensor digitorum longus muscle were analyzed by immunofluorescence for Pax7 (green) and dystrophin (red). Nuclei were stained with 4′, 6-diamino-2-phenylindole (DAPI, blue). Scale bars, 20 µm. g Quantification of the number of Pax7+ cells. Arrowheads indicate Pax7+ cells identified as double-positive color (light blue) of Pax7 and a nucleus. Five non-overlapping fields per animal were counted (n = 4, per group per timepoint). Values are expressed as mean ± standard error, and statistical significance was assessed with two-way analysis of variance. The main effects for group (*) and time (§) are significant. The interaction between group and time is not significant. *P < 0.05; statistical difference between groups. §P < 0.05; statistical difference between times
Icing after skeletal muscle injury does not alter M2 macrophage accumulation and IGF-1 expression patterns
To identify M2 macrophages in the injured area, double immunolabeling for CD68 (green) and CD206 (red) was performed (Fig. 7). M2 macrophages were identified by their double-positive (yellow) coloration surrounding a nucleus (blue). M2 macrophages were barely detectable 6 h after injury in the non-icing and icing groups (Fig. 7a, f, k, l). Subsequently, M2 macrophages were observed in both groups at 12 h and gradually increased in both groups by 3 days following injury (Fig. 7b–d, g–i, k, l). Between 4 and 7 days after injury, no significant changes in the number of M2 macrophages were observed in either group (Fig. 7e, j–l). Two-way ANOVA demonstrated a significant effect of time but not group (Fig. 7k), suggesting that icing immediately following muscle injury does not affect M2 macrophage accumulation.
Chronological changes in the number of M2 macrophages after skeletal muscle injury. a–j Representative images of M2 macrophages in the injured sites in non-icing (a–e) and icing (f–j) groups at 6 h (a, f), 1 day (b, g), 2 days (c, h), 3 days (d, i), and 4 days (e, j) after injury. Transverse sections of the extensor digitorum longus muscle were analyzed by immunofluorescence for CD68 (green) and CD206 (red). Nuclei were stained with 4′, 6-diamino-2-phenylindole (DAPI, blue). Scale bars, 20 µm. k Quantification of the number of M2 macrophages after crush injury. l Percentages of CD68+CD206+ cells in total CD68+ cells after injury. Arrowheads indicate M2 macrophages identified as double-positive color (yellow) of CD68 and CD206 surrounding a nucleus (blue). Five non-overlapping fields per animal were counted (n = 5, per group per timepoint). Values are expressed as mean ± standard error, and statistical significance was assessed with two-way analysis of variance. The main effect is significant for time, not group, in k and l
IGF-1 is a cytokine that is expressed by both M1 and M2 macrophages and is known to cause phenotypic transition of macrophages from M1 to M2 (Tonkin et al. 2015). In this study, IGF-1 protein expression levels were examined at the period when the number of M2 macrophages started to increase (i.e., 1–3 days after injury). IGF-1 expression levels were comparable between the non-icing and icing groups at 1–3 days after injury. Two-way ANOVA indicated that neither time nor group had significant effects (Fig. 8a, b). Thus, the findings support our observation that M2 macrophage accumulation was not altered by icing after muscle injury.
Chronological changes in the expression level of insulin-like growth factor-1 (IGF-1) after skeletal muscle injury. Representative blots (a) and quantification (b) of IGF-1 at 1, 2, and 3 days after injury in non-icing and icing groups. The expression levels were normalized to the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels and expressed as fold change (arbitrary unit, a.u.) from the values in the non-icing group at 1 day after injury. Values are expressed as mean ± standard error (n = 4, per group per timepoint), and statistical significance was assessed with two-way analysis of variance. The main effects for time and group are not significant
Discussion
The dynamics of M1/M2 macrophages and the expression levels of macrophage-related cytokines were investigated in this study to better understand the key phenomena associated with the inflammatory response that is indispensable for optimal muscle regeneration, underlying icing-induced retardation of muscle regenerative process. The present study established that icing applied immediately after injury for 20 min (1) decreased the accumulation of M1 macrophages; (2) decreased the expression of TNF-α; (3) decreased the accumulation of Pax7+ cells at the regenerating site; and (4) eventually resulted in poor muscle regeneration. In our previous studies, icing after skeletal muscle injury delayed the accumulation of macrophages and impaired muscle regeneration (Takagi et al. 2011; Kawashima et al. 2021a, b). In addition, before retardation of macrophage accumulation, inhibition of the migration of the monocyte chemoattractant protein-1 (MCP-1) positive cells was occurred (Miyakawa et al. 2020). In the previous study using eccentric contraction-induced muscle damage by electrical stimulation that causes the sustained occurrence of muscle damage over several days, M1 macrophage accumulation and TNF-α expression were retarded by icing (Kawashima et al. 2021a, b). However, this study used muscle crush injury, which causes acute damage, and firstly showed that icing after acute muscle injury may affect muscle regeneration through reduction, not retardation, of M1 macrophage accumulation (Figs. 3, 4) and TNF-α expression (Fig. 5) during the early stages of regeneration.
M1 macrophages are one of the most common leukocytes infiltrating injured sites following muscle injury (Arnold et al. 2007) and are critical for muscle regeneration (Chazaud 2020). In this study, icing applied shortly after muscle injury decreased M1 macrophage accumulation (Figs. 3, 4) and TNF-α expression at 3 days after injury (Fig. 5). Given that M1 macrophages are the primary source of TNF-α during the muscle regeneration process (Tracey and Cerami 1994; Tidball and Villalta 2010; Liu et al. 2017), the decreased accumulation of M1 macrophages in this study could be attributed to the decreased TNF-α expression during the icing-treated muscle regeneration. TNF-α is a key factor in promoting the proliferation of myoblasts (Li 2003; Arnold et al. 2007; Chen et al. 2007). Between day 3 and 5 after injury, the icing group had fewer Pax7+ cells at the injured sites than the non-icing group (Fig. 6). Therefore, decreased M1 macrophage accumulation and decreased TNF-α expression in the icing group may account for the suppression of myoblast proliferation in the injured or regenerating sites. These findings suggest that icing-induced retardation of muscle regeneration is caused, at least in part, by disruption of M1 macrophage accumulation and TNF-α expression during the early stages of the regenerative process.
At 2 days after injury, icing reduced the accumulation of M1 macrophages, which are reported to be the major source of TNF-α (Figs. 3, 4). Nonetheless, the icing group had a significantly lower level of TNF-α protein expression at 3 days after injury (Fig. 5a, b), implying a time-course gap between M1 macrophage accumulation and TNF-α protein expression. A previous study reported that TNF-α expression was observed in the macrophage-invading area and interstitial spaces at 3 days after muscle injury (Ramos et al. 2016). Similarly, in the present study, TNF-α positive area was widely detected not only in the macrophage and adjacent area, but also in the interstitial space (Fig. 5c, d), indicating that the residue of TNF-α derived from M1 macrophages accumulated at 2 days after injury may have contributed to the decreased TNF-α expression in the icing group at 3 days after injury. Additionally, TNF-α is expressed by cells involved in muscle regeneration, such as myoblasts and myotubes (Cassatella 1999; Collins and Grounds 2001). In the present study, TNF-α-positive area was observed adjacent to Pax7+ cells (Fig. 5d), and the number of Pax7+ cells was significantly lower in the icing group than in the non-icing group at 3–5 days after injury (Fig. 6). Furthermore, some myotubes emerged at 3 days after injury in the group that did not receive icing (Fig. 1). Cumulatively, our data suggest that the decrease in TNF-α expression at 3 days after injury in the icing group is due to an altered accumulation of TNF-α-expressing cells such as M1 macrophages, myoblasts, and myotubes. Future studies will elucidate the effect of icing on the capacity of macrophages and myogenic cells to express TNF-α during muscle regeneration.
Additionally, this study showed that icing after muscle injury did not alter M2 macrophage accumulation despite decreased M1 macrophage recruitment (Figs. 3, 4, 7). IGF-1 has been shown to cause a shift in the macrophage phenotype from M1 to M2 (Tonkin et al. 2015). No difference in IGF-1 expression levels was observed between the non-icing and icing groups, implying that icing disrupts M1 but not M2 macrophage accumulation. M1 macrophages play important roles in muscle regeneration, including the expression of pro-inflammatory cytokines, phagocytosis of necrotic muscle debris, and proliferation of myoblasts (Tidball 2011; Juban and Chazaud 2017). Admittedly, a previous study reported that depletion of M1 macrophages during the early stages of muscle regeneration impairs regeneration (Wang et al. 2014). These previous findings support our notion that icing-induced impaired muscle regeneration is associated with decreased M1 macrophage accumulation and TNF-α expression during the early stages of regeneration.
A limitation of this study is that we did not perform confocal microscopy or observation of thinner sections to obtain more precise data of M1/M2 macrophage detection. In this study, M1 macrophages were identified as the cells that were double-positive for iNOS and CD68 within 3 days after injury. CD68+iNOS+ cells and CD68+CD206− cells had similar spatial distributions at 1 day after injury in the non-icing group (Supplementary Fig. 3). Ratios of iNOS+CD68+ and CD206+CD68+ cells at 2 days after injury of the non-icing group were 84.27% and 23.09%, respectively (Figs. 3j, 7l). By contrast, these ratios of the icing group were 70.06% and 29.04%, respectively (Figs. 3j, 7l). At 3 days after injury, the ratios of these cells of the non-icing group were 50.63% and 55.51%, respectively, while in the icing group, these were 62.32% and 39.84%, respectively (Figs. 3j, 7l). From these data, double-positive or double-negative cells of iNOS and CD206 might exist. However, these cells were few in the total data we obtained. Therefore, statistical differences between the non-icing and icing groups in this study were not altered because of these cells. To detect these polarizations of macrophages and to obtain precise data, future investigations will include experiments with flow cytometry or confocal microscopy using thinner sections.
Another limitation is the effect of icing is variable depending on the depth in the tissues. In our previous study, muscle surface and rectal temperatures were measured by using the same subject and icing method as in this study (Takagi et al. 2011). As mentioned in the study, the icing application could decrease local muscle surface temperature by more than 10 °C, but cause little decrease in rectal temperature (Takagi et al. 2011). However, there is a possibility that some regional variances may affect the results. In this study, every icing application was performed by one person (A.M.) with the same technique throughout the experiments to minimize technical error. Therefore, if the effect of cooling differs depending on the depth, the results of this study would not markedly change because of this effect (depth of cooling).
In conclusion, we demonstrated that icing disturbs M1 macrophage accumulation and its related phenomena during the early stages of muscle regeneration. This study may contribute to a better understanding of the mechanism underlying the icing-caused retardation of skeletal muscle regeneration.
Data availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
References
Arnold L, Henry A, Poron F et al (2007) Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med 204:1057–1069. https://doi.org/10.1084/jem.20070075
Bencze M, Negroni E, Vallese D et al (2012) Proinflammatory macrophages enhance the regenerative capacity of human myoblasts by modifying their kinetics of proliferation and differentiation. Mol Ther 20:2168–2179. https://doi.org/10.1038/mt.2012.189
Bleakley C, McDonough S, MacAuley D (2004) The use of ice in the treatment of acute soft-tissue injury: a systematic review of randomized controlled trials. Am J Sports Med 32:251–261. https://doi.org/10.1177/0363546503260757
Cassatella MA (1999) Neutrophil-derived proteins: selling cytokines by the pound. Adv Immunol 73:369–509. https://doi.org/10.1016/s0065-2776(08)60791-9
Chazaud B (2020) Inflammation and skeletal muscle regeneration: leave it to the macrophages! Trends Immunol 41:481–492. https://doi.org/10.1016/j.it.2020.04.006
Chen S, Jin B, Li Y (2007) TNF-α regulates myogenesis and muscle regeneration by activating p38 MAPK. Am J Physiol Cell Physiol 292:1660–1671. https://doi.org/10.1152/ajpcell.00486.2006
Collins RA, Grounds MD (2001) The role of tumor necrosis factor-alpha (TNF-α) in skeletal muscle regeneration: studies in TNF-α (−/−) and TNF-α (−/−)/LT-α(−/−) mice. J Histochem Cytochem 49:989–1001. https://doi.org/10.1177/002215540104900807
Fan Y, Hao Y, Gao D et al (2021) Phenotype and function of macrophage polarization in monocrotaline-induced pulmonary arterial hypertension rat model. Physiol Res 70:213–226. https://doi.org/10.33549/physiolres.934456
Freitas LS, Freitas TP, Silveira PC et al (2007) Effect of therapeutic pulsed ultrasound on parameters of oxidative stress in skeletal muscle after injury. Cell Biol Int 31:482–488. https://doi.org/10.1016/j.cellbi.2006.11.015
Fujita N, Ono M, Tomioka T et al (2014) Effects of hyperbaric oxygen at 1.25 atmospheres absolute with normal air on macrophage number and infiltration during rat skeletal muscle regeneration. PLoS One 9:e115685. https://doi.org/10.1371/journal.pone.0115685
Goetsch SC, Hawke TJ, Gallardo TD et al (2003) Transcriptional profiling and regulation of the extracellular matrix during muscle regeneration. Physiol Genomics 14:261–271. https://doi.org/10.1152/physiolgenomics.00056.2003
Hatade T, Takeuchi K, Fujita N et al (2014) Effect of heat stress soon after muscle injury on the expression of MyoD and myogenin during regeneration process. J Musculoskelet Neuronal Interact 14:325–333
Huard J, Li Y, Fu FH (2002) Muscle injuries and repair: current trends in research. J Bone Joint Surg Am 84:822–832
Jarvinen TAH, Jarvinen TLN, Kaariainen M et al (2007) Muscle injuries: optimising recovery. Best Pract Res Clin Rheumatol 21:317–331. https://doi.org/10.1016/j.berh.2006.12.004
Juban G, Chazaud B (2017) Metabolic regulation of macrophages during tissue repair: insights from skeletal muscle regeneration. FEBS Lett 591:3007–3021. https://doi.org/10.1002/1873-3468.12703
Kaliman P, Canicio J, Tester X et al (1999) Insulin-like growth factor-II, phosphatidylinositol 3-kinase, nuclear factor-kB and inducible nitric-oxide synthase define a common myogenic signaling pathway. J Biol Chem 274:17437–17444. https://doi.org/10.1074/jbc.274.25.17437
Kawashima M, Miyakawa M, Sugiyama M et al (2020) Unloading during skeletal muscle regeneration retards iNOS-expressing macrophage recruitment and perturbs satellite cell accumulation. Histochem Cell Biol 154:355–367. https://doi.org/10.1007/s00418-020-01897-3
Kawashima M, Kawanishi N, Tominaga T et al (2021a) Icing after eccentric contraction-induced muscle damage perturbs the disappearance of necrotic muscle fibers and phenotypic dynamics of macrophages in mice. J Appl Physiol 130:1410–1420. https://doi.org/10.1152/japplphysiol.01069.2020
Kawashima M, Iguchi S, Fujita N et al (2021b) Structural changes in skeletal muscle fibers after icing or heating on downhill running in mice. Kobe J Med Sci 67:48–54
Le Moal E, Juban G, Bernard AS et al (2018) Macrophage-derived superoxide production and antioxidant response following skeletal muscle injury. Free Radic Biol Med 120:33–40. https://doi.org/10.1016/j.freeradbiomed.2018.02.024
Li Y (2003) TNF-α is a mitogen in skeletal muscle. Am J Physiol Cell Physiol 285:370–376. https://doi.org/10.1152/ajpcell.00453.2002
Li Q, Bai Y, Xu Y et al (2010) Autografting satellite cells to repair damaged muscle induced by repeated compression: an animal model. Foot Ankle Int 31:706–711. https://doi.org/10.3113/fai.2010.0706
Liu X, Liu Y, Zhao L et al (2017) Macrophage depletion impairs skeletal muscle regeneration: the roles of regulatory factors for muscle regeneration. Cell Biol Int 41:228–238. https://doi.org/10.1002/cbin.10705
Lovering RM, De Deyne PG (2004) Contractile function, sarcolemma integrity, and the loss of dystrophin after skeletal muscle eccentric contraction-induced injury. Am J Physiol Cell Physiol 286:230–238. https://doi.org/10.1152/ajpcell.00199.2003
Mantovani A, Sica A, Sozzani S et al (2004) The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 25:677–686. https://doi.org/10.1016/j.it.2004.09.015
Miyakawa M, Kawashima M, Haba D et al (2020) Inhibition of the migration of MCP-1 positive cells by icing applied soon after crush injury to rat skeletal muscle. Acta Histochem 122:151511. https://doi.org/10.1016/j.acthis.2020.151511
Oyaizu T, Enomoto M, Yamamoto N et al (2018) Hyperbaric oxygen reduces inflammation, oxygenates injured muscle, and regenerates skeletal muscle via macrophage and satellite cell activation. Sci Rep 8:1288. https://doi.org/10.1038/s41598-018-19670-x
Ramos GV, Pinheiro CM, Messa SP et al (2016) Cryotherapy reduces inflammatory response without altering muscle regeneration process and extracellular matrix remodeling of rat muscle. Sci Rep 6:18525. https://doi.org/10.1038/srep18525
Rayagiri SS, Ranaldi D, Raven A et al (2018) Basal lamina remodeling at the skeletal muscle stem cell niche mediates stem cell self-renewal. Nat Commun 9:1075. https://doi.org/10.1038/s41467-018-03425-3
Santa FD, Vitiello L, Torcinaro A et al (2019) The role of metabolic remodeling in macrophage polarization and its effect on skeletal muscle regeneration. Antioxid Redox Signal 30:1553–1598. https://doi.org/10.1089/ars.2017.7420
Sciorati C, Rigamonti E, Manfredi AA et al (2016) Cell death, clearance and immunity in the skeletal muscle. Cell Death Differ 23:927–937. https://doi.org/10.1038/cdd.2015.171
Shibaguchi T, Sugiura T, Fujitsu T et al (2016) Effects of icing or heat stress on the induction of fibrosis and/or regeneration of injured rat soleus muscle. J Physiol Sci 66:345–357. https://doi.org/10.1007/s12576-015-0433-0
Smith C, Kruger MJ, Smith RM et al (2008) The inflammatory response to skeletal muscle injury. Sports Med 38:947–969. https://doi.org/10.2165/00007256-200838110-00005
Srikuea R, Pholpramool C, Kitiyanant Y et al (2010) Satellite cell activity in muscle regeneration after contusion in rats. Clin Exp Pharmacol Physiol 37:1078–1086. https://doi.org/10.1111/j.1440-1681.2010.05439.x
Swenson C, Sward L, Karlsson J (1996) Cryotherapy in sports medicine. Scand J Med Sci Sports 6:193–200. https://doi.org/10.1111/j.1600-0838.1996.tb00090.x
Takagi R, Fujita N, Arakawa T et al (2011) Influence of icing on muscle regeneration after crush injury to skeletal muscles in rats. J Appl Physiol 110:382–388. https://doi.org/10.1152/japplphysiol.01187.2010
Takeuchi K, Hatade T, Wakamiya S et al (2014) Heat stress promotes skeletal muscle regeneration after crush injury in rats. Acta Histochem 116:327–334. https://doi.org/10.1016/j.acthis.2013.08.010
Taylor PR, Martinez-Pomares L, Stacey M et al (2005) Macrophage receptors and immune recognition. Annu Rev Immunol 23:90–144. https://doi.org/10.1146/annurev.immunol.23.021704.115816
Tian Z, Jiang S, Zhang M et al (2016) Detection of satellite cells during skeletal muscle wound healing in rats: time-dependent expressions of Pax7 and MyoD in relation to wound age. Int J Legal Med 130:163–172. https://doi.org/10.1007/s00414-015-1251-x
Tidball JG (1995) Inflammatory cell response to acute muscle injury. Med Sci Sports Exerc 27:1022–1032. https://doi.org/10.1249/00005768-199507000-00011
Tidball JG (2011) Mechanisms of muscle injury, repair, and regeneration. Compr Physiol 1:2029–2062. https://doi.org/10.1002/cphy.c100092
Tidball JG (2017) Regulation of muscle growth and regeneration by the immune system. Nat Rev Immunol 17:165–178. https://doi.org/10.1038/nri.2016.150
Tidball JG, Villalta SA (2010) Regulatory interactions between muscle and the immune system during muscle regeneration. Am J Physiol Regul Integr Comp Physiol 298:1173–1187. https://doi.org/10.1152/ajpregu.00735.2009
Tidball JG, Dorshkind K, Wehling-Henricks M (2014) Shared signaling systems in myeloid cell-mediated muscle regeneration. Development 141:1184–1196. https://doi.org/10.1242/dev.098285
Tonkin J, Temmerman L, Sampson RD et al (2015) Monocyte/macrophage-derived IGF-1 orchestrates murine skeletal muscle regeneration and modulates autocrine polarization. Mol Ther 23:1189–1200. https://doi.org/10.1038/mt.2015.66
Tracey KJ, Cerami A (1994) Tumor necrosis factor: a pleiotropic cytokine and therapeutic target. Annu Rev Med 45:491–503. https://doi.org/10.1146/annurev.med.45.1.491
Vannella KM, Wynn TA (2016) Mechanisms of organ injury and repair by macrophages. Annu Rev Physiol 79:593–617. https://doi.org/10.1146/annurev-physiol-022516-034356
Villalta SA, Rinaldi C, Deng B et al (2011) Interleukin-10 reduces the pathology of mdx muscular dystrophy by deactivating M1 macrophages and modulating macrophage phenotype. Hum Mol Genet 20:790–805. https://doi.org/10.1093/hmg/ddq523
Wang H, Melton DW, Porter L et al (2014) Altered macrophage phenotype transition impairs skeletal muscle regeneration. Am J Pathol 184:1167–1184. https://doi.org/10.1016/j.ajpath.2013.12.020
Watanabe S, Alexander M, Misharin AV et al (2019) The role of macrophages in the resolution of Inflammation. J Clin Invest 129:2619–2628. https://doi.org/10.1172/jci124615
Yin H, Price F, Rudnicki MA (2013) Satellite cells and the muscle stem cell niche. Physiol Rev 93:23–67. https://doi.org/10.1152/physrev.00043.2011
Acknowledgements
The authors thank the members of our laboratory for cooperation. We also would like to thank Enago (www.enago.jp) for the English language review.
Funding
Financial support for this study was provided by JSPS KAKENHI (grant number 21K11238).
Author information
Authors and Affiliations
Contributions
Conceptualization: AM, MK, and TA; methodology: AM, MK, MMiyakawa, MS, and TA; formal analysis and investigation: AM, MK, IN, MMiyakawa, MMiyoshi, TSakuraya, and TSonomura; writing—original draft preparation: AM; writing—review and editing: AM, MK, IN, and TA; funding acquisition: TA; supervision: TA All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical approval
This study was approved by the Institutional Animal Care and Use Committee and was conducted in accordance with the Kobe University Animal Experimentation Regulations (approval numbers P180706 and P200906).
Informed consent
N/a (animal experiment).
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Miyazaki, A., Kawashima, M., Nagata, I. et al. Icing after skeletal muscle injury decreases M1 macrophage accumulation and TNF-α expression during the early phase of muscle regeneration in rats. Histochem Cell Biol 159, 77–89 (2023). https://doi.org/10.1007/s00418-022-02143-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-022-02143-8