Abstract
In alleged sexual assault cases, identification of the presence of spermatozoa at the crime scene, or on items of eventual significance, or associated with the body of the victim, is integral to the forensic investigation to support or refute the proposition that sexual act has occurred. A 3-plex MSRE-PCR (methylation-sensitive restriction enzyme-PCR) system has been developed previously to identify spermatozoa based on the presence or absence of DNA methylation. This assay showed that 0.1 ng of DNA from a semen extract was sufficient to identify the presence of spermatozoa even when there was excessively more DNA isolated from vaginal fluid than DNA from a semen extract (80 ng/0.1 ng) or a mix of the menstrual blood/semen DNA (5 ng/0.1 ng). In this study, we combine spermatozoa detection with co-amplification of 23 Y-STR loci. We perform standard validation steps to present a novel test that saves time and uses the same sample for both DNA typing and spermatozoa detection in the same reaction. The combined assay can identify Y-STR and spermatozoa simultaneously using just 0.1 ng semen DNA, even in the presence of 5 ng of DNA from a female (male/female:1/50). No other body fluid tested, such as saliva, gave a result for the presence of spermatozoa. A total of 9 non-probative forensic samples from 7 sexual assault cases were tested by this co-amplification system. In all cases, the same sperm-positive data were obtained, concordant with our previous study analyzed by only 3-plex MSRE-PCR, and the Y-STR results were also consistent with that analyzed by only PowerPlex® Y23 kit. The co-amplification will be beneficial for the limited samples in many criminal cases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Forensic investigation of cases of alleged sexual assault includes the identification of semen and then the recording of spermatozoa. If a positive result is obtained from the presence of male-specific material, then STR profiling can provide the source attribution. All too often there is a delay in reporting such a crime with collection of a sample from the victim; biological evidence such as the protein used to detect semen and the protein coat covering the spermatozoa can be adversely affected in such instances. A test is needed that has high sensitivity and will target male-specific biological evidence in the presence of relatively excessive amounts of materials from the female victim.
Common tests for the presence of semen include acid phosphatase or immunological tests such as Prostate Specific Antigen (PSA) and Semenogelin (Sg). The last two tests can provide a positive identification for semen up to 47 h and 72 h, respectively, after a sexual assault [1, 2]. In two reports, a few spermatozoa could still be identified 96 h after intercourse [3, 4]. Microscopic examination can however lead to a false-negative result for samples with deformed and, or, limited number of spermatozoa. This scenario is frequently encountered in cases where there is a long time interval between the alleged event and sample collection, or when there are few spermatozoa for reasons of male health. A DNA Y-STR profile can still be identified from samples collected 48–96 h after intercourse [3, 5] indicating that perhaps DNA from the spermatozoa can still be detected after spermatozoa are no longer identified by microscopy. DNA from the spermatozoa rather than the spermatozoa themselves may therefore be a better target to detect any male DNA associated with a vaginal swab.
A 3-plex MSRE-PCR system was reported previously by our laboratory [6]. This system utilized the methylation-sensitive restriction enzyme, HhaI, selectively digesting the non-methylated recognition sequence 5′-GCGC-3′. In contrast, if the DNA sequence 5′-GCGC-3′ is methylated, then it is resistant to enzymatic digestion and can be amplified in the following PCR. Three markers were analyzed in this system: DC, containing the HhaI recognition sequences which were without, or with, low methylation in a wide range of body fluids; these were used as a digestive control to monitor the digestion. PCR products should be absent when the digestion is effective and complete. SP, exclusively hypermethylated in spermatozoa, was utilized as an indicator for sperm. Resultant PCR products should only be observed in samples which contain sperm cells. SRY, a region on Y chromosome, was used to determine that the DNA came from a male. The validation study demonstrated the specificity and sensitivity of the 3-plex MSRE-PCR assay and the usefulness of its application in spermatozoa identification. The advantages of this assay are as follows: generation of an unambiguous profile; well-defined guideline for interpretation; high sensitivity for trace detection of DNA from spermatozoa; a positive result even when the isolated DNA contains massive amounts of DNA from the female (80 ng vaginal or 5 ng menstrual blood DNA) where approximately 0.1 ng of DNA from a semen extract could still be identified; and readily integrating to the current DNA analysis process.
The application of autosomal STR DNA typing is often impractical in these cases if DNA from the male is at trace levels in a mixed with large amounts of DNA from a female. Instead, Y-STR typing is an ideal tool in such circumstance. It has been reported that Y-STR profile can still be identified from evidential material in which no spermatozoa were observed [7,8,9]. Recently, rapidly mutating Y-chromosomal short tandem repeats (RM Y-STRs) have been adopted to improve differentiation of unrelated males and also to enable separating related males in human identity testing [10, 11]. The commercial PowerPlex® Y23, including RM Y-STRs, is widely used in many crime laboratories and reported to be significantly more sensitive than the other kits [12, 13].
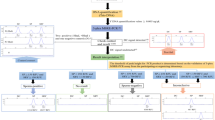
A comprehensive workflow of biological stains in cases of alleged sexual assault usually includes a presumptive test for the body fluid, DNA extraction (differential extraction [14, 15] or non-differential extraction [16]), DNA quantification, and STR (or Y-STR) DNA typing. To save sample and time, simultaneous amplification of autosomal STR loci with an assay for DNA methylation-based body fluid identification (such as MSRE-PCR) has been reported [17, 18]. An issue with these reports is that either the recommended amount of template DNA needed to be lower than 2 ng (the tested upper limit) [18] or a differential extraction needs to be performed for DNA isolation from forensic samples [17]. However, typically vaginal swabs contain relatively high amounts of DNA from the female compared to any DNA from the spermatozoa or semen. Differential extraction may not be able to separate the male and female DNA sufficiently so as to detect the trace amounts of male DNA. Instead, non-differential DNA extraction for DNA isolation from trace amounts of semen [16] can be followed by Y-STR assay. We therefore report on a novel co-amplification system for simultaneous amplification of 23 Y-STR and spermatozoa identification. We report on standard validation steps allowing a streamlined workflow for biological stain identification for sexual assault cases.
Materials and methods
Sample collection
Samples used in this study were collected after informed consent and following the procedures approved by the Institutional Review Board (IRB) of Tao-Yuan General Hospitals (IRB No. TYGH102011) and Antai-Tian-Sheng Memorial Hospital (IRB No. 18–074-B) in Taiwan. A total of 65 samples were collected including 12 semen (9 from healthy non-vasectomized donors, and 3 from vasectomized donors), 7 peripheral blood samples (6 males and 1 female), 7 saliva (5 males and 2 females), 5 sweat (4 males and 1 female), 7 urine (6 males and 1 female), 7 feces (5 males and 2 females), 10 vaginal secretions, and 10 menstrual blood samples.
DNA extraction and quantification
Genomic DNA was isolated by using the Qiagen Mini kit (Qiagen, Hilden, Germany) following the manufacturer’s recommendations for “DNA purification from tissues.” An exception was for semen DNA extraction where the ATL buffer (180 µL) was modified by the addition of 7 μL of DTT (1 M). The isolated DNA was quantified by using the Quantifiler™ Trio DNA Quantification Kit (Life Technologies, CA, USA) using a 7500 real-time PCR machine following the manufacturer’s recommendation (Life Technologies).
Primers for the 3-plex MSRE-PCR
The sequence of the primers for the 3-plex MSRE-PCR used in this study is the same as our previous study [6]; however, for consistency in the co-amplification with the Y-STRs, the primers were labelled with Fluorescein dye instead of FAM, as used in the previous study. Sequences of the primers and their optimal concentrations for the reactions of co-amplifications are listed in Table 1.
Co-amplifications of the 3-plex MSRE-PCR with Y-STR
To combine the 3-plex MSRE-PCR into the PowerPlex® Y23 (Y-STR, Promega, WI, USA), the PCR preparation was modified to a total volume of 25 μL containing 0.5 ng of DNA template, 30 unit of HhaI (New England Biolabs, MA, USA), 1.5 µL of 360 GC enhancer (Thermo Fisher Scientific, MA, USA), optimal primer concentrations of the 3-plex MSRE-PCR (Table 1), 5 μL of PowerPlex® Y23 mixture, and 2.5 μL of PowerPlex® Y23 10X primer set. The reactions were conducted in a GeneAmp® PCR System 9700 (Life Technologies). Before the cycling reaction, the first step of the thermal program was for DNA digestion at 37 °C for 30 min. This was followed by incubation at 96 °C for 2 min, for both heat inactivation of the HhaI enzyme and PCR initiation, and then followed by 30 cycles of 94 °C for 10 s, 61 °C for 1 min, and 72 °C for 30 s, with a final extension at 60 °C for 20 min. Except for the initial step of 37 °C for 30 min, the other steps are performed following the recommendations of the manufacturers of the PowerPlex® Y23 [19]. PCR products were analyzed using the ABI PRISM 3500 Genetic Analyzer and GeneMapper® ID-X v1.4 software (Life Technologies). The threshold for a positive peak was set at 150 Relative Fluorescence Unit (RFU) according to the threshold of our laboratory for Y-STR typing and the previous study for 3-plex MSRE-PCR [6].
Validation tests
Specificity
Specificity to spermatozoa was tested using 0.5 ng DNA from semen samples and also from the other types of body fluids or tissue samples (peripheral blood, saliva, sweat, urine, feces, vaginal secretion, and menstrual blood).
Sensitivity and repeatability
The isolated DNA from semen samples taken from healthy donors was diluted to 1, 0.5, 0.25, 0.1, and 0.05 ng. All testing was in triplicate and CV (coefficient of variation) for the triplicate was calculated by Excel 2010 to assess the repeatability.
Influence of the presence of excessive amounts of DNA from a female
To evaluate the influence of an excessive amount of DNA from a female on the co-amplification system, DNA was isolated from 10 vaginal secretions and 10 menstrual blood samples. According to the results of our previous study [6], the most tolerable amount of female DNA for the 3-plex MSRE-PCR to prevent the generation of any non-specific SP peak was 80 ng and 5 ng DNA isolated from vaginal secretions and menstrual blood respectively. It was observed that more than 100 ng DNA of vaginal secretions and 10 ng DNA of menstrual blood could generate non-specific SP peaks. Therefore, in this study, initially 10 ng DNA for the vaginal secretion and menstrual blood was used to evaluate the influence of an excessive amount of DNA from a female. If it showed the expected result (no peak detected) on the electropherogram, double amount of input DNA was further used to test this higher tolerance. Finally, the input DNA tested was 10 ng and 20 ng from 10 vaginal secretions and 10 menstrual blood samples.
Simulated mixture tests
Two types of the mixtures were prepared to evaluate the interference of DNA from other body fluids in Y-STR analysis and spermatozoa identification. One was for a mixture of semen DNA (0.1 ng) and excessive amounts of DNA from a female (5 ng for vaginal secretion or menstrual blood), totally ten sets of simulated mixtures were prepared (5 sets for vaginal secretion DNA with semen DNA, and 5 sets for menstrual blood DNA with semen DNA); and the other was a mix of semen DNA and saliva DNA from 2 males at a variety of ratios (1:0, 1:1, 1:3, 1:9, and 0:1 for semen DNA:saliva DNA) in five sets, the amount of DNA used was 1 ng DNA in total for these dilutions, and five mixtures for each set were generated.
Non-probative forensic samples
A total of 9 non-probative forensic samples from 7 sexual assault cases were tested by the co-amplification system of Y-STR and 3-plex MSRE-PCR. These 9 samples were selected as their Y DNA quantification values were more than 0.005 ng/μL and male/female DNA ratio more than 1/50; these are the suggested limitations to unambiguously record the presence of spermatozoa using the 3-plex MSRE-PCR in the previous study [6].
Results
Co-amplifications of the 3-plex MSRE-PCR and Y-STR
Three samples from extracts of semen DNA (0.5 ng) were used to test the co-amplification system of the 3-plex MSRE-PCR and Y-STR. Each sample was performed in triplicate. All 23 Y-STR loci were successfully identified and the SP and SRY signals for spermatozoa identification were observed at the expected locations in the electrophoretogram. The electropherogram of one example for the co-amplification system is shown (Fig. 1). Based on the information in STRBase (Short Tandem Repeat DNA Internet DataBase, https://strbase.nist.gov/index.htm) and the NCBI database (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/), none of the 23 Y-STR loci and their flanking regions contains the recognition sequence for HhaI (5′-GCGC-3′) [20, 21]. As a result, the 23 Y-STR amplicons were not digested by the restriction enzyme and were amplified as expected. It was noted that the RFU for 13 loci (in 23 Y-STR) decreased in all three samples when tested by the co-amplification system compared when the amplification was conducted by the PowerPlex® Y23 kit with no enzyme and digestion step (Online Resource 1). Among the 13 decreased loci, the decreased ratio ranged from − 5.6% of DYS438 to − 70.5% of DYS390. However, all the peaks were more than 3000 RFU and can be interpreted unambiguously and accurately.
An electropherogram generated from DNA obtained from a semen sample collected from a healthy male. The Y-STRs are from the PowerPlex® Y23 kit. Additionally, in the blue channel, there is SP that indicates the presence of spermatozoa and SRY that detects the SRY gene on Y chromosome, indicating the
Validation
Specificity
A total of 65 samples were used to test the specificity of the co-amplification system for the 3-plex MSRE-PCR and Y-STR. These are described under the “Sample collection” section. All 65 results were exactly as predicted: DC signal was not observed for all the samples; SP signal was only observed for the DNA from semen taken from healthy non-vasectomized donors; SRY signal and complete 23 Y-STR amplifications were observed for all male samples. All 27 samples taken from the female donors returned no signal. An example of an electropherogram from each of the body fluid types is shown in Fig. 2.
Electropherogram for one example of displaying the specificity of detection. Data are presented after co-amplification of the 3-plex MSRE-PCR and Y-STR from a range of DNA extracts: semen (a), vasectomized semen (b), male blood (c), male saliva (d), male sweat (e), male urine (f), female feces (g), vaginal secretion (h), and menstrual blood (i). SP should only be detected in the semen sample and no other body fluid. SRY should be detected in samples from males and not from females. Only fluorescein columns were shown in the figure
Sensitivity and repeatability
The semen from five donors (healthy and non-vasectomized) was collected and the isolated DNA was diluted to 1, 0.5, 0.25, 0.1, and 0.05 ng. All samples for each input of DNA were tested in triplicate resulting in 75 tests and 15 results for each DNA amount. Complete 23 Y-STR profiles and sperm-positive results were observed in all samples when semen DNA was not less than 0.1 ng. It was only when the DNA from the semen extract was 0.05 ng that the incomplete data were obtained: one sample returned a complete Y-STR profile and was clearly identified as “sperm positive” in all three results; the other four samples exhibited allele drop-out at 1 to 3 Y-STR loci (out of 23); and one reported a sperm-positive result as expected in 2 of the triplicates but failed to return this positive result in one of the triplicates. In summary, when the input DNA was 0.05 ng, dropout was observed at 15 alleles out of a possible 345 (23 × 15) alleles; therefore, the detection ratio of Y-STR alleles was 95.65% ((345–15)/345); SP was not observed for one time in the 15 tests; therefore, spermatozoa were identified with a success of 93.33% ((15–1)/15).
Analysis of the peak heights is shown in Fig. 3. The peak heights reduced with decreasing input DNA, as expected. When 1 ng of DNA from semen was used, the average peak height for Y-STRs was 12,164 RFU, and 13,644 and 14,011 RFU for SP and SRY respectively; when semen DNA was 0.5 ng, the average peak height for Y-STRs, SP, and SRY was 7511, 8152, and 8247 RFU respectively. Most CV (coefficient of variation) of each locus for triplicate analysis was less than 50% for these two amounts of inputs. When semen DNA was 0.25 ng, the average peak height for Y-STRs was 3422 RFU, and 3634 and 3443 RFU for SP and SRY respectively; however, the CV of 3 loci (DYS576, DYS390, and DYS439) was more than 75% in one of the five samples. When semen DNA was 0.1 ng, the average peak height for Y-STRs was 1570 RFU, and 1670 and 1787 RFU for SP and SRY respectively; all alleles were still clearly obviously identified. One sample showed CV higher than 75% for DYS570 and the other four samples showed CV less than 50% for most loci. When reduced to 0.05 ng, 4 of 5 extracts of semen DNA showed allele dropout in 1 to 3 tests of the triplicates. Based on the PowerPlex® Y23 System technical manual, the test was optimized and balanced for 0.5-ng DNA template [19]. Therefore, the results tested with a higher CV in some Y-STR loci for DNA template less than 0.5 ng are acceptable. These data clearly indicated that the template DNA for the co-amplification system when ranged between 0.1 and 1 ng resulted in unambiguous and concordant amplification of all 23 Y-STR loci and spermatozoa identification; to respond the technical manual of PowerPlex® Y23 System, 0.5 ng is suggested for this co-amplification system.
Comparison within the same in-put of DNA showed little variation between the relative peak heights for Y-STR, SP, and SRY; all were approximately similar (Online Resource 2). The ratios (relative peak heights) of SP/Y-STR AVE ranged from 106.2% (0.25 ng) to 112.2% (1 ng); and SRY/Y-STR AVE ranged from 100.6% (0.25 ng) to 124.6% (0.05 ng). It showed that no obviously preferential amplification for the loci of Y-STR and 3-plex MSRE-PCR occurred in the range of 0.05 to 1 ng DNA.
Influence of an excessive amount of DNA from a female
Our previous study showed that no non-specific results were obtained when DNA from vaginal secretions was not more than 80 ng and DNA from menstrual blood not more than 5 ng [6]. Using 10 ng of DNA from the 10 vaginal secretion and the 10 menstrual blood samples resulted in no non-specific signal in any of the 20 samples. An increase to 20 ng of input DNA was also used to evaluate the influence of the excessive female DNA on the co-amplification system. Nine of the DNA extracts from vaginal secretions gave no signal (as expected), and one sample showed a small peak at SP (244 RFU). Eight of the DNA extracts from menstrual blood gave no signal, and as expected, two samples showed small peaks at SP with peak height of 156 and 177 RFU respectively. This indication is that less than 10 ng of DNA from the female will still give unambiguous and reproducible results.
Simulated mixture tests
Mixtures of semen and female body fluids, especially vaginal secretion and menstrual blood, are frequently encountered and collected in cases of alleged sexual assault. Therefore, simulated mixtures of semen DNA and excessive female DNA were tested in this study. According to the sensitivity test, 0.1 ng of semen DNA could be unambiguously identified, and the tests of excessive female DNA, input female DNA less than 10 ng was recommended. Therefore, ten sets of simulated mixtures containing 5 ng female DNA from vaginal secretion (5 sets) or menstrual blood (5 sets) with 0.1 ng semen DNA were prepared and tested by the co-amplification system. The results showed a tendency in a decrease in the peak heights of the average of Y-STR alleles, SP, and SRY in all sets of semen/vaginal secretion mixtures and 3 sets of semen/menstrual blood mixtures comparing with that in only 0.1 ng semen DNA (Online Resource 3). However, complete Y-STR profile and spermatozoa positive results were still clearly observed for all sets of the mixtures (Fig. 4).
Simulated mixtures of DNA from semen and female DNA tested by the co-amplification system for the 3-plex MSRE-PCR and Y-STR. Five sets of mixtures numbered from 1 to 5 are listed along the x axis. The y axis represents the peak height (RFU). Y-STR AVE represents the average peak height of all the Y-STR alleles in PowerPlex® Y23
Saliva from a male could also be collected as evidence in alleged cases of sexual assault, particularly when mixed with the other body fluids, such as semen. Five sets of mixtures of DNA from semen and saliva, from a different male, were prepared to test the influence of male saliva DNA. Five mixtures for each set were generated following the ratios of 1:0, 1:1, 1:3, 1:9, and 0:1 (semen DNA:saliva DNA), and for each ratio, a total of 1 ng DNA was used as the template in a test. The results showed that when the ratios were 1:1 and 1:3, complete Y-STR profile and sperm positive result were observed for all the 5 sets of the mixtures. When the ratio was 1:9, the mixtures still clearly returned a spermatozoa positive result, however, the peak heights of SP decreased to 4 ~ 22% of SRY (peak height ratio; PHR); in this scenario, SP signal can still be clearly identified without confusion with any artifact, which might cause confusion when interpreting STRs in the stutter area for a 1:9 mixture of two sources of DNA. For Y-STR, an allele of DYS448 from the donor of semen dropped out, only showing the profile matching that of the donor of saliva. No SP was detected in the saliva only sample (0:1) as expected. An electropherogram of one example is shown in Fig. 5.
Non-probative forensic sample tests
A total of 9 non-probative forensic samples from 7 sexual assault cases were tested by the co-amplification system. These were selected as their Y DNA quantification value was more than 0.005 ng/μL and male/female DNA ratio more than 1/50 [6] (Table 2). The results for 3-plex MSRE-PCR were all sperm-positive and concordant with our previous study [6]. Except one sample (sample 10), which showed a mixed Y-STR profile with a PHR (SP/SRY) of 58% in SP and SRY loci, the other 8 samples were all with the single-source Y-STR profile. The Y-STR results were also consistent with that analyzed by only PowerPlex® Y23 kit. The result also showed both Y-STR and spermatozoa could be detected simultaneously even in the presence of excessive amounts of DNA from the female (male/female: 1/41 for sample 7–1).
Discussion
In cases of alleged sexual assault, associating a male DNA profile with the presence of semen and spermatozoa provides high confidence as to the source of these biological materials. Traditionally, one part of sample may be used in a presumptive test and another part of the sample set aside for microscopic examination. A further part of the sample needs to be submitted for DNA extraction. Many types of evidential material, such as vaginal swabs or semen stains, may contain large amounts of DNA from the female relative to the male’s DNA and here the use of Y-STR has the advantage of targeting the male-specific markers. It was reported that complete Y-STR profiles can be identified using the PowerPlex® Y23 kit from mixed samples with 62.5 pg of male DNA in a background of 400 ng of female DNA [12]. In our previous study, 0.1 ng of DNA from semen was sufficient to identify the presence of spermatozoa even when 5 ng of female DNA was present [6]. Although the two systems focus on 2 different targets, Y-STR and spermatozoa DNA, they have one thing in common: to identify any trace of the contribution from a male within the biological material from the victim. To address this, the co-amplification system of PowerPlex® Y23 and 3-plex MSRE-PCR was developed to amplify Y-STR and identify spermatozoa simultaneously in a single reaction. The advantage of this co-amplification system is not only saving time but also reduces the consumption of sample. In some cases, it is greatly crucial when the sample is very limited. The only alteration to the current practice when using the PowerPlex® Y23 kit is addition of an enzyme and an extra 30-min digestion step performed in the beginning of the thermal program and the addition of three primer sets. The addition of detecting spermatozoa along with Y-STR loci is designed to be easy to introduce into current workflows for easy implementation by forensic laboratories.
Regarding the influence of excessive female DNA, both vaginal secretion and menstrual blood DNA would not produce any non-specific SP peak when the template DNA was not more than 10 ng in this co-amplification system. It was slightly different from our previous study results from by only the 3-plex MSRE-PCR assay that vaginal secretion DNA not more than 80 ng and menstrual blood DNA not more than 5 ng would not produce non-specific signals [6]. Since the commercial kit PowerPlex® Y23 could improve the detection ability of trace amount of DNA in PCR analysis [12, 13], the difference of the most tolerable amount of female DNA between this study and our previous study is possible due to the increased detection ability of non-specific trace DNA and stochastic effect in this co-amplification test. Reliable and reproducible results were recorded if less than 5 ng of DNA (from the female) is used as template in this study. Therefore, the female DNA is suggested to be less than 5 ng from both vaginal secretions and menstrual blood to cautiously avoid the occurrence of non-specific SP peak when performing the co-amplification system in practice.
The primers to amplify DC, SP, and SRY when added to the PCR, for the identification of spermatozoa and male DNA, did not affect the detection of the 23 Y-STR typing. The assignment of the correct alleles was not affected also when amplifying semen DNA extract of mixtures containing other male body fluid or when large amount DNA from females was present. The use for the co-amplification system and interpretation of spermatozoa identification were similar to those in only 3-plex MSRE-PCR [6]: the human and male DNA quantification (such as Quantifiler™ Trio DNA quantification kit) needs to be simultaneously conducted to evaluate the male and female DNA amount in the extract. A positive result for the presence of spermatozoa is based on a profile containing peaks at SP, SRY, and Y-STR loci and no peak at DC locus; the recommendation is not to conduct the assay if more than 10 ng of vaginal DNA or menstrual blood DNA are present to prevent the generation of non-specific SP signal; the amount of male DNA as template should range from 0.1 ng to less than 1.0 ng (0.5 ng is suggested) and the female DNA should be less than 10 ng (less than 5 ng is better) from vaginal secretions and menstrual blood.
This co-amplification system was used to test samples for the presence of spermatozoa and Y-STR profiles of 9 non-probative forensic samples; they had been previously analyzed by the 3-plex MSRE-PCR in our previous study [6]. The results were consistent with the previous study. The tests on non-probative forensic samples showed the applicability of identifying both Y-STR and spermatozoa simultaneously on trace male DNA with excessive female DNA in the background (male/female: 1/41).
Conclusions
This study demonstrated a co-amplification system by combining PowerPlex® Y23 and 3-plex MSRE-PCR in a single reaction. The co-amplification system can identify Y-STR and spermatozoa DNA simultaneously using just 0.1 ng DNA from semen, even in the presence of 5 ng of DNA from a female. This study demonstrates a new concept for the adaption of a Y-STR kit to also detect the presence of spermatozoa. By adding the markers for identification of spermatozoa DNA and slight modification, the spermatozoa can be identified at the same time while the Y-STR is analyzed. The co-amplification will be beneficial for the forensic investigations where there are limited samples.
References
Graves HC, Sensabaugh GF, Blake ET (1985) Postcoital detection of a male-specific semen protein: application to the investigation of rape. N Engl J Med 312(6):338–343
Suttipasit P, Wongwittayapanich S (2018) Detection of prostate specific antigen and semenogelin in specimens from female rape victims. J Forensic Leg Med 54:102–108
Casey DG, Domijan K, MacNeill S, Rizet D, O’connell D, Ryan J (2017) The persistence of sperm and the development of time since intercourse (TSI) guidelines in sexual assault cases at forensic science Ireland, Dublin, Ireland. J Forensic Sci 62(3):585–592
van Oorschot RAH, Szkuta B, Meakin GE, Kokshoorn B, Goray M (2019) DNA transfer in forensic science: a review. Forensic Sci Int Genet 38:140–166
Sibille I, Duverneuil C, De La Grandmaison GL, Guerrouache K, Teissiere F, Durigon M (2002) Y-STR DNA amplification as biological evidence in sexually assaulted female victims with no cytological detection of spermatozoa. Forensic Sci Int 125(2–3):212–216
Liu KL, Tsai LC, Lin YC, Huang NE, Yang LJ, Su CW, Lee JCI, Linacre A, Hsieh HM (2020) Identification of spermatozoa using a novel 3-plex MSRE-PCR assay for forensic examination of sexual assaults. Int J Legal Med 134(6):1991–2004
Sibille I, Duverneuil C, Lorin De La Grandmaison G, Guerrouache K, Teissiere F, Durigon M, De Mazancourt P (2002) Y-STR DNA amplification as biological evidence in sexually assaulted female victims with no cytological detection of spermatozoa. Forensic Sci Int 125(2–3):212–216
Johnson CL, Giles RC, Warren JH, Floyd JI, Staub RW (2005) Analysis of non-suspect samples lacking visually identifiable sperm using a Y-STR 10-plex. J Forensic Sci 50(5):1116–1118
McDonald A, Jones E, Lewis J, O’Rourke P (2015) Y-STR analysis of digital and/or penile penetration cases with no detected spermatozoa. Forensic Sci Int Genet 15:84–89
Alghafri R, Goodwin W, Ralf A, Kayser M, Hadi S (2015) A novel multiplex assay for simultaneously analysing 13 rapidly mutating Y-STRs. Forensic Sci Int Genet 17:91–98
Ralf A, Lubach D, Kousouri N, Winkler C, Schulz I, Roewer L, Purps J, Lessig R, Krajewski P, Ploski R, Dobosz T, Henke L, Henke J, Larmuseau MHD, Kayser M (2020) Identification and characterization of novel rapidly mutating Y-chromosomal short tandem repeat markers. Hum Mutat 41(9):1680–1696
Thompson JM, Ewing MM, Frank WE, Pogemiller JJ, Nolde CA, Koehler DJ, Shaffer AM, Rabbach DR, Fulmer PM, Sprecher CJ, Storts DR (2013) Developmental validation of the PowerPlex® Y23 System: a single multiplex Y-STR analysis system for casework and database samples. Forensic Sci Int Genet 7(2):240–250
Ferreira-Silva B, Fonseca-Cardoso M, Porto MJ, Magalhães T, Cainé L (2018) A comparison among three multiplex Y-STR profiling kits for sexual assault cases. J Forensic Sci 63(6):1836–1840
Sambrook J, Fritsch ER, Maniatis T (1989) Cold Spring Harbor Laboratory Press; New York. Molecular cloning: a laboratory manual:9.14–9.19.
Clark C, Turiello R, Cotton R, Landers JP (2021) Analytical approaches to differential extraction for sexual assault evidence. Anal Chim Acta 1141:230–245
Rodriguez JJRB, Calacal GC, Laude RP, De Ungria MCA (2017) Non-differential DNA extraction of post-coital samples submitted as evidence for investigating sexual assault cases in the Philippines. Philipp Sci Lett 10(1):14–21
Wasserstrom A, Frumkin D, Davidson A, Shpitzen M, Herman Y, Gafny R (2013) Demonstration of DSI-semen—a novel DNA methylation-based forensic semen identification assay. Forensic Sci Int Genet 7(1):136–142
Lin YC, Tsai LC, Lee JCI, Su CW, Tzen JTC, Linacre A, Hsieh HM (2016) Novel identification of biofluids using a multiplex methylation sensitive restriction enzyme-PCR system. Forensic Sci Int Genet 25:157–165
PowerPlex® Y23 System, technical manual, instructions for use of products DC2305 and DC2320, Revised 5/17 TMD036, Revised 4/17 TMD035
Butler JM (2003) Recent developments in Y-short tandem repeat and Y-single nucleotide polymorphism analysis. Forensic Sci Rev 15(2):91–111
Geppert M, Edelmann J, Lessig R (2009) The Y-chromosomal STRs DYS481, DYS570, DYS576 and DYS643. Leg Med (Tokyo) 11(Suppl 1):S109–S110
Funding
This study was supported by the Ministry of Science and Technology (NSC 102–2628-B-015–001-MY2), and Ministry of the Interior (108–0805-05–17-01 and 109–0805-05–17-01) in Taiwan.
Author information
Authors and Affiliations
Contributions
All authors contributed to the conception of this study and the experimental design. Material preparation, data collection, and analyses were performed by Yu-Chih Lin, Li-Chin Tsai, Kuo-Lan Liu, Nu-En Huang, and Chih-Wen Su. The first draft of the manuscript was written by Lih-Jing Yang, James Chun-I Lee, Adrian Linacre, and Hsing-Mei Hsieh, and all authors commented on subsequent versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
In this study, samples were collected after informed consent and following the procedures approved by the Institutional Review Board (IRB) of Tao-Yuan General Hospitals (IRB No. TYGH102011) and Antai-Tian-Sheng Memorial Hospital (IRB No. 18–074-B) in Taiwan. Neither the authors are affiliated to these hospitals nor the study was carried out in these hospitals. And the studies have been performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lin, YC., Tsai, LC., Liu, KL. et al. A novel co-amplification system for simultaneous amplification of 23 Y-STR and identification of spermatozoa. Int J Legal Med 136, 73–84 (2022). https://doi.org/10.1007/s00414-021-02723-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-021-02723-8