Abstract
Identification of semen and then spermatozoa is essential to verify that sexual activity has occurred in alleged cases of sexual assault. Microscopic examination commonly used for spermatozoa identification is however time-consuming and can often lead to false-negative results for samples with deformed and, or, limited number of spermatozoa. To address this limitation, we report on a novel 3-plex MSRE-PCR (methylation-sensitive restriction enzyme-PCR) assay to specifically identify spermatozoa. This assay is comprised of 3 markers: a digestive control marker (DC), sperm-specific marker (SP), and Y chromosome marker (SRY). A total of 214 samples from 10 body fluids or tissues were analyzed. Specificity testing showed that all the normal semen samples were unambiguously identified as being sperm-positive, and no other body fluid (or tissues) showed a sperm-specific signal in the electropherogram. Testing for sensitivity showed that 0.1 ng of DNA from a semen extract was sufficient to identify the presence of spermatozoa by this assay. Mixture analyses illustrated the sensitivity of the assay when the vaginal/semen DNA ratio (80/0.1) was under 800 or the menstrual blood/semen DNA ratio (5/0.1) was under 50, the trace amounts (approximately 0.1 ng) of DNA from semen can still be identified by this 3-plex MSRE-PCR assay. This assay was also applied to the identification of 31 non-probative forensic samples from 18 sexual assault cases. The case studies showed that the 3-plex MSRE-PCR assay was an improvement in the sensitivity of spermatozoa detection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Determination of the source of a body fluid encountered on crime scene exhibits can be pivotal in many criminal investigations. This is especially the case in alleged sexual assaults where there may be dispute over the source, transfer, and persistence of such evidential material [1]. The assay for acid phosphatase (ACP) activity is the most frequently used chemical test as a presumptive assay for semen [2]. The immunological test for the detection of Sg (Semenogelin) and the microscopic examination for sperm cells are the most common confirmatory tests [3, 4]. For any vasectomized or sperm-free samples, it is common practice to use an immunological test for a specific antigen for semen, such as P30 (or PSA, prostate-specific antigen) and Sg. It should be noted that P30 can also be detected in body fluids other than semen [5, 6]. The advantages of the semen-identification assays currently adopted are that they are convenient, simple to use, inexpensive, and fast; however, they usually suffer from the limitations of specificity and sensitivity in practical applications.
All too often, rape cases are not reported immediately to the authorities and any subsequent delay in the collection of evidence can lead to the loss of useful forensic evidence. It has been reported that spermatozoa can be identified by microscopy up to 72 h, and PSA and Sg can be identified up to 47 h and 72 h, respectively, after a sexual assault [7, 8]. Further, it was reported that the persistence of spermatozoa in vagina and anus of a victim significantly declines 48 h after intercourse [9] but that a DNA Y-STR profile can still be identified from samples collected 5 to 6 days after intercourse [10].
More recently, methods for semen identification have been reported that include spermatozoa cell purification using affinity reagents [11], spermatozoa laser microdissection [12], Raman spectroscopy analysis [13], and specific mRNA and microRNA (miRNA) detection [14,15,16], as well as assessing DNA methylation [17, 18]. DNA methylation-based assays have been regarded as a promising assay for semen testing due to their high specificity and feasibility of merging into current forensic casework processes [2]. DNA methylation is generally related to gene regulation and correlated to cell differentiation [19,20,21]. The tissue-specific differentially methylated regions (tDMRs) exhibit different DNA methylation profiles according to cell or tissue types by restriction landmark genomic scanning [22].
The kit known as DNA source identifier (DSI-semen) has been reported for confirmation of semen with semen-specific methylation patterns by methylation-sensitive restriction enzyme-PCR (MSRE-PCR) [23]. In this system, DNA template from forensic casework samples was obtained by using the differential extraction method [24]. However, vaginal swabs from the sexual assault cases usually contain high levels of female epithelial cells and only a limited number of spermatozoa, making it unfavorable to apply a differential extraction method to recover sufficient male DNA. Other methylation-based assays for spermatozoa identification use a single-base extension system [25], and pyrosequencing [26] for analysis of bisulfite converted DNA. However, the genomic DNA can be degraded during bisulfite treatment [27].
A 10-plex MSRE-PCR system to identify various biofluids (including semen) was reported previously by our laboratory [28]. While it is an effective assay for body fluid typing, non-specific signals including those markers for the semen were observed in the presence of excessive DNA from vaginal fluid (e.g., 25 ng). In order therefore to overcome such artifacts created by excessive DNA from the female, a 3-plex MSRE-PCR assay was established with high specificity for DNA from spermatozoa as well as low sensitivity for DNA from other body fluids, and a validation study was also performed.
Materials and methods
Sample collection
A total of 214 samples were collected from 55 male and 63 female adults in this study, including semen (both healthy and vasectomized donors), vaginal secretion, menstrual blood, peripheral blood, saliva, nasal secretion, sweat, urine, feces, and breast milk. The age of semen donors ranged from 20 to 52 years old. Details of the samples are listed in Table 1. Samples were collected after informed consent and following the procedures approved by the Institutional Review Board (IRB) of Taoyuan General Hospitals (IRB No. TYGH102011) and Antai-Tian-Sheng Memorial Hospital (IRB No. 18-074-B) in Taiwan.
DNA extraction and quantification
Genomic DNA was isolated from the collected samples using the Qiagen Mini kit (Qiagen, Hilden, Germany) following the manufacturer’s suggestions (for DNA purification from Tissues), and it is notable that this protocol was modified by adding 7 μL of DTT (1 M) to the ATL buffer for semen samples. The isolated DNA was quantified by using the Quantifiler™ Trio DNA Quantification Kit (Life Technologies, CA, USA) in combination with a 7500 Real-Time PCR machine (Life Technologies).
Marker selection
Candidate CpG loci were selected from the database of the Infinium HumanMethylation450 BeadChip Kit in the GPL13534 platform of the Gene Expression Omnibus at NCBI (http://www.ncbi.nlm.nih.gov/geo). These included 8 spermatozoa samples [29], 11 peripheral blood samples [30], 20 cervical tissues [31, 32], and 22 saliva samples [33]. For each candidate CpG locus, mean and standard deviation of the beta-values (the calibration ratio of methylation) for each biofluid or tissue were calculated using Microsoft Office Excel 2007. The candidate loci were selected with the criterion of a high beta-value (at least 0.9) for DNA from spermatozoa, however also a low (near zero) value for the DNA from other body fluids. Additionally, they must contain as many recognition sequences (5′-GCGC-3′) for methylation-sensitive restriction enzyme HhaI as possible.
For confirmation of the restriction enzyme digestion, “digestive control” markers were selected with the criteria of containing as many recognition sequences for HhaI as possible, and no or low methylation in any biofluid or tissue DNA. The male-specific sex-determining region Y (SRY) without the HhaI recognition sequences was used to confirm the DNA from male.
Preliminary MSRE-PCR tests
Primers were designed using the Primer3 software for amplification of the selected markers [34, 35], and the Tm (melting temperature) ranged from 60 to 63 °C. The predicted amplicon size was from 395 to 420 bp. The preliminary MSRE-PCR test was performed in a total volume of 10 μL containing DNA template (0.5 ng of semen DNA [23, 28], 25 ng of vaginal DNA, or 25 ng of menstrual blood DNA), 10 unit of HhaI (New England Biolabs, MA, USA), 0.5 unit of AmpliTaq Gold® 360 DNA Polymerase (Thermo Fisher Technologies), 1 μL of AmpliTaq Gold 360 10× PCR Buffer, 1.5 mM of Mg2+, 200 μM for each dNTP, 1 μL of 360 GC enhancer, and 300 nM of each primer. The reactions were conducted in a GeneAmp® PCR System 9700 (Life Technologies). Before the cycling reaction, the first step of the thermal program was for DNA digestion using HhaI at 37 °C for 60 min. This was followed by 95 °C for 11 min for both heat inactivation and PCR initiation, and then 30 cycles of 94 °C for 20 s and 61.5 °C for 2 min, with a final extension at 72 °C for 30 min. The amplification products were checked on a 2% agarose gel.
A 3-plex MSRE-PCR assay
Three markers were selected according to the results of preliminary tests. A 3-plex MSRE-PCR system including these markers was established. They were a sperm-specific marker cg26763284 (named SP in this study), a digestive control marker cg21784498 (DC), and a Y chromosome marker (SRY). This 3-plex MSRE-PCR was performed in a total volume of 10 μL with the compositions as described in the preceding preliminary MSRE-PCR tests except for the DNA amount (0.2–0.5 ng in this study) and primer concentrations. The optimal concentrations and sequences for the primers are shown in Online Resource 1. These reactions were conducted in a GeneAmp® PCR System 9700 with the conditions as previously described in the preliminary MSRE-PCR tests. PCR products were analyzed using the ABI PRISM 3500 Genetic Analyzer and GeneMapper® ID-X v1.4 software (Life Technologies). For each sample tested, only the plus HhaI reaction (no minus HhaI reaction) needs to be conducted; however, for a batch of samples, a plus and minus HhaI reactions were both performed as the controls by using 0.5 ng semen DNA for each reaction. The threshold for a positive signal was 150 RFU (Relative fluorescence unit) under considerations of the LOQ (limit of quantitation, average + 10 SD of noise) and the peak heights of other artifacts. The sizes of the signals from the ladders (composed of DC, SP, and SRY peaks) from 15 batches for the 3-plex MSRE-PCR assay were collected to determine the definition of the size bins (± 3 SD).
Validation tests for the 3-plex MSRE-PCR assay
Specificity
In addition to the semen samples, vaginal secretion and menstrual blood samples (which are encountered frequently in sexual assault cases), and other commonly encountered specimens were used in the specificity tests using 0.2–1 ng of DNA template [23, 28] for the 3-plex MSRE-PCR assay. Furthermore, DNA from breast milk was also tested due to reports of the presence of PSA in breast milk by highly sensitive immunoassays [36, 37].
Sensitivity
DNA from semen was diluted serially to 0.2, 0.1, 0.05, and 0.025 ng/μL for the sensitivity tests of this 3-plex MSRE-PCR assay. DNA (1 μL) from each dilution was used as the template in a reaction volume of 10 μL.
Excessive female DNA and mixture
To test the effects of excessive female DNA, a large amount of female DNA from vaginal secretions and menstrual blood was tested by the 3-plex MSRE-PCR assay. Each DNA extract from 20 vaginal secretions was tested at templates of 5, 10, 20, 40, 80, and 100 ng, and from 20 menstrual blood samples using 5, 10, 20, and 40 ng. These tests were used to evaluate the effects of female DNA in different amounts and assay for any individual variation.
Furthermore, simulated mixtures were prepared to evaluate the influence of excessive vaginal or menstrual blood DNA on spermatozoa identification within these mixtures. Each body fluid was collected from 10 individuals. Each combination of the mixture composed of 0.1 ng DNA from semen with 80 ng DNA from vaginal fluid, or 0.1 ng DNA from semen with 5 ng menstrual blood DNA, with preparation of both combinations to create 10 sets of samples.
Other mixed sample types encountered in sexual assault cases, such as saliva DNA mixed with sperm DNA, have also been tested in our study. Five semen and five female saliva DNA samples were collected to prepare five mixture samples. Each sample was mixed by adding 0.1 ng semen DNA with 80 ng female saliva DNA. Furthermore, five semen and five male saliva DNA were also collected to prepare mixture samples. Each sample was mixed at the ratios of 1:0, 1:1, 1:3, 1:9, and 0:1 (semen to saliva), and the total DNA input was 1 ng in the 3-plex MSRE-PCR assay.
Degraded DNA
Samples to mimic degraded DNA were prepared and tested in our study. Semen DNA was aliquoted into 8 separate microtubes for each of the three donors, and then damaged by UVC radiation (Philips, TUV 15 W, wavelength 100–280 nm) for 1, 2, 5, 10, 20, 30, 60, and 120 min respectively in a laminar flow. Each microtube (50 μL DNA, 0.5 ng/μL) was about 54 cm away with 32° (angle) from UVC radiation. These artificially degraded semen DNA samples were then quantified with trio DNA quantification kit and tested with the 3-plex MSRE-PCR assay.
Non-probative forensic sample
For comparison of the 3-plex MSRE-PCR assay with the currently used methods for semen identification in forensic practice, 31 non-probative forensic samples (from 18 alleged sexual assault cases) were collected that included low-vaginal swabs (collected from the position near and around vaginal orifice, including vulva and perineum), high-vaginal swabs (collected from the position between cervix to posterior vaginal fornix with the help of a disposable vaginal speculum), underpants, tissue papers, and T-shirt (Table 2). Stains and swab heads were tested by Kastle-Meyer test [38], acid phosphatase test [39], PSA test (SERATEC® GmbH, Goettingen, Germany), RSID-Semen (Independent Forensics, Hillside, IL, USA), microscopy, Trio DNA quantification (Quantifiler™ Trio DNA Quantification Kit, Life Technologies), STR typing, and the 3-plex MSRE-PCR assay (this study). For the PSA test, if the color intensities were equal to or more than that for the control line of 4 ng/μL, then it was recorded as positive, and less than that then recorded as a weak positive. For RSID-Semen, a clear and definite color on test line was identified as a positive result and faint color as weak positive. STR typing was performed with the AmpFLSTR Identifiler Plus PCR Amplification Kit (Life Technologies) for autosomal STR or PowerPlex® Y23 System (Promega, WI, USA) for Y chromosomal STR.
Results
Marker selection
A search was conducted successfully for 2 digestive control markers and 2 sperm-specific markers based on the strategies and criteria as previously described in “Materials and methods” (Marker selection). Furthermore, the primers for the digestive control and semen-specific markers (SE-I and SE-II) of 10-plex MSRE-PCR assay in our previous study [28] were redesigned to extend their amplicon size to contain more recognition sequences for HhaI digestion. Totally, there were 3 digestive control and 4 sperm-specific loci selected and evaluated in the following preliminary tests (Table 3). The beta value was low for all the digestive control markers in all biofluids and tissues; for the sperm-specific markers, the beta value was high in spermatozoa and low in both the other body fluids and tissues.
Preliminary MSRE-PCR tests
To evaluate the specificity and efficiency for semen DNA and extracts from large amounts of vaginal DNA, the candidate markers were preliminarily tested by using 0.5 ng semen DNA and 25 ng vaginal DNA (from which non-specific signals for semen markers have been observed in our previous 10-plex MSRE-PCR assay) respectively for each of 3 samples (Online Resource 2), where the ratio of vaginal to seminal DNA (25/0.5) was 50. All of the 4 sperm-specific candidate markers could generate PCR products in 3 semen samples; however, only CpG ID cg26763284 (renamed SP in this study) did not generate any detectable PCR products for all of the 3 samples of 25 ng vaginal DNA. According to the database, this locus showed a beta value of 97.20% for spermatozoa (higher than the other markers) and only 0.51% for cervical tissue (Table 3). Furthermore, it contained 6 HhaI recognition sequences in the amplicon and was more accessible for HhaI digestion for any non-methylation fragments.
To evaluate the efficiency of HhaI digestion in the presence of excessive amounts of female DNA, the 3 candidate digestive control markers were tested using 25 ng of vaginal DNA and menstrual blood DNA for each of 3 samples (Online Resource 2). Only CpG ID cg21784498 (renamed DC in this study) did not generate any detectable PCR products (complete digestion) for any of the female DNA samples. This locus contained 8 recognition sequences for HhaI (Table 3) and therefore was used as the control marker for HhaI digestion in this study.
SRY is a sex-determining gene on the Y chromosome, and selected as an indicator to confirm that the source of DNA was from a male. The SRY amplicon did not contain any HhaI recognition sequences (Online Resource 3), and therefore, it cannot be digested by HhaI; thus, PCR products should always be generated in the presence of DNA from a male. PCR products were observed as expected from all the semen DNA extracts and not observed for all the samples of vaginal DNA (Online Resource 2).
A 3-plex MSRE-PCR assay
A digestive control (DC, cg21784498), a sperm-specific marker (SP, cg26763284), and a Y chromosome marker (SRY) were combined to create a 3-plex MSRE-PCR assay for spermatozoa identification. The electropherogram of an example DNA from semen, vaginal secretion, and menstrual blood identified by this 3-plex MSRE-PCR assay is shown in Fig. 1. Reactions without HhaI digestion were also performed for comparison. Without HhaI digestion (HhaI-), the peaks of DC and SP loci were observed in all samples; the peak of SRY marker was only observed in DNA from semen as expected. After the HhaI digestion (HhaI+), no peaks were observed for DNA samples from vaginal secretion and menstrual blood samples at these three loci due to their un-methylated recognition sequences within the DC and SP markers and a deficiency of the SRY gene. In contrast, peaks for both the SP and SRY loci were observed for DNA extracts from semen due to the methylated SP fragment and the presence of the male-specific SRY gene. The signals of the ladders from 15 batches were collected for definition of the size bins for precise sizing in the 3-plex MSRE-PCR assay. The average ± SD of size for each signal is calculated as 400.68 ± 0.19 bp for DC, 407.23 ± 0.27 bp for SP, and 410.03 ± 0.20 bp for SRY respectively. The largest SD was 0.27 bp, and thus, 3 SD was 0.81 bp. Therefore, the size bins for each signal could be determined as ± 0.81 bp in the panel management of GeneMapper® ID-X v1.4 software for run to run comparison.
Validation tests for the 3-plex MSRE-PCR assay
Specificity
In addition to semen and female body fluids encountered frequently in sexual assault cases, other biofluids occasionally observed in crime scenes were also collected to assess the specificity of the 3-plex MSRE-PCR assay. These comprised peripheral blood, saliva, nasal secretion, sweat, urine, and feces. Furthermore, 2 semen samples from vasectomized males and 5 breast milk samples were also collected and tested (Table 1). The expected results were observed for all samples using the 3-plex MSRE-PCR assay (Online Resource 4). For all the female samples (Online Resource 4c, e, g, i, k, m, and o), no peaks were observed. For all the male samples but the healthy non-vasectomized semen, only the SRY peak was observed (Online Resource 4b, d, f, h, j, l, and n). The SP peak was observed only for semen samples collected from the non-vasectomized donors (Online Resource 4a), but not for those that had a vasectomy (Online Resource 4b). The results showed that the 3-plex MSRE-PCR assay was highly specific for DNA from spermatozoa rather than DNA from seminal fluid.
Sensitivity
DNA from 20 semen samples (donated by non-vasectomized males) was used to determine the sensitivity of the 3-plex MSRE-PCR assay. For each semen DNA extract, 0.2, 0.1, 0.05, and 0.025 ng were used (Fig. 2). None of the samples showed any peaks at the DC locus indicating complete digestion by the restriction enzyme HhaI. When 0.1 and 0.2 ng DNA were used (in Fig. 2c, d), all of the samples exhibited more than 40% peak height ratio (PHR, the lower peak height divided by the higher peak height) between SP and SRY loci (sample 19 has the lowest PHR of 47% in SP and SRY loci in Fig. 2d). Though the PHR of sample 19 was the lowest, the peak height of the lower peak (SP) of sample 19 was close to 5000 RFU. When 0.05 ng DNA was used (in Fig. 2b), samples 01 and 13 have a lower PHR between 20 and 40% in SP and SRY loci (35 and 32% respectively); moreover, sample 18 has a PHR lower than 20% in SP and SRY loci (13%). When 0.025 ng DNA was used (in Fig. 2a), 2 out of the 20 samples have lower PHR (samples 12 and 13, by 21% and 36% respectively); moreover, the SRY peaks dropped out in 3 of the 20 samples (samples 06, 09, and 18) and SP peak in one sample (sample 03). The result also illustrates that stochastic effects will occur, such as lower PHR or peak drop-out, in some samples at the trace amounts of DNA. For interpretation of the results, regardless if the sperm DNA is over 0.1 ng or not, if both SRY and SP peaks are detected (≧150 RFU) with no DC peak, the interpretation is “sperm-positive.” For interpretation of the sperm negative, the threshold of SRY needs to be determined. On the basis of sensitivity testing (only SRY detected with peak height of 609 RFU for sample 03, Fig. 2a), the threshold for determining sperm-negative was considered as 700 RFU. Therefore, when only SP is detected, or only SRY is detected with peak height from 150 RFU to less than 700 RFU, the results would be identified as inconclusive. In addition, a recommendation is that more than 0.1 ng but less than 1 ng of DNA from semen should be used as template for the 3-plex MSRE-PCR assay since definite results were obtained using 0.1 ng of DNA from semen; however, pull-up signals were observed with more than 1 ng of semen DNA (data not shown).
Sensitivity test of the 3-plex MSRE-PCR assay for semen. The 20 semen samples were from different donors and are numbered from 01 to 20 on the x axis. The y axis is the RFU data. For 0.025 ng DNA (a), the empty arrows indicate that the SRY has dropped out (samples 06, 09 and 18) and the diagonally striped arrow indicates that the SP has dropped out (sample 03) in the electropherogram. The dotted frames indicate the inter-locus peak height imbalance with a peak height ratio (PHR) lower than 40% in SP and SRY loci (samples 12 and 13, by 21% and 36% respectively). For 0.05 ng DNA (b), the dotted frames indicate the inter-locus peak height imbalance with a PHR lower than 40% in SP and SRY loci (samples 01 and 13, by 35% and 32% respectively) and sample 18 shows a significant inter-locus peak height imbalance with PHR lower than 20% in SP and SRY loci (13%)
Excessive female DNA and mixture
Each of 20 samples of DNA from vaginal secretion and menstrual blood was used to evaluate the effects of excessive amounts of female DNA in the 3-plex MSRE-PCR assay. The results showed that when the DNA from the female was less than 80 ng, no peaks were observed as expected for all of the 20 vaginal samples (Online Resource 5). However, when the vaginal DNA was 100 ng, the SP peak was observed in 2 of the 20 samples (No. 11 and 12) with 208 and 170 RFU, respectively, and a DC peak was observed in one sample (No. 11) with 187 RFU (Online Resource 5f). No peaks were observed for these DNA extracts from vaginal secretions at SRY locus. The same tests for the menstrual blood DNA were performed (Online Resource 6). No peaks were observed when the DNA template was 5 ng for all of the 20 menstrual blood samples. However, when DNA was increased to 10 ng, a signal of 165 RFU at the SP locus for the No. 12 sample was observed and at 20 ng DNA, 2 signals (169 and 180 RFU) at the SP locus for the No. 01 and 12 samples were recorded. When DNA was 40 ng, there were 7 samples (No. 01, 04, 11, 12, 14, 17, and 18) that showed signals at either the DC or SP locus or both loci. No SRY signal was observed for all DNA extracts from only menstrual blood. In the presence of excessive amounts of DNA from female, incomplete enzymatic digestion using HhaI occurred as indicated by the DC signal. The results showed incomplete digestion of HhaI when the vaginal DNA was 100 ng, and the menstrual blood DNA was 40 ng, and in this scenario, the result would be interpreted as “failed” in the 3-plex MSRE-PCR assay.
It should be noted that the 3-plex MSRE-PCR assay could be invalidated if incomplete digestion of DNA template by HhaI occurred. The recommended template DNA for this assay was therefore suggested to be not more than 80 ng from vaginal secretions and 20 ng from menstrual blood. A positive result for the presence of DNA from semen is therefore based on a profile containing peaks at both the SP and SRY loci and no peak at DC locus. A profile where only the SP peak is detected could be the result of excessive female DNA, and therefore, not more than 80 ng of vaginal secretion DNA and 5 ng of menstrual blood DNA were recommended for the 3-plex MSRE-PCR assay to prevent the generation of any non-specific SP peak.
A further 10 samples of semen, vaginal secretion, and menstrual blood DNA were used to test the sensitivity of the assay for the mixture (Fig. 3). A dilution of 0.1 ng of semen DNA gave results in all 10 samples and all with high RFU value for both the SP and SRY peaks. Data are also shown when mixing 0.1 ng of semen DNA and 80 ng of vaginal DNA, and also 0.1 ng of semen DNA and 5 ng of menstrual blood DNA (Fig. 3). In each mixed body fluid sample, both the SP and SRY signals were clearly detected for spermatozoa DNA (Fig. 3c, e). An example of the electropherogram from the mixture F in Fig. 3 is shown in Fig. 4. The peak heights of both SP and SRY were all higher than 1000 RFU in the mixed samples even when the semen DNA was only 0.1 ng. Only 2 (0.1 ng semen DNA mixed with 80 ng vaginal DNA, Fig. 3c, mixtures B and H) out of the 10 mixtures exhibited peak heights under 2000 RFU for both SP and SRY loci, which were lower than the peak heights of 0.1 ng semen DNA tested alone. These data indicated that although the peak heights for some mixtures were lower than the semen tested alone, the detection of spermatozoa using this 3-plex MSRE-PCR was therefore not affected by the excessive female DNA. This detection of semen DNA was effective even when there was 800 times more vaginal secretion DNA than semen DNA (ratio 80/0.1) or 50 times more menstrual blood than semen DNA (ratio 5/0.1).
Furthermore, other mixed sample types encountered in sexual assault cases, such as saliva DNA mixed with sperm DNA, have also been tested in our study. Five semen and five female saliva DNA samples were collected to prepare five mixture samples. Each sample was mixed by adding 0.1 ng semen DNA with 80 ng female saliva DNA (ratio 80/0.1), yet all samples resulted as sperm-positive in the 3-plex MSRE-PCR assay. These results are similar to those of sperm and vaginal DNA mixtures. Furthermore, five semen and five male saliva DNA samples were also collected to prepare mixture samples. Each sample was mixed at the ratios of 1:0, 1:1, 1:3, 1:9, and 0:1 (semen to saliva) and the total DNA input was 1 ng in the 3-plex MSRE-PCR assay. The results also showed all (excepting 0:1) were sperm-positive even for the mixture with 0.1 ng semen DNA and 0.9 ng male saliva DNA (1:9). However, in this scenario, a significant inter-locus peak height imbalance (PHR < 5%) was observed.
Degraded DNA
From the results of DNA quantification, the semen DNA samples damaged by UVC exposure for 1, 2, 5, and 10 min were recorded as “non-degraded” with a DI (degradation index) lower than 1.5, according to the report [40]. In this report, four arbitrary degradation categories are provided: 0 < DI < 1.5 for non-degraded, 1.5 < DI < 4 for mildly degraded, 4 < DI < 10 for degraded, and 10 < DI for severely degraded. Following these categories, in our study, samples exposed for 20 and 30 min were recorded as “mildly degraded,” samples exposed for 60 min recorded as “degraded,” and samples exposed for 120 min recorded as “severely degraded” (Online Resource 7). From the results of the 3-plex MSRE-PCR assay, the semen DNA samples exposed for 1, 2, 5, 10, 20, and 30 min were identified as “sperm-positive” following the user guideline (Online Resource 8), though the mildly degraded semen samples exposed for 20 and 30 min were detected with a lower PHR. One (semen DNA 2) out of three degraded semen DNA samples exposed for 60 min was interpreted to be sperm-positive with a significant inter-locus peak height imbalance (PHR 4.8%), and the other two samples recorded only a SP signal but no SRY. The three severely degraded semen DNA samples exposed for 120 min were identified as inconclusive due to only SP signal but no SRY (Online Resource 8). The results showed that the mildly degraded semen DNA samples can be used in the 3-plex MSRE-PCR assay, the degraded semen DNA samples may still be detected (such as the semen DNA 2), and the severely degraded semen DNA samples were identified as inconclusive.
Non-probative forensic sample tests
A total of 31 samples were collected from 18 sexual assault cases. These comprised low-vaginal swabs (collected from the position near and around vaginal orifice, including vulva and perineum), high-vaginal swabs (collected from the position between cervix to posterior vaginal fornix with the help of disposable vaginal speculum), underpants, tissue paper, and T-shirt (Table 2). When the samples were collected, they were put in evidence storage boxes for air-dry and stored at room temperature in evidence room. The time interval between the sample collections to testing is approximately 1 month (~ 30 days). As part of previous forensic examinations, these items had been tested by Kastle-Meyer test, acid phosphatase test, PSA test, RSID-semen test, microscopic examination, Trio DNA Quantification, and STR typing (autosomal STR and Y-STR), and in this study, the 3-plex MSRE-PCR assay was performed. The results for these tests are listed in Table 4. Sixteen out of 31 samples were identified as sperm-positive by both microscopy and the 3-plex MSRE-PCR assay. Another 11 samples of the 31 samples were identified as sperm-negative by the microscopy and “inconclusive” or “no result” by the 3-plex MSRE-PCR assay; additionally, STR typing (Y-STR) of these samples also gave no result as the male DNA in the extracts was undetected (UD) or less than 0.005 ng/μL (detection limit of the quantification kit). For the other four out of the 31 samples (No. 2, 9–2, 9–4, and 10), no spermatozoon was observed by microscopy; however, the results were spermatozoa-positive by using the 3-plex MSRE-PCR assay and male STR profiles had been obtained (autosomal STR or Y-STR). One of the 4 samples (No. 2) was a high-vaginal swab reported to have been collected 3–4 days after an alleged sexual assault; the swab gave a weak positive result for ACP test and negative results for RSID-semen and PSA tests. A male STR profile had been obtained from the high-vaginal swab and a fair assumption was that the DNA originated from sperm cells due to the spermatozoa with the longest persistence and highest detection rate [8, 9]. Another of the four samples (No.10) was reported to be from tissue paper used after an alleged sexual assault; the tissue paper gave a positive result for ACP and PSA tests and a negative result for RSID-semen test. The other two samples (Nos. 9–2 and 9–4) were a low-vaginal swab and a T-shirt. The former (No. 9–2) gave a weak positive result for ACP and RSID-semen tests, and a negative result for PSA test; the latter (No. 9–4) gave a weak positive result for ACP test, a negative result for RSID-semen test, and failed in the PSA test.
Among these 31 samples, sample no. 17–2 was recorded as having the longest time interval between the alleged sexual assault and evidence collection; this item was a high-vaginal swab labeled as collected up to 6–7 days after the alleged offense. Only 4 heads of spermatozoa were recorded by microscopy, which was corroborated by the 3-plex MSRE-PCR assay giving a spermatozoa-positive result. The ratio of female/male DNA was approximately 833 (4.5 μL of DNA template, 78.786/0.0945 = 833.714). Sample no. 13–2 was a high-vaginal swab stained with menstrual blood based on victim’s statement and recorded as collected up to 3–4 days after the alleged offense. This swab gave a positive result for blood using Kastle-Meyer test and spermatozoa recorded by microscopy and the 3-plex MSRE-PCR assay. The ratio of female/male DNA was approximately 113 (1 μL of DNA template, 18.6358/0.1641 = 113.5637). The DNA amount taken for the 3-plex MSRE-PCR assay was following the above validation results as not more than 80 ng DNA from vaginal secretions and 20 ng DNA from menstrual blood.
From the results of these tests for forensic samples (Table 4), RSID-semen tests were observed predominantly concordant with the microscopic examinations except for two samples (Nos. 5–2 and 9–2). Both gave weak positive results for RSID-semen test and negative results for microscopic examination. One of the two samples (No. 9–2) was positive for spermatozoa based on the 3-plex MSRE-PCR assay and male STR profile was obtained; the other sample (No. 5–2) gave no result for both 3-plex MSRE-PCR assay and male STR typing. Additionally, results of the 3-plex MSRE-PCR assay for all these forensic samples matched with the results for male STR typing.
The process of differential extraction is common practice in forensic casework; in this study, four evidential samples identified as sperm-positive by microscopic examination were selected. Differential DNA extraction was performed, and thus, eight DNA samples (four sperm fraction DNA and four non-sperm fraction DNA) were quantified by the trio DNA quantification and tested by the 3-plex MSRE-PCR assay. Six DNA samples (four sperm fraction DNA and two non-sperm fraction DNA extracts) returned quantification data of more than 0.01 ng/μL for male DNA and interpreted as sperm-positive by the 3-plex MSRE-PCR assay, and Y-STR typing generated the expected alleles. The results indicated that all sperm fraction DNA extracts were interpreted as sperm-positive for the 3-plex MSRE-PCR assay, and some of non-sperm fraction DNA extracts containing more female DNA and less male DNA also have limited sperm DNA to be detected.
These case studies illustrated how the newly designed 3-plex MSRE-PCR has higher sensitivity, and is better corroborated, than spermatozoa identification by microscopy. The new assay is a further method for the effective identification of spermatozoa, particularly if the spermatozoa are deformed, or they are in very limited number and hard to find.
Discussion
Unambiguous identification of spermatozoa is central to the investigation of alleged sexual assault cases. Commonly, swabs and stains are tested first with a presumptive test for semen and, if positive, followed by microscopy to identify spermatozoa. Genetic testing to confirm the presence of spermatozoa would be greatly aided by the identification of a spermatozoa-specific marker (SP). Such a genetic marker was the subject of a search in this study and based on the analysis of 214 DNA samples from 10 body fluids (or tissues), only semen containing spermatozoa exhibited a spermatozoa-specific pattern with both SP and SRY peaks generated by the 3-plex MSRE-PCR assay. No SP peak was ever observed if spermatozoa were not present (such as body fluids other than semen) even for instances when the male vasectomized. Males with a vasectomy will produce semen, and associated epithelial cells from the male, but no spermatozoa, thus confirming that the SP marker was spermatozoa-specific and not able to generate a result from other male-specific cell types. Sensitivity testing showed that even 0.1 ng of DNA from semen (containing spermatozoa) was sufficient to identify the presence of spermatozoa. A positive result for the 3-plex MSRE-PCR assay can be observed using around 35 sperm cells (for 100 pg); therefore, about 550 sperm cells in the extract (100 μL) are necessary to robustly have a positive result. If the sperm cells are insufficient in the extract, so that the total input DNA will be less than 100 pg, the results may show a drop-out of SP and/or SRY peaks or an imbalance of PHR of SP and SRY loci due to the influences of stochastic effects. In fact, it is difficult to accurately determine how many sperm cells are present in an extract, since the male DNA from DNA quantification includes both sperm and non-sperm male DNA. In practical cases, the concentration of DNA extracts can be adjusted in the process of extraction. For instance, use less amount of H2O (such as 50 μL) to elute the DNA in the final step of extraction.
However, the complete digestion for methylation-sensitive restriction enzyme (e.g., HhaI) is crucial to the MSRE-PCR assay. Inhibitors for HhaI and excessive amounts of DNA template can result in the incomplete digestion of the DNA template making the results invalid. In this study, amplicons of DC and SP fragments were designed to cover more GCGC recognition sequences to assist in complete digestion of the DNA. When DNA from vaginal fluids was more than 80 ng and menstrual blood DNA more than 20 ng, the 3-plex MSRE-PCR assay could still fail due to the incomplete digestion of the DNA. These results indicated that the restriction enzyme was less efficient in digestion of DNA from menstrual blood than the vaginal DNA. This finding is possible due to the presence of hemoglobin in menstrual blood, which is a well-known inhibitor for some enzymes, such as the DNA polymerase for PCR amplification [41].
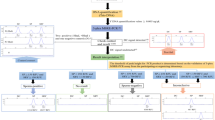
For more convenient use of the 3-plex MSRE-PCR assay, a recommended workflow and user guideline is shown in the online resource 8. Steps are as the following: The human and male DNA can be simultaneously quantified by the Quantifiler™ Trio DNA quantification kit (b in the flow chart) to evaluate the male and female DNA in the extract. If the samples contain male DNA not less than 0.005 ng/μL, then the 3-plex MSRE-PCR assay (c in the flow chart) can be applied. For the total input DNA, the recommendation is not to use the assay if more than 80 ng of vaginal DNA and 5 ng of menstrual blood DNA are present to prevent the generation of non-specific SP signal. Further, the male DNA should be ranged from 0.1 ng to less than 1.0 ng (0.2–0.5 ng is appropriate). When only the trace DNA is extracted, the maximum volume (5.7 μL) is suggested (the acceptable amount of female DNA also needs to be concerned). The absence of DC peak in each sample needs to be confirmed to make sure the complete digestion of HhaI (d in the flow chart).
Interpretations of the results for the 3-plex MSRE-PCR assay include the following 4 situations (e in the flow chart): (i) sperm-positive indicates that SP and SRY peaks both show the peak height not less than 150 RFU (f in the flow chart); (ii) No result is recorded when the SP and SRY peaks are both undetected (peak height less than 150 RFU); (iii) sperm-negative indicates that any SP peak is undetected and any SRY peak is at a peak height not less than 700 RFU (g in the flow chart); (iv) inconclusive is when only either the SP is detected, or only SRY is detected with peak height from 150 RFU to less than 700 RFU.
The SP signals were also observed for 100 ng DNA from vaginal fluids and 10 ng, 20 ng, and 40 ng menstrual blood DNA. The original attempt in this study was to explore sperm-specific markers on Y chromosome, however, only the 456 CpG loci on the Y chromosome were present in the Infinium HumanMethylation450 BeadChip, and none of the loci have been shown to be spermatozoa-specific. If a sperm-specific CpG locus on Y chromosome can be identified then this will increase greatly the specificity as excessive amounts of female DNA should not produce any PCR product from a marker on Y chromosome.
Applying the 3-plex MSRE-PCR assay to forensic exhibits showed that the assay had the greatest sensitivity for the detection of spermatozoa in semen, followed by the RSID-semen test and microscopic examination, both of which shared a similar detection rate for semen identification. The PSA test can exhibit reduced specificity for semen [5, 6, 36, 37], and therefore, a recommendation is that PSA test should be combined with other tests for semen identification. These results are compliance with a previous report on the persistence and detection rate of semen using various tests [8]. However, spermatozoa cannot always be detected in once semen has been detected, such as semen from vasectomized men, and under this situation, the RSID-semen and PSA tests remain a good choice for semen detection.
The 3-plex MSRE-PCR is easily transferrable to forensic practices because it uses the same equipment already in use at crime laboratories. As the same DNA sample for STR typing can also be used for the 3-plex MSRE-PCR assay, no more stain is consumed. The STR typing and 3-plex MSRE-PCR assay could be performed at the same time after DNA quantification. The only additional requirement is a 60-min incubation for DNA digestion by HhaI for the 3-plex MSRE-PCR assay. It is noted that the time taken in this assay is more than the immunological test (PSA, Sg), typically only 10 min of running time. For the 3-plex MSRE-PCR assay, the reagent cost is about US$0.85 per reaction, which is cheaper than the cost of immunological tests (more than US$5 per PSA test and US$15 per RSID test in Taiwan). Therefore, in addition to its higher sensitivity and specificity for normal semen as validated in the study, the 3-plex MSRE-PCR assay has great advantages of less cost, no more stain consumption, and easy transfer to forensic practice. However, immunological tests have some benefits, such as being fast, convenient, and important for sperm-free semen sample.
Conclusions
This study reported a specific and sensitive 3-plex MSRE-PCR assay for spermatozoa identification. The sensitivity tests showed that 0.1 ng of DNA from semen was sufficient to identify the spermatozoa, and no more than 80 ng of vaginal DNA and 5 ng of menstrual blood DNA for the mixtures are recommended. This 3-plex MSRE-PCR assay was shown to be particularly valuable for spermatozoa identification in cases where the spermatozoa are deformed, or in very limited number. The methodology described is readily transferrable to current forensic practice as it uses the same equipment already in use at crime laboratories and with the advantages of less cost and no more stain consumption.
References
Malsom S, Flanagan N, McAlister C, Dixon L (2009) The prevalence of mixed DNA profiles in fingernail samples taken from couples who co-habit using autosomal and Y-STRs. Forensic Sci Int Genet 3(2):57–62
An JH, Shin KJ, Yang WI, Lee HY (2012) Body fluid identification in forensics. BMB Rep 45(10):545–553
Old J, Schweers BA, Boonlayangoor PW, Fischer B, Miller KW, Reich K (2012) Developmental validation of RSID™-semen: a lateral flow immunochromatographic strip test for the forensic detection of human semen. J Forensic Sci 57(2):489–499
Allery JP, Telmon N, Mieusset R, Blanc A, Rougé D (2001) Cytological detection of spermatozoa: comparison of three staining methods. J Forensic Sci 46(2):349–351
Henky H, Budiningsih Y, Widiatmaka W (2011) The validity of rapid test to detect prostate-specific antigen (PSA) in seminal fluid. Med J Indonesia 20(4):278–282
Sato I, Sagi M, Ishiwari A, Nishijima H, Ito E, Mukai T (2002) Use of the “SMITEST” PSA card to identify the presence of prostate-specific antigen in semen and male urine. Forensic Sci Int 127(1–2):71–74
Graves HC, Sensabaugh GF, Blake ET (1985) Postcoital detection of a male-specific semen protein: application to the investigation of rape. N Engl J Med 312(6):338–343
Suttipasit P, Wongwittayapanich S (2018) Detection of prostate specific antigen and semenogelin in specimens from female rape victims. J Forensic Legal Med 54:102–108
Casey DG, Domijan K, MacNeill S, Rizet D, O'connell D, Ryan J (2017) The persistence of sperm and the development of time since intercourse (TSI) guidelines in sexual assault cases at forensic science Ireland, Dublin, Ireland. J Forensic Sci 62(3):585–592
Hanson EK, Ballantyne J (2016) Enhanced DNA profiling of the semen donor in late reported sexual assaults: use of Y-chromosome-targeted pre-amplification and next generation Y-STR amplification systems. In: Forensic DNA Typing Protocols. Springer, pp 185–200
Katilius E, Carmel AB, Koss H, O’Connell D, Smith BC, Sanders GM, LaBerge GS (2018) Sperm cell purification from mock forensic swabs using SOMAmer™ affinity reagents. Forensic Sci Int Genet 35:9–13
Costa S, Correia-de-Sá P, Porto MJ, Cainé L (2017) The use of laser microdissection in forensic sexual assault casework: pros and cons compared to standard methods. J Forensic Sci 62(4):998–1006
Muro CK, Doty KC, de Souza Fernandes L, Lednev IK (2016) Forensic body fluid identification and differentiation by Raman spectroscopy. Forensic Chem 1:31–38
Roeder AD, Haas C (2013) mRNA profiling using a minimum of five mRNA markers per body fluid and a novel scoring method for body fluid identification. Int J Legal Med 127(4):707–721
Ingold S, Dørum G, Hanson E, Berti A, Branicki W, Brito P, Elsmore P, Gettings K, Giangasparo F, Gross T (2018) Body fluid identification using a targeted mRNA massively parallel sequencing approach–results of a EUROFORGEN/EDNAP collaborative exercise. Forensic Sci Int Genet 34:105–115
Sirker M, Fimmers R, Schneider P, Gomes I (2017) Evaluating the forensic application of 19 target microRNAs as biomarkers in body fluid and tissue identification. Forensic Sci Int Genet 27:41–49
Frumkin D, Wasserstrom A, Budowle B, Davidson A (2011) DNA methylation-based forensic tissue identification. Forensic Sci Int Genet 5(5):517–524
Jung SE, Cho S, Antunes J, Gomes I, Uchimoto ML, Oh YN, Di Giacomo L, Schneider PM, Park MS, van der Meer D (2016) A collaborative exercise on DNA methylation based body fluid typing. Electrophoresis 37(21):2759–2766
Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13(7):484–492
Hashimshony T, Zhang J, Keshet I, Bustin M, Cedar H (2003) The role of DNA methylation in setting up chromatin structure during development. Nat Genet 34(2):187–192
Miranda TB, Jones PA (2007) DNA methylation: the nuts and bolts of repression. J Cell Physiol 213(2):384–390
Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, Held WA (2005) Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci U S A 102(9):3336–3341
Wasserstrom A, Frumkin D, Davidson A, Shpitzen M, Herman Y, Gafny R (2013) Demonstration of DSI-semen—a novel DNA methylation-based forensic semen identification assay. Forensic Sci Int Genet 7(1):136–142
Sambrook J, Fritsch E, Maniatis T (1989) Molecular cloning: a laboratory manual:9.14–19.19. Cold Spring Harbor Laboratory Press, New York
Yu-Chih L, Li-Chin T, James Chun-I L, Kuo-Lan L, Jason Tze-Cheng T, Adrian L, Hsing-Mei H (2016) Novel identification of biofluids using a multiplex methylation-specific PCR combined with single-base extension system. Forensic Sci Med Pathol 12(2):128–138
Balamurugan K, Bombardi R, Duncan G, McCord B (2014) Identification of spermatozoa by tissue-specific differential DNA methylation using bisulfite modification and pyrosequencing. Electrophoresis 35(21–22):3079–3086
Tanaka K, Okamoto A (2007) Degradation of DNA by bisulfite treatment. Bioorg Med Chem Lett 17(7):1912–1915
Yu-Chih L, Li-Chin T, James Chun-I L, Chih-Wen S, Jason Tze-Cheng T, Adrian L, Hsing-Mei H (2016) Novel identification of biofluids using a multiplex methylation sensitive restriction enzyme-PCR system. Forensic Sci Int Genet 25:157–165
Krausz C, Sandoval J, Sayols S, Chianese C, Giachini C, Heyn H, Esteller M (2012) Novel insights into DNA methylation features in spermatozoa: stability and peculiarities. PLoS One 7(10):e44479
Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén SE, Greco D, Söderhäll C, Scheynius A, Kere J (2012) Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One 7(7):e41361
Farkas SA, Milutin-Gašperov N, Grce M, Nilsson TK (2013) Genome-wide DNA methylation assay reveals novel candidate biomarker genes in cervical cancer. Epigenetics 8(11):1213–1225
Souren NY, Lutsik P, Gasparoni G, Tierling S, Gries J, Riemenschneider M, Fryns J-P, Derom C, Zeegers MP, Walter J (2013) Adult monozygotic twins discordant for intra-uterine growth have indistinguishable genome-wide DNA methylation profiles. Genome Biol 14(5):R44
Slieker RC, Bos SD, Goeman JJ, Bovée JV, Talens RP, van der Breggen R, Suchiman HED, Lameijer E-W, Putter H, van den Akker EB (2013) Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics Chromatin 6(1):26
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3--new capabilities and interfaces. Nucleic Acids Res 40(15):e115. https://doi.org/10.1093/nar/gks596
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23(10):1289–1291
Yu H, Diamandis EP (1995) Prostate-specific antigen in milk of lactating women. Clin Chem 41(1):54–58
Lövgren J, Valtonen-André C, Marsal K, Liua H, Lundwall Å (1999) Measurement of prostate-specific antigen and human glandular kallikrein 2 in different body fluids. J Androl 20(3):348–355
Cox M (1991) A study of the sensitivity and specificity of four presumptive tests for blood. J Forensic Sci 36(5):1503–1511
Kaye S (1949) Acid phosphatase test for identification of seminal stains. J Lab Clin Med 34(5):728–732
Vernarecci S, Ottaviani E, Agostino A, Mei E, Calandro L, Montagna P (2015) Quantifiler® Trio Kit and forensic samples management: a matter of degradation. Forensic Sci Int Genet 16:77–85
Sidstedt M, Hedman J, Romsos EL, Waitara L, Wadsö L, Steffen CR, Vallone PM, Rådström P (2018) Inhibition mechanisms of hemoglobin, immunoglobulin G, and whole blood in digital and real-time PCR. Anal Bioanal Chem 410(10):2569–2583
Funding
This study was supported by the Ministry of Science and Technology (NSC 102–2628-B-015-001-MY2), and Ministry of the Interior (108–0805-05-17-01) in Taiwan.
Author information
Authors and Affiliations
Contributions
All the authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Kuo-Lan Liu, Li-Chin Tsai, Yu-Chih Lin, Nu-En Huang, Lih-Jing Yang, and Chih-Wen Su. The first draft of the manuscript was written by James Chun-I Lee, Adrian Linacre, and Hsing-Mei Hsieh, and all the authors commented on previous versions of the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
In this study, samples were collected after informed consent and following the procedures approved by the Institutional Review Board (IRB) of Taoyuan General Hospitals (IRB No. TYGH102011) and Antai-Tian-Sheng Memorial Hospital (IRB No. 18–074-B) in Taiwan. Neither the authors are affiliated to these hospitals nor the study was carried out in these hospitals. And the studies have been performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Online Resource 1
(DOCX 19 kb)
Online Resource 2
(DOCX 42 kb)
Online Resource 3
(DOCX 18 kb)
Online Resource 4
(DOCX 309 kb)
Online Resource 5
(DOCX 155 kb)
Online Resource 6
(DOCX 137 kb)
Online Resource 7
(DOC 52 kb)
Online Resource 8
(DOC 154 kb)
Rights and permissions
About this article
Cite this article
Liu, KL., Tsai, LC., Lin, YC. et al. Identification of spermatozoa using a novel 3-plex MSRE-PCR assay for forensic examination of sexual assaults. Int J Legal Med 134, 1991–2004 (2020). https://doi.org/10.1007/s00414-020-02285-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-020-02285-1