Abstract
Interpreting mixed DNA samples containing material from multiple contributors has long been considered a major challenge in forensic casework, especially when encountering low-template DNA (LT-DNA) or high-order mixtures that may involve missing alleles (dropout) and unrelated alleles (drop-in), among others. In the last decades, extraordinary progress has been made in the analysis of mixed DNA samples, which has led to increasing attention to this research field. The advent of new methods for the separation and extraction of DNA from mixtures, novel or jointly applied genetic markers for detection and reliable interpretation approaches for estimating the weight of evidence, as well as the powerful massively parallel sequencing (MPS) technology, has greatly extended the range of mixed samples that can be correctly analyzed. Here, we summarized the investigative approaches and progress in the field of forensic DNA mixture analysis, hoping to provide some assistance to forensic practitioners and to promote further development involving this issue.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In forensics, mixed DNA samples are those in which two or more individual body fluids or secretions are mixed, which often occurs in the cases of sexual crimes or large disaster scenes, as well as in products of conception and fingernail cuttings taken by police or at autopsy [1]. Due to the different types of materials, the number of donors and the different proportions of each component in the mixture, identification, and interpretation are much more complicated for mixtures than for single-sourced samples. DNA mixtures occur routinely in our forensic investigations, and through years of efforts, there have been some relevant developments and improvements in the aspects of DNA separation/extraction methods, useful genetic markers, detection platforms, and analytical tools for mixtures [2,3,4,5]. As is widely known, despite such advances, forensic analysis of complicated mixtures remains a process that requires too much time and energy. In this review, we summarized the development and progress of DNA mixture analysis and put forward the existing problems, which may provide a reference for future forensic research and practical work.
Separation and DNA extraction from mixtures
For the mixed samples of sperm and vaginal fluid collected to investigate sexual crimes, Gill et al. [6] proposed a two-step differential extraction (DE) method based on the characteristics of sperm nuclei, which are ramified with cross-linked thiol-rich proteins. Female epithelial cells are first preferentially lysed by preliminary incubation in SDS/proteinase K buffer, releasing DNA into suspension, and then the sperm components were obtained by centrifugation and lysed in a second buffer containing SDS/proteinase K/DTT, which can break the protein disulfide bridges of the sperm nuclear membrane. At present, this method is most commonly used in forensic laboratories. However, when the specimen is older, the sperm component is very low or there are multiple male contributors mixed within a complex sample, and the separation efficiency of the differential lysis method becomes poor; moreover, repeated washing steps during the process can cause the loss of sperm components, or the precipitate may include female components due to incomplete lysis. For the past two decades, researchers have made many attempts to improve the efficiency of the DE method [7,8,9], and several novel techniques, such as laser capture microdissection (LCM), immunomagnetic separation (IMS), and microfluidic chip analysis, were developed to separate and extract DNA in complex DNA mixtures [10,11,12].
Exploration of DE method
Despite the widespread application of DE, the DNA typing efficiency of sperm in a male-female mixture after sexual intercourse is still limited by the small number of sperm in the context of a wide range of female-contributed materials. Therefore, many researchers are committed to improving the effectiveness of the DE method through a variety of different approaches. Wiegand et al. proposed a modified DE method that uses a mild preferential lysis to avoid further loss of sperm DNA in a low sperm content mixture [13]. Later, Yoshida et al. developed a method in which the temperature for incubation was elevated and the concentration of proteinase K was increased in the DE process [14].
In sexual assault and rape cases, genetic analysis of the perpetrator and victim DNA from vaginal cotton swabs is a well-established forensic technique for investigation [6, 15]. Voorhees et al. focused on a cellulose-based enzyme mixture obtained from Aspergillus niger, Trichoderma reesei, and Trichoderma viride, which come from different fungal sources, and indicated its potential to improve the release of sperm and epithelial cells from a common cotton swab compared with the buffer elution of DE alone, permitting forensic DNA analysis [7]. Afterwards, their team focused on the development of a method that chemically induced enhancement of cell elution and recovery from cotton swabs. They found that different detergents, as well as proteinase K, affected the sperm cell yield, with anionic detergent (e.g., SDS) and suitable use of proteinase K having the greatest effect [16]. Later, Benschop and his colleagues indicated that although nylon flocked swabs dried much slower than cotton swabs for post-coital vaginal sampling, which may promote microbial growth, the cellular eluate from the nylon flocked swabs contained a higher number of and more intact cells; additionally, less cell material was retained after DNA extractions, and at the same time, the yield of the extracted DNA was higher [8]. However, cotton swabs are still the most commonly used collection tools in sexual assault cases, possibly because the results from the two kinds of swabs are not so different in practical work compared to investigations.
Most recently, a semi-quantitative ratio-based analysis, termed Separation Potential Ratio of the Extraction Differential (SPRED), was conducted to evaluate the separation efficiency of both sperm DNA recovery and non-sperm DNA removal in the DE methods [3]. A higher SPRED ratio indicates a higher potential for obtaining a primarily male component in the sperm fraction. The SPRED value of the two-step DE could be improved significantly by performing a second non-sperm lysis step, which reduced the non-sperm cell DNA carryover with no concomitant reduction in sperm DNA recovery.
LCM
LCM technology has provided a significant breakthrough for the separation of a limited number of sperm cells from an overwhelming quantity of female epithelial cells [10]. It combines laser beam technology with light microscopic devices and targets specific cells or tissue sections that need to be isolated from others. Under direct microscopic visualization, the cells of interest are isolated by infrared (IR) capture systems [17] or ultraviolet (UV) cutting systems [18,19,20], and then separated cells or tissue regions are placed into independent tubes for DNA extraction and analysis [21,22,23].
Lucy et al. claimed that spermatozoa contain only half the genetic material of a contributor, because they are haploid cells, and that the theoretical number of sperm that need to be pooled for a full representation of the alleles comprising the contributor profile is 15–20 intact and non-degraded sperm. [24]. However, manual screening of microscope slides for sperm morphological identification is time-consuming and labor intensive. Vandewoestyne et al. introduced an automated screening method to detect spermatozoa stained with Sperm HY-LITER, and then LCM was used for isolation. The DNA analysis results showed that a minimum of 30 spermatozoa recovered from post-coital samples could generate a robust DNA profile without allelic dropout [25]. There have been other studies proposing staining methods to perform sex-specific labeling of cells for LCM [26,27,28]. Fluorescence in situ hybridization (FISH) was also conducted using Y-chromosome-specific probes to separate male cells from male-female mixtures [29].
For a long period, researchers used multiple techniques to extract DNA from cells obtained from LCM. Vandewoestyne and his colleagues compared three different DNA isolation methods for laser microdissected blood cells and found that the PicoPure DNA extraction kit (Arcturus, Mountain View, CA, USA) performed better than the DNA IQ™ system extraction kit (Promega Corporation, Madison, WI, USA) and the short alkaline DNA extraction method. The cell collection, lysis, and PCR could all be conducted in the same tube when using the PicoPure DNA extraction kit, which reduced the contamination risk, and as few as 10 cells could be used to detect the full DNA profiles [11]. Han et al. used a strategy for sperm isolation and short tandem repeat (STR) typing from multidonor sperm mixtures in which they applied LCM and low volume-PCR (LV-PCR) for single sperm isolation and detection [30]. In their study, the platform was so sensitive that the profiling of a single sperm cell could generate a minimum of 13–16 loci in nearly three quarters of the Y-chromosome STR (Y-STR) assays.
IMS
The IMS of eukaryotic and prokaryotic cells has been widely explored. Cell separation by IMS involves two steps: the first step is the specific binding of immunomagnetic beads and cells, the core of which is the specific binding reaction of the antigen-antibody. The second step is the application of immune complexes (antigen-antibody and magnetic beads) in an appropriate magnetic field [31]. In 2002, three monoclonal anti-sperm antibodies (MHS-1O, NUH-2 and HS-21), after attachment to magnetic beads, were evaluated and applied by Eisenberg et al. to capturing sperm from mixtures consisting of varying numbers of vaginal epithelial cells and sperm. The results proved that none of the three antibodies bound the vaginal epithelial cells and that the MHS-IO antibody bound as much as 90–95% of the input sperm [32]. In Anslinger’s study, another three monoclonal anti-sperm antibodies, 1E10, 4E3, and 4E10, were selected against the testicular isoform of angiotensin-converting enzyme (tACE), among which 4E3 was proven to be the most effective [33]. However, the vaginal swabs were required to be stored in PBS buffer to provide a sufficient amount of intact sperm with mid-piece and flagellum, which is a drawback for forensic application. In the study of Zhao et al., sperm were isolated from different mixtures of sperm cells and buccal cells with the help of anti-SPAG8 antibody that can combine a special antigenic protein located on the head of the sperm [34]. Wang et al. applied the complex of biotin-labeled rabbit anti-human sperm antibody and avidin-labeled magnetic beads to separate sperm from the mixture, which indicated that polyclonal antibodies interact with multiple surface proteins of sperm and thus have a greater likelihood of binding with sperm and a stronger ability to form more stable sperm-antibody-biotin complexes [35].
Microfluidic chip/LOC
With multiple laboratory techniques, including cell sorting, DNA extraction, DNA quantitation, and DNA amplification, all integrated in a chip of a few tens of square centimeters, microfluidic devices can provide fast genetic analysis for forensic application. The microfluidic chip, also called lab-on-a-chip (LOC), can also reduce the risk of cross-contamination and provide the possibility of direct analysis at the crime scene, which minimizes sample handling in a sealed microfluidic environment [36,37,38]. There have been many studies making improvements on the microfluidic chip [39]. Due to the differences in cell size, chemical composition, and membrane structure between sperm and epithelial cells, they respond differently to a non-uniform electric field, resulting in different motion in dielectrophoresis (DEP) that may be used to separate them. Buoncristiani’s group applied a commercially available Silicon Biosystems DEPSlide™ System that could separate sperm and epithelial cells in a microfluidic chip. However, they found that the purity or yield of the separated cells was not as good as expected based on visual observation and that the quality of the DEP was no better than that of standard DE [40]. In further developmental efforts, the DEP platform was integrated into the microfluidic system [41], which may allow the inexpensive, fast, highly sensitive, and label-free detection and analysis of sperm and epithelial cells for sexual assault cases. In addition, an acoustic differential extraction (ADE) analysis was developed on a microfluidic device, which relied on the acoustic trapping of sperm cells, while transferring the female components into a separate outlet [42, 43]. Because it involves similar lysis steps as the DE method, there will inevitably be problems with typing failure caused by over/incomplete digestion. Furthermore, Fontana et al. applied an image-based and microfluidic digital system, DEPArray™ Technology (Menarini Silicon Biosystems, Italy, MSB), to recover and detect pure homogeneous cells from imitated blood/saliva and semen/saliva mixtures as well as casework mixtures with an outstanding precision [44]. Although their research was not a current standard, it could promote the investigation and development of this technology in forensic biological mixtures.
The LOC has been proven to be a versatile technology that is fast, efficient, and integrated, with low sample input volume and low reagent consumption. The chip not only can carry out multithroughput operations but also is lightweight and small in size such that it is portable. Therefore, the LOC is very conducive to the realization of automated separation of sperm from challenging biological mixtures. However, the technology of microfluidic chips is still in development, and before it can truly be applied in forensic scenes, several problems need to be overcome: the cost must be reduced, additional DNA or RNA analysis techniques must be integrated for challenging samples, and uniform standardization for microfluidic interconnection, chip dimension, and vocabulary needs to be established to fully exploit the superiorities provided by LOC [37].
Other improved methods
In 2003, Garvin et al. created a vacuum-driven filtration system that allowed the DNA of digested diploid cells to pass through while sperm were stuck on the membrane surface. In this way, sperm were separated from mixtures containing a large number of female epithelial cells [45]. This method was considered to be more automatable and efficient than standard DE, but for old or severely degraded samples, cell recovery was not ideal due to the sperm clogging or adhering within the filtration membranes.
Schneider et al. described a new procedure in their work to evaluate the combination of the sensitive staining method Sperm Hy-Liter™, which allows staining even if the sperm morphology has disintegrated, with micromanipulation using the aureka® cell isolation system to isolate a low number of sperm on a microscope slide (as few as 20 sperm) to generate full STR profiles. Typically, laser microdissection (LMD) technology needs transparent objects, whereas aureka® is an open system that can handle various kinds of materials on different surfaces at different heights [46, 47]. Subsequently, Moors et al. also indicated that the Sperm Hy-Liter™ assay is highly specific and sensitive for human spermatozoa and reliable and simple to use for the detection of spermatozoa in a variety of sexual assault samples [48].
The high specificity and sensitivity of flow cytometry provides a possible method to sort and analyze the DNA of sperm cells [49, 50], which is also known as fluorescence assisted cell sorting (FACS). Technological advancement has given FACS the potential for more promising applications, enabling it not only to separate sperm from the mixture containing vaginal epithelial cells [51, 52] but also to sort other cells of forensic interest, such as the separation of saliva-derived epithelial cells from blood-derived leukocytes [53]. Of course, the loss of a certain number of cells in the process of separation and staining also requires attention.
In other research directions, Aptamer technology has also been used to capture and separate intact sperm cells from the presence of female epithelial cells and other non-sperm semen components [54, 55]. The haplotype-specific extraction (HSE) method was used for the separation of Y-chromosomal DNA from two-male mixtures to extract the haploid fractions and in the separation/analysis of the mitochondrial DNA (mtDNA) of samples containing DNA from two individuals, while situations with more than two donors or low DNA quality made it difficult to apply [56,57,58,59]. Koukouvinos et al. demonstrated a sensitive and accurate biosensor based on white light reflectance spectroscopy to identify prostate-specific antigen (PSA) in forensic samples containing semen, which could be completed within 10 min and had a low detection limit of 0.5 ng/mL PSA [60].
Another article proposed a robust real-time PCR system, termed InnoQuant® HY, which provided accurate quantitation of the DNA of a male from challenging male-female mixtures with a male to female ratio of as low as 1:128,000, thus allowing better decision-making about the appropriate DNA input amount for PCR [61]. The system provides significant data on the male to female ratio, degradation state, and the presence or absence of PCR inhibitors to support a more efficient and clear downstream workflow involving complex biological specimens.
Blood, saliva, or other body fluids can also be common components of mixtures from forensic scenes, and several studies have been devoted to their identification [62, 63]. For example, Yano and his colleagues used anti-human leukocyte CD45 and ABO blood group antibodies to separate leukocytes from mixed bloodstains involving different contributors, and the target DNA could be detected accurately with a ratio of as low as 1:512 [64]; this was only applicable for discriminating from a mixture containing different blood types.
Detection technology and genetic markers for mixture analysis
Detection technology
Capillary electrophoresis (CE) is still the most prevalent technology in contemporary forensic investigation on mixed DNA samples, and most of the available calculation methods and statistical software are based on the STR genotype generated by CE technology. It has some limitations for mixture analysis on CE platform, such as the limited number and length of each fluorescently labeled PCR fragments in a multisystem [65], the low efficiency of handling degraded or trace samples, and the difficulty in distinguishing genuine alleles provided by minor contributors from stutter alleles of major contributors [66]. Compared with the CE platform, many forensic analysts believe that the rapidly developing massively parallel sequencing (MPS) technology may shed some light on mixture analysis. For example, Morling’s team compared traditional PCR-CE and MPS at three STR loci among the Danish population and observed that approximately 30% of the samples identified as having a homozygous genotype with CE turned out to be heterozygous when sequenced by MPS [67]. STR alleles can be recognized not only by repeat number but also by sequence variation, which may simplify mixture interpretation [68]. Moreover, on the MPS platform, it is possible to detect a large number of different types of genetic markers with overlapping sizes, including STRs, single nucleotide polymorphism (SNPs), and insertions or deletions (indels), and the short PCR fragment may help in dealing with degraded or trace mixtures [5, 68]. Last, but not least, MPS is more sensitive than CE, and the former can detect the minor donor in a 1:100 mixture, while the latter is unable to handle ratios of less than 1:10 [5, 69, 70]. However, population genetic data generated from the MPS platform is still at a disadvantage due to the problems in popularization and normalization, which should be improved by further study. In general, MPS technology has the absolute advantage that the detailed sequence information provided by MPS may facilitate mixture interpretation and increase the statistical weight of the evidence, which offers new possibilities for forensic genetic casework [71].
Genetic markers
Many genetic markers, including STRs, SNPs, and indels of autosomes and Y-chromosomes, are commonly used for mixture detection and analysis [30, 72, 73]. Y-STR analysis can detect male components directly to avoid the loss of DNA. The number of donors in a mixture can be estimated on the basis of the number of Y-STR alleles, which is of great significance in the analysis of gang rape cases [74, 75]. In addition, mtDNA plays an important role in degraded specimens and can also be used to determine the number of individuals in a mixture [76, 77].
STRs
STR markers have been used in the analysis of mixed samples for many years, while allelic dropout and drop-in often occur due to the unbalanced DNA of the contributors, which makes it difficult to establish consensus guidelines [78,79,80]. Limited numbers of STRs available in commercial kits provide low statistical strength of inclusion in mixtures with more than three contributors or in cases where the minor contributor component is extremely low. From that point of view, the combination of autosomal and Y-chromosomal STR analysis was conducted, which provided an additional means for the investigation of mixtures in sexual assault cases, because Y-STR is highly valuable in some cases in which the minor male proportion in the DNA mixtures is undetectable by autosomal STR analysis [30, 81, 82]. Recently, nine novel pentameric STRs with lower stutter peak ratios were strongly proposed to aid in the analysis of mixed DNA profiles when minor donor alleles may coincide with stutter peaks from the major components [83].
The collaborative RNA/DNA co-analysis exercise results of the European DNA Profiling Group (EDNAP) illustrated special mRNA profiling as a reliable body fluid identification method that can easily be combined with standard STR typing technology [84,85,86,87]. Uchimoto et al. applied an in-house miRNA analysis to stains containing a mixture of blood and saliva that could determine the presence of more than one body fluid and major/minor contributors with a lower limit of detection than the enzymatic equivalent [88]. Future work will be expected to explore the use of the abovementioned methods with more body fluids, including seminal fluid, menstrual blood, and vaginal fluid.
SNPs
Voskoboinik et al. [89] indicated that if an individual carries some rare alleles and a complex DNA mixture also contains these particular alleles, then this individual is represented in the mixture. Based on this rationale, they proposed a theoretical framework of a method with a large number of SNPs in 2011 and presented implementation utilizing a panel of 3000 SNPs with relatively low minor allele frequency (MAF), below 0.25, in 2015 [72]. The later study could robustly identify individuals who contribute at minimum 5% to a mixture and examine the usefulness of the whole genome amplification (WGA) of complex mixtures before allelotyping to handle low DNA quantities. However, this method has some shortcomings; specifically, it can only be applied in cases with a known suspect and is not compatible with common STR databases. As well as establishing a SNP database, combining it with existing STR databases requires arduous work. Further research should be carried out to validate the proposed panel and to invest forensically focused SNP microarrays on DNA mixtures.
A study by Liu et al. used a sensitive system containing primer-specific alleles of 14 indel markers to detect ratios as low as 1:500 to 1:1000 minor components in two-source mixtures [73]. Recently, Hwa and his colleagues established a 1204 SNP/indel panel optimized for MPS consisting of a different number of SNPs and indels located on the autosomes, the X/Y chromosomes, and the mitochondria, combined with a scoring system, which could accurately identify minor contributors contributing 1% or more to DNA mixtures [5]. This panel enabled the successful simultaneous analysis of numerous different markers and was believed to be more sensitive and flexible than traditional CE approaches. The large number of loci in the panel could also increase the statistical power of the discrimination, which could be applied in individual identification for forensic DNA mixtures as a primary method or as a supplement to STR analysis.
mtDNAs
The Spanish and Portuguese working group of the International Society for Forensic Genetics (GEP-ISFG) has performed mtDNA collaborative exercises over many years [90,91,92,93,94,95]. They undertook extraordinary efforts to improve the quality and standardization of mtDNA analysis, including the analysis of different mixtures (semen-saliva, hair-saliva, saliva-saliva mixture), from both methodological and theoretical perspectives. The success rate for analyzing mixed samples by mtDNA was moderate, and they noted that errors in mtDNA testing occur mainly due to the lack of a solid devised experimental approach. In some cases involving mixed semen stains, it is more difficult to identify individuals than to just exclude unrelated ones. Zhang et al. first applied the LCM system to choose single sperm cells to target. Then, the mtDNA hypervariable region I (HVR I), which shows genetic polymorphisms in different matrilineal-related individuals, was amplified from each cell; by combining the cellular DNA of the same HVR I sequence from multiple cells, enough nuclear DNA for STR typing (reamplified) was obtained [96]. They indicated that a collection of 20 sperm could be typed correctly, but more than 30 sperm cells should be collected to avoid errors during the final typing process. Zander et al. performed haplotype-specific extraction (HSE) to separate the mtDNA of each donor from a two-source mixture and subsequently sequenced the mtDNA to reveal the underlying individual haplotypes successfully; the limits of this method are used in cases with more than two donors or low-quality DNA, which could make separating mtDNA haplotypes using HSE difficult [59]. However, mtDNA is not sufficient to differentiate individuals in the same maternal line and, as such, using other markers is necessary to identify individuals from a mixture. Therefore, Hwa et al. combined 129 mitochondrial SNPs and other markers into their 1204-marker panel to detect non-degraded and highly degraded DNA mixtures [5].
New markers
DIP-STRs
There have been several novel sets of genetic markers proposed in recent years, such as pairing deletion/insertion polymorphisms (DIP) with standard STR, called DIP-STR. Hall’s team proposed DIP-STR as an innovative genetic marker composed of a DIP tightly adjacent to an STR polymorphism, detected by a specially designed pair of PCR primers (L-long allele for insertion and S-short allele for deletion). These compound markers are able to target a minor component in the presence of a 1000-fold excess of background DNA from an unbalanced DNA mixture [97]. They compared DIP-STRs with traditional STRs and Y-STRs from a statistical and forensic perspective, which indicated that each method has its own advantages and that the use of DIP-STR markers could be of interest for all kinds of DNA mixtures [98]. They then applied the first set of 10 DIP-STR markers to estimate the allelic frequencies of a group from the Swiss population and the presence of informative alleles, as well as to calculate the random match probability of the minor DIP-STR profile detected across a large number of DNA mixtures in silico [99]. Six of these 10 DIP-STRs were also powerful for use in unbalanced DNA mixture investigations of the southwest Chinese Han population [100]. Recently, Hall et al. used another set of six DIP-STR markers for the analysis of unbalanced mixtures from challenging “touch” DNA samples to detect the minor donor and found an analogous sensitivity and similar occurrence of allelic dropout in comparison with Y-STRs [4]. They also reported the first use of 18 validated DIP-STRs in eight real cases, in which these markers were found to be more sensitive and specific and performed well on challenging DNA samples of both sexes [101].
It is noteworthy that DIP-STR can be used in DNA mixtures containing low levels of DNA or extremely unbalanced major/minor ratios, irrespective of the sexes of the donors. When male suspects from the same male lineage or from a highly inbred population are involved, DIP-STRs are more powerful. With regard to same-sex DNA mixtures or a female perpetrator, it can be practical to use DIP-STRs instead of Y-STRs in further investigation. In fact, DIP-STR markers can complement Y-STR data and help to provide new clues for investigation. In addition, DIP-STR data may be helpful in estimating the number of contributors and strengthening the DNA evidence from a mixed sample. Even so, these improved DIP-STR markers have several limits. For example, it can be used only when the major contributor is homozygous for the DIP allele and the minor contributor is heterozygous or homozygous for the opposite allele. Multiplexed assays for DIP-STRs have not yet been established, so the genotyping of these markers still requires several hundred picograms of DNA template, which is frequently not available in routine forensic practice. Lastly, DIP-STR markers cannot be used to analyze a DNA mixture of more than two individuals.
Microhaplotypes
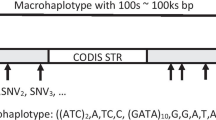
To achieve the purposes of human familial identification and ancestry inference, Kidd’s team defined a mini-haplotype containing three or more SNPs within the expanse of 10 kilobase pairs (KB) [102]. Similarly, they identified many loci with two or more SNPs extending over a smaller molecular interval of 200 base pairs in the human genome, and when linkage disequilibrium is not complete, these loci are regarded as microhaplotype loci (microhaps) [103]. They illustrated that microhaps, with high heterozygosity and regional or global Fst, should be capable of providing information for ancestry inference, lineage-clan-family relationships, and individual identification [104]. Additionally, microhaps have great value for disentangling mixtures. When the microhaplotype loci are genotyped by sequencing, three or more genotypes with a sufficient number of reads appeared at each locus of a DNA mixture that can also be detected qualitatively according to different numbers of reads for each allele. MPS makes it realizable to integrate a cluster of SNPs and detect microhaps. Kidd and Speed defined the term “Ae” as the effective number of alleles at a microhaplotype locus, with which greater than three microhaps will be useful in routine forensic practice, and three-SNP microhaps will sometimes meet this criterion. They indicated that the Ae value and the ability of microhaps to discriminate mixed samples showed a positive correlation. When five microhaps with an average Ae value of four were detected, the cumulative probability of qualitative detection of a mixture was greater than 99% [105]. Later, Kidd proposed the nomenclature for the microhaps [106], and Zhu et al. designed the novel software FLfinder to type microhaps [107]. The great potential of optimally selected microhaps in forensic casework encouraged the search for novel loci that were better suited for forensic applications. Of course, there are some aspects that need to be improved in the development of microhaps. For example, the applicability of more than 100 existing loci proposed by Kidd’s team in other populations remains to be explored. MPS technology is less prevalent in forensic laboratories, which is also one of the factors limiting its development.
Interpretation of mixture profiles
Calculation models
Due to the extensive use of STR markers in mixture analysis on the CE platform, researchers have established several computational models for explaining the STR typing of mixed samples. In forensic casework, DNA mixture interpretation using STR loci is initially based on a binary model, which may be a qualitative binary model or a semi-quantitative binary model; it is simple to apply but not involved in peak heights in the computation of two-source mixtures [108, 109]. The peak height information is of benefit for analyzing mixed profiles. For example, four peaks were detected at the D16S539 locus for a two-donor mixture (Fig. 1), and if peak height is not considered, six possible genotype combinations will be given the same weight (Table 1). Although the binary model has served well for a great many years in forensic cases, it is not suitable to handle questionable low-level or LT-DNA samples, which often contain loci showing non-concordances, and it thus has a risk of being misused. In addition, the binary model cannot consider multiple replicates and has difficulty handling multiperson mixtures [110].
The motivator for change is the shortfalls of binary models mentioned above. Like the binary model, a semi-continuous model does not make use of peak heights themselves either, while the model parameters were informed and different dropout rates per contributor were allowed [111, 112]. The drop-in can be distinct from contamination in the semi-continuous model and can also handle multiple replicates. The probabilities of all possible genotype groups in the mixture profile and the likelihood ratio (LR) for a series of propositions can be rapidly calculated in software employing this model [113]. A semi-continuous model makes improvements that can be used in the analysis of complex mixtures and LT-DNA, while ignoring peak height information, and the implementation always needs specialized software.
The continuous model, proposed a few years ago, incorporates peak heights and utilizes this information to assess all possible genotype combinations of the contributors by calculating their probabilities. For instance, use of the peak height and mixture ratio (based on data from multiple loci) information can help eliminate possible genotype combinations in Table 1. In situations with a 3:1 mixture, the correct genotypes are 10,14 for contributor 1 (the major one) and 9,13 for contributor 2 (the minor one) (shown in bold font). The continuous model has the potential to cope with any kinds of non-concordance and may assess multiple replicates without preprocessing or information loss [110]. PCR stochastic effects and potential stutters are taken into account, and the quantitative information taken by this model makes it relatively objective [114, 115]. The continuous method also requires specialized software and must be calibrated for different STR systems and conditions.
Among the three calculation models, the binary model is the simplest to apply or explain, and the continuous model is the most complex for the best use of all available information from the mixture profiles. Moreover, model choice dictates the probabilistic calculation, and different models may result in different results, particularly for complex DNA mixtures [116, 117]. The relatively simple models may descend into the misunderstood category due to their ignoring several useful pieces of information, and the risk in the continuous model is that our analysts may not be fully clear about the limitations of these complicated calculation programs in computer software, which result in inappropriate applications. There is no so-called gold standard for the use of these computing models, which depend on different circumstances, while the ideal continuous model is advocated for its reliability and objectivity. Additionally, Bayesian networks and the Monte Carlo Markov Chain (MCMC) method have been implemented with different statistical approaches for probabilistically resolving DNA mixtures in several studies [115, 118,119,120,121].
Statistical approach
In the daily work of forensic science, although statistical approaches for reporting DNA mixtures vary, two calculations are the most commonly used by forensic communities for evaluation. One is the LR, which is also the preferred method to determine the weight of evidence according to the ISFG DNA commission and several other forensic communities [122,123,124,125]. In the forensic evidence (E), a classical analysis involves the prosecution hypothesis (Hp) and the defense hypothesis (Hd), two of which are competing hypotheses evaluating the strength of evidence provided by genetic analysis. For a DNA profile with more than one donor, the Hp represents the probability that the suspect (S) and one unknown person (U) were the donors (Hp = S + U), whereas the Hd represents the probability that there were two unknown donors U1 and U2 (Hd = U1 + U2). The LR formula is represented as: LR = Pr (E | Hp) / Pr (E | Hd). When the LR is greater than 1, the evidence favors Hp; when it is less than 1, the evidence favors Hd. [122]. Due to its ability to handle PCR stochastic effects that occur frequently in mixtures, such as dropout and stutter, the LR approach can maximize the usage of genetic information obtained from the mixed profiles. Consequently, many forensic practitioners recognize the mathematical efficacy of LR and apply it for the analysis of mixtures [126, 127]. The abilities and limitations of the LR approach in the analysis of complex mixtures have been characterized in many studies. Marsden et al. indicated that the strength of the evidence could be extracted from complex mixtures containing a maximum of five donors on the condition involving no dropout [128]. The effect of incorrect estimation of the number of donors (caused by allele sharing) to the LR value was examined by Benschop and his colleagues and was illustrated to exert a great effect on the LR [129]. Prof. Slooten used the standard statistical technique of integrating the LR, by which he could avoid estimating the number of donors or their probabilities of dropout in his study [130]. The assessment depended on the allele frequencies and the mixture data, which was considered to be more objective as a practical advantage. The complex calculation process for LR makes it difficult to explain in court, and thus, several computer software programs have been designed for LR calculation and mixture interpretation [131, 132].
Another calculation method is the combined probability of inclusion/exclusion (CPI/CPE). The CPI evaluates the proportion of a population that would occur as a potential donor in the DNA mixture. CPI also refers to the mathematical complement of random man not excluded (RMNE), which is the probability that a random person has the same DNA profile as the evidence profile or considers that a random person cannot be excluded by the evidence. When applying the RMNE method in the mixture calculation, the main advantage is that it is straightforward to implement and explainable in court because it does not need the assumption of the possible number of contributors, the peak heights, or the genotype of known donors to a mixture; on the other hand, for these same reasons, the RMNE statistic is also deemed to underestimate the strength of the evidence and waste information that should be utilized [133]. However, the results assessed according to a mixture from the RMNE method are still conceptually correct and constantly improved by researchers [134, 135]. The CPI evaluation includes three steps: profile assessment; comparison with reference profiles and inclusion/exclusion determination, and calculation of the statistic [1]. Nevertheless, the two alleles at each locus of a donor being considered must exceed the threshold for analysis, which restricts the CPI approach to unambiguous DNA profiles. For example, low-level DNA mixture may occur with allele dropouts or when the distinction between minor alleles and stutters of major alleles becomes difficult [122, 136]. Thus, the CPI approach has less flexibility than the LR method under the condition of allele dropout in challenging mixtures and needs to be performed by more experienced practitioners under strict guidelines. Bieber et al. described a protocol as a guideline for applying the CPI approach that could lead the forensic communities to reduce variations in DNA mixture interpretation and promote a more defensible application of the CPI [1].
Slooten and Egeland indicated that the RMNE could be revealed as a certain average of the LR, implying that the expected value of the LR is at least equal to the inverse RMNE probability in the case of an actual donor to the mixture. In a great many of the examples for mixtures without dropout, LR even revealed a smaller weight of evidence than 1/RMNE based on the binary model [137]. Meanwhile, the mixture profiles are often characterized by artifacts such as dropout/drop-in and stutter, which makes the concept of exclusion hard to define in the RMNE. Therefore, although it takes more skill to correctly interpret the complicated evidence, LR is considered to be a more powerful method with which to address these situations. Beyond these two kinds of statistical approaches, Bille and his colleagues presented an extension to the concept of random match probability (RMP), which is utilized in standard single-source samples, which can be applied to analyze mixtures [138]. They proposed RMP as an intermediate method, as it is the probability that a random person is included in the mixture (similar to the RMNE) and requires profiling information, including the number of donors and the peak heights into analysis; the latter property is analogous to the LR method.
Interpretation software
Both the computational models and the LR approach are too complicated for routine calculations on mixed samples, so the use and optimization of appropriate software is highly encouraged by Forensic Science International Genetics (FSIG) [139] to avoid hand-calculation errors. Thus, a series of probabilistic DNA profile interpretation software programs have come into being. The study by Curran et al. proposed a set theory, based on which an expert interpretation system, named LoComatioN, was made available [140]. It is the first feasible expert system to rapidly evaluate different explanations in an LR approach, considering the factors of allelic dropout and drop-in [113]. The first open-source bio-statistical tool, Forensim, is a package for R statistical software that is dedicated to forensic DNA evidence interpretation, including that of mixtures [141]. It can simulate mixed DNA profiles and conduct common statistical calculations, which have been used in the research on the efficiency of the maximum-likelihood approach for DNA mixture interpretation [142, 143]. What is more, it was in the Forensim package that LRmix, an open-source software program, was applied by Gill and Haned to evaluate complex mixtures. They illustrated that there is no need for the known contributor and the mixed stain to have (any) matching alleles, and the calculations of strength of evidence are simplified [144]. Based on LRmix, Haned et al. quantified the relative risk of analyzing high-order mixtures by the comparison of the “gold standard” LR (having the known genotypes and number of donors) with the LR obtained in the casework (unknown donors are assumed). They presented some of the exploratory approaches used when encountering complex sample analyses and encouraged the forensic communities to evaluate the risk before applying any LR-based interpretation software [131]. Later, it was demonstrated by Gill and his colleagues that this LRmix program can not only interpret multiallelic STRs but also be extended to bi-allelic SNPs generated from Life Technologies’ HID-Ion AmpliSeq™ Identity Panel v2.2 using the Ion PGM™ MPS system [145]. However, when faced with complex SNP profiles containing three or more donors, LRmix is generally inefficient. The open-source EuroForMix software could significantly improve the analysis of complex SNP mixtures by incorporating the “sequence read” coverage value into the quantitative model and showed a great benefit over the qualitative approach [146]. The EuroForMix program could also interpret STR profiles from a mixture of contributors with artifacts based on the continuous model presented by Cowell and his colleagues [147], which is presented in the R-package euroformix and is freely accessible at www.euroformix.com [148]. Not long ago, the LRmix model was further modified and translated into a user-friendly software, SmartRank, that was recently utilized by Benschop and his colleagues to search several national DNA databases with mixed DNA profiles [132]. They deduced the profile types for which SmartRank can be complementary to CODIS and provided guidelines, as well as defined the applicable domains for the SmartRank software.
There have been other software programs employed for genotype determination and mixed DNA profiles interpretations, such as GeneMapper® ID-X, TrueAllele®, LikeLTD, STRmix™, Lab Retriever, and so on [149,150,151,152,153]. GeneMapper® ID-X and TrueAllele® could conduct complex quantitative analysis of DNA mixtures, LikeLTD may remain useful as a robust method for the analysis of LT-DNA profiles, Lab Retriever applies the semi-continuous model, and STRmix™ and TrueAllele® use a fully continuous model. The ISFG has established validation guidelines and presented recommendations for bio-statistical software to be used in forensic genetics [154]. We also advocate that forensic DNA profile interpretation software should be cross-validated and demonstrate performance under relevant guidelines, in order to facilitate improved evaluation of the complex evidence in court.
Conclusion
Mixed DNA samples are a common biological material in forensic crime cases, and the analysis and interpretation of the results is one of the difficulties in forensic casework. The analytic approaches and detection technologies described herein are intended to provide some instructions for forensic practitioners about how to apply different methods in the evaluation of forensic DNA mixtures. In addition to the two-step DE method used in routine work, other separation and extraction methods have achieved certain advancements in the past years. Additionally, some novel genetic markers, such as DIP-STRs and microhaps, have been defined and applied to improve the ability to present legally powerful evidence when handling DNA mixtures. What is more, much open-source or commercial software has been proposed to facilitate mixture analysis and reduce manually calculated errors.
High-throughput sequencing has accelerated investigation in many fields of biological science. In the last several decades, the use of MPS in forensic genetics has been questioned, and now, we know that it can be applied specifically in forensic casework, including human identification, phenotypic trait determination, and mixture detection [68, 155]. MPS technology has many advantages when compared with a traditional CE method, especially the capability to simultaneously detect many different kinds of genetic markers and to export detailed sequence information, which significantly enhances the forensic investigation of complex DNA mixtures. As a result, it is increasingly implemented and used in forensic laboratories. Currently, the validation of software solutions and the cost of instruments and kits are key factors in the introduction of MPS into forensic genetics.
In general, the interpretation of mixed DNA profiles obtained from multiple contributors has proven to be a particularly difficult problem in forensic science in terms of providing legal evidence, while the development of various appropriate methods, software, and detection techniques over the years has indeed significantly improved the ability to address this type of data. Increased detection sensitivity also means more challenges for mixture interpretation. From our point of view, varieties of DNA mixtures are too miscellaneous to establish a unified standard for detection and interpretation, which need extensive experience and careful training. The future advanced investigation should be forced to quantitatively and entirely assess mixture results, as well as to develop a reliable sequencing system with corresponding analysis software.
References
Bieber FR, Buckleton JS, Budowle B, Butler JM, Coble MD (2016) Evaluation of forensic DNA mixture evidence: protocol for evaluation, interpretation, and statistical calculations using the combined probability of inclusion. BMC Genet 17(1):125. https://doi.org/10.1186/s12863-016-0429-7
Bright JA, Taylor D, Gittelson S, Buckleton J (2017) The paradigm shift in DNA profile interpretation. Forensic Sci Int Genet 31:e24–e32. https://doi.org/10.1016/j.fsigen.2017.08.005
Klein SB, Buoncristiani MR (2017) Evaluating the efficacy of DNA differential extraction methods for sexual assault evidence. Forensic Sci Int Genet 29:109–117. https://doi.org/10.1016/j.fsigen.2017.03.021
Oldoni F, Castella V, Grosjean F, Hall D (2017) Sensitive DIP-STR markers for the analysis of unbalanced mixtures from “touch” DNA samples. Forensic Sci Int Genet 28:111–117
Hwa HL, Chung WC, Chen PL, Lin CP, Li HY, Yin HI, Lee JC (2018) A 1204-single nucleotide polymorphism and insertion-deletion polymorphism panel for massively parallel sequencing analysis of DNA mixtures. Forensic Sci Int Genet 32:94–101. https://doi.org/10.1016/j.fsigen.2017.11.002
Gill P, Jeffreys A, Werrett D (1985) Forensic application of DNA ‘fingerprints’. Nature 318(6046):577–579
Voorhees JC, Ferrance JP, Landers JP (2006) Enhanced elution of sperm from cotton swabs via enzymatic digestion for rape kit analysis. J Forensic Sci 51(3):574–579. https://doi.org/10.1111/j.1556-4029.2006.00112.x
Benschop C, Wiebosch D, Kloosterman A, Sijen T (2010) Post-coital vaginal sampling with nylon flocked swabs improves DNA typing. Forensic Sci Int Genet 4(2):115–121
Lounsbury JA, Nambiar SM, Karlsson A, Cunniffe H, Norris JV, Ferrance JP, Landers JP (2014) Enhanced recovery of spermatozoa and comprehensive lysis of epithelial cells from sexual assault samples having a low cell counts or aged up to one year. Forensic Sci Int Genet 8(1):84–89. https://doi.org/10.1016/j.fsigen.2013.06.015
Elliott K, Hill DS, Lambert C, Burroughes TR, Gill P (2003) Use of laser microdissection greatly improves the recovery of DNA from sperm on microscope slides. Forensic Sci Int 137(1):28–36
Vandewoestyne M, Nieuwerburgh FV, Hoofstat DV, Deforce D (2012) Evaluation of three DNA extraction protocols for forensic STR typing after laser capture microdissection. Forensic Sci Int Genet 6(2):258–262. https://doi.org/10.1016/j.fsigen.2011.06.002
Zhao XC, Wang L, Sun J, Jiang BW, Zhang EL, Ye J (2016) Isolating sperm from cell mixtures using magnetic beads coupled with an anti-PH-20 antibody for forensic DNA analysis. PLoS One 11(7):e0159401. https://doi.org/10.1371/journal.pone.0159401
Wiegand P, Schurenkamp M, Schutte U (1992) DNA extraction from mixtures of body fluid using mild preferential lysis. Int J Legal Med 104(6):359–360
Yoshida K, Sekiguchi K, Mizuno N, Kasai K, Sakai I, Sato H, Seta S (1995) The modified method of two-step differential extraction of sperm and vaginal epithelial cell DNA from vaginal fluid mixed with semen. Forensic Sci Int 72(1):25–33
Iwasaki M, Kubo S, Ogata M, Nakasono I (1989) A demonstration of spermatozoa on vaginal swabs after complete destruction of the vaginal cell deposits. J Forensic Sci 34(3):659–664
Norris JV, Manning K, Linke SJ, Ferrance JP, Landers JP (2007) Expedited, chemically enhanced sperm cell recovery from cotton swabs for rape kit analysis. J Forensic Sci 52(4):800–805. https://doi.org/10.1111/j.1556-4029.2007.00453.x
Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta LA (1997) Laser capture microdissection: molecular analysis of tissue. Science 278(5342):1481–1483
Schutze K, Posl H, Lahr G (1998) Laser micromanipulation systems as universal tools in cellular and molecular biology and in medicine. Cell Mol Biol 44(5):735–746
Kolble K (2000) The LEICA microdissection system: design and applications. J Mol Med (Berl) 78(7):B24–B25
Micke P, Ostman A, Lundeberg J, Ponten F (2005) Laser-assisted cell microdissection using the PALM system. Methods Mol Biol 293:151–166
EmmertBuck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA (1996) Laser capture microdissection. Science 274(5289):998–1001
Sanders CT, Sanchez N, Ballantyne J, Peterson DA (2006) Laser microdissection separation of pure spermatozoa from epithelial cells for short tandem repeat analysis. J Forensic Sci 51(4):748–757. https://doi.org/10.1111/j.1556-4029.2006.00180.x
Vandewoestyne M, Deforce D (2010) Laser capture microdissection in forensic research: a review. Int J Legal Med 124(6):513–521. https://doi.org/10.1007/s00414-010-0499-4
Lucy D, Curran J, Pirie A, Gill P (2007) The probability of achieving full allelic representation for LCN-STR profiling of haploid cells. Sci Justice 47(4):168–171
Vandewoestyne M, Hoofstat DV, Nieuwerburgh FV, Deforce D (2009) Automatic detection of spermatozoa for laser capture microdissection. Int J Legal Med 123(2):169–175
Anslinger K, Mack B, Bayer B, Rolf B, Eisenmenger W (2005) Digoxigenin labelling and laser capture microdissection of male cells. Int J Legal Med 119(6):374–377
Anslinger K, Bayer B, Mack B, Eisenmenger W (2007) Sex-specific fluorescent labelling of cells for laser microdissection and DNA profiling. Int J Legal Med 121(1):54–56
Murray C, McAlister C, Elliott K (2007) Identification and isolation of male cells using fluorescence in situ hybridisation and laser microdissection, for use in the investigation of sexual assault. Forensic Sci Int Genet 1(3–4):247–252
Vandewoestyne M, Hoofstat DV, Nieuwerburgh FV, Deforce D (2009) Suspension fluorescence in situ hybridization (S-FISH) combined with automatic detection and laser microdissection for STR profiling of male cells in male/female mixtures. Int J Legal Med 123(5):441–447. https://doi.org/10.1007/s00414-009-0341-z
Han JP, Yang F, Xu C, Wei YL, Zhao XC, Hu L, Ye J, Li CX (2014) A new strategy for sperm isolation and STR typing from multi-donor sperm mixtures. Forensic Sci Int Genet 13:239–246. https://doi.org/10.1016/j.fsigen.2014.08.012
Safarík I, Safaríková M (1999) Use of magnetic techniques for the isolation of cells. J Chromatogr B Biomed Sci Appl 722(1–2):33–53
Eisenberg A (2002) Spermatozoa capture during the differential extraction process for STR typing of sexual assault evidence. Bureau of Justice Statistics
Anslinger K, Bayer B, Danilov SM, Metzger R (2008) Application of sperm-specific antibodies for the separation of sperm from cell mixtures. Forensic Sci Int Genet Suppl Ser 1(1):394–395
Zhao XC, Jiang BW (2012) Preliminary study on a high specific method for directional capture and separation of sperm cells from forensic samples. Foren Sci Technol 1:14–17
Wang Q, Ning S, Li X, Hong W, Lin Z (2013) Isolation of sperm cells from mixed stains by immunomagnetic bead. Chin J Forensic Med 28(4):317–319
Bienvenue JM, Landers JP (2010) DNA extraction on microfluidic devices. Forensic Sci Rev 22(2):187–197
Wu J, Kodzius R, Cao W, Wen W (2013) Extraction, amplification and detection of DNA in microfluidic chip-based assays. Microchim Acta 181(13–14):1611–1631. https://doi.org/10.1007/s00604-013-1140-2
Bruijns B, Asten AV, Tiggelaar R, Gardeniers H (2016) Microfluidic devices for forensic DNA analysis: a review. Biosensors 6(3). https://doi.org/10.3390/bios6030041
Kim YT, Heo HY, Oh SH, Lee SH, Kim DH, Seo TS (2015) Microchip-based forensic short tandem repeat genotyping. Electrophoresis 36(15):1728–1737
Buoncristiani MR, Timken MD (2009) Development of a procedure for dielectrophoretic (DEP) separation of sperm and epithelial cells for application to sexual assault case evidence. Bureau of Justice Statistics
Khoshmanesh K, Nahavandi S, Baratchi S, Mitchell A, Kalantar-zadeh K (2011) Dielectrophoretic platforms for bio-microfluidic systems. Biosens Bioelectron 26(5):1800–1814. https://doi.org/10.1016/j.bios.2010.09.022
Evander M, Horsman KM, Easley CJ, Landers JP, Nilsson J, Laurell T (2006) Using acoustic differential extraction to enhance analysis of sexual assault evidence on a valveless glass microdevice. Appl Environ Microbiol 64(2):575–580
Norris JV, Evander M, Horsman-Hall KM, Nilsson J, Laurell T, Landers JP (2009) Acoustic differential extraction for forensic analysis of sexual assault evidence. Anal Chem 81(15):6089–6095. https://doi.org/10.1021/ac900439b
Fontana F, Rapone C, Bregola G, Aversa R, de Meo A, Signorini G, Sergio M, Ferrarini A, Lanzellotto R, Medoro G, Giorgini G, Manaresi N, Berti A (2017) Isolation and genetic analysis of pure cells from forensic biological mixtures: the precision of a digital approach. Forensic Sci Int Genet 29:225–241
Garvin AM (2003) Filtration based DNA preparation for sexual assault cases. J Forensic Sci 48(5):1084–1087
Brück S, Evers H, Heidorn F, Müller U, Kilper R, Verhoff M (2011) Single cells for forensic DNA analysis-from evidence material to test tube. J Forensic Sci 56(1):176–180
Schneider C, Muller U, Kilper R, Siebertz B (2012) Low copy number DNA profiling from isolated sperm using the aureka®-micromanipulation system. Forensic Sci Int Genet 6(4):461–465. https://doi.org/10.1016/j.fsigen.2011.10.001
De Moors A, Georgalis T, Armstrong G, Modler J, Fregeau CJ (2013) Sperm Hy-Liter: an effective tool for the detection of spermatozoa in sexual assault exhibits. Forensic Sci Int Genet 7(3):367–379. https://doi.org/10.1016/j.fsigen.2013.02.011
Schoell WMJ, Klintschar M, Mirhashemi R, Pertl B (1999) Separation of sperm and vaginal cells with flow cytometry for DNA typing after sexual assault. Obstet Gynecol 94(4):623–627
Schoell WMJ, Klintschar M, Mirhashemi R, Strunk D, Giuliani A, Bogen-sberger G et al (1999) Separation of sperm and vaginal cells based on ploidy, MHC class I-, CD45-, and cytokeratin expression for enhancement of DNA typing after sexual assault. Cytometry 36(4):319–323
Di Nunno N, Melato M, Vimercati A, Di Nunno C, Costantinides F, Vecchiotti C, Frezzini C, Cina S, Vimercati F (2003) DNA identification of sperm cells collected and sorted by flow cytometry. Am J Forensic Med Pathol 24(3):254–270. https://doi.org/10.1097/01.paf.0000070224.58005.ac
Kovacs T, Bekesi G, Fabian A, Rakosy Z, Horvath G, Matyus L, Balazs M, Jenei A (2008) DNA flow cytometry of human spermatozoa: consistent stoichiometric staining of sperm DNA using a novel decondensation protocol. Cytometry A 73(10):965–970. https://doi.org/10.1002/cyto.a.20618
Verdon TJ, Mitchell RJ, Chen W, Xiao K, Van Oorschot RAH (2015) FACS separation of non-compromised forensically relevant biological mixtures. Forensic Sci Int Genet 14:194–200
Katilius E, Sanders G, Carmel A, Gold L, Laberge GS (2014) Sperm capture using aptamer based technology. Bureau of Justice Statistics
Sanders G, Katilius E, Gold L (2012) Method for purification and identification of sperm cells. United States, Patent, SomaLogic Inc. (Boulder, CO, US) 8703416. http://www.freepatentsonline.com/8703416.html
Rothe J, Watkins NE Jr, Nagy M (2012) New prediction model for probe specificity in an allele-specific extension reaction for haplotype-specific extraction (HSE) of Y chromosome mixtures. PLoS One 7(9):e45955
Rothe J (2014) Establishment of a Y-chromosome specific extraction method for the separation of Y-chromosomal haplotypes from male DNA mixtures. Social Science Electron Publ 12(6):132–133
Rothe J, Nagy M (2015) Separation of Y-chromosomal haplotypes from male DNA mixtures via multiplex haplotype-specific extraction. Forensic Sci Int Genet 19:223–231
Zander J, Rothe J, Dapprich J, Nagy M (2017) New application for haplotype-specific extraction: separation of mitochondrial DNA mixtures. Forensic Sci Int Genet 29:242–249
Koukouvinos G, Metheniti A, Karachaliou CE, Goustouridis D, Livaniou E, Misiakos K, Raptis I, Kondili A, Miniati P, Petrou P (2017) White light reflectance spectroscopy biosensing system for fast quantitative prostate specific antigen determination in forensic samples. Talanta 175:443–450
Loftus A, Murphy G, Brown H, Montgomery A, Tabak J, Baus J, Carroll M, Green A, Sikka S, Sinha S (2017) Development and validation of InnoQuant® HY, a system for quantitation and quality assessment of total human and male DNA using high copy targets. Forensic Sci Int Genet 29:205–217. https://doi.org/10.1016/j.fsigen.2017.04.009
Fujita Y, Tokunaga I, Shin-Ichi K (2003) Forensic identification of a vaginal fluid and saliva mixture through DNA analysis. Acta Criminologiae Et Medicinae Legalis Japonica 69(2):48–52
Honda K, Yano S, Nishi T, Iwabuchi Y, Kurosu A, Sugano Y (2013) Selective blood-DNA extraction from mixed stain using ABO antibody for short tandem repeat typing. Forensic Sci Int Genet Suppl Ser 4(1):e326–e327
Yano S, Honda K, Kaminiwa J, Nishi T, Iwabuchi Y, Sugano Y, Kurosu A, Suzuki Y (2014) DNA extraction for short tandem repeat typing from mixed samples using anti-human leukocyte CD45 and ABO blood group antibodies. Forensic Sci Int Genet 10:17–22. https://doi.org/10.1016/j.fsigen.2014.01.001
Westen AA, Kraaijenbrink T, Ea RDM, Harteveld J, Willemse P, Zuniga SB, Kj VDG, Weiler NE, Warnaar J, Kayser M (2014) Comparing six commercial autosomal STR kits in a large Dutch population sample. Forensic Sci Int Genet 10(3):55–63
Budowle B, Onorato AJ, Callaghan TF, Della MA, Gross AM, Guerrieri RA, Luttman JC, Mcclure DL (2009) Mixture interpretation: defining the relevant features for guidelines for the assessment of mixed DNA profiles in forensic casework. J Forensic Sci 54(4):810–821
Gelardi C, Rockenbauer E, Dalsgaard S, Børsting C, Morling N (2014) Second generation sequencing of three STRs D3S1358, D12S391 and D21S11 in Danes and a new nomenclature for sequenced STR alleles. Forensic Sci Int Genet 12:38–41. https://doi.org/10.1016/j.fsigen.2014.04.016
van der Gaag KJ, de Leeuw RH, Hoogenboom J, Patel J, Storts DR, Laros JFJ, de Knijff P (2016) Massively parallel sequencing of short tandem repeats—population data and mixture analysis results for the PowerSeq system. Forensic Sci Int Genet 24:86–96. https://doi.org/10.1016/j.fsigen.2016.05.016
Isaacson J, Schwoebel E, Shcherbina A, Ricke D, Harper J, Petrovick M, Bobrow J, Boettcher T, Helfer B, Zook C, Wack E (2015) Robust detection of individual forensic profiles in DNA mixtures. Forensic Sci Int Genet 14:31–37. https://doi.org/10.1016/j.fsigen.2014.09.003
Børsting C, Fordyce SL, Olofsson J, Mogensen HS, Morling N (2014) Evaluation of the Ion Torrent™ HID SNP 169-plex: a SNP typing assay developed for human identification by second generation sequencing. Forensic Sci Int Genet 12:144–154
Goodwin S, Mcpherson JD, Mccombie WR (2016) Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17(6):333–351
Voskoboinik L, Ayers SB, Lefebvre AK, Darvasi A (2015) SNP-microarrays can accurately identify the presence of an individual in complex forensic DNA mixtures. Forensic Sci Int Genet 16:208–215
Liu J, Wang J, Zhang X, Li Z, Yun K, Liu Z, Zhang G (2017) A mixture detection method based on separate amplification using primer specific alleles of INDELs-a study based on two person’s DNA mixture. J Forensic Legal Med 46:30–36. https://doi.org/10.1016/j.jflm.2017.01.002
Prinz M, Ishii A, Coleman A, Baum HJ, Shaler RC (2001) Validation and casework application of a Y chromosome specific STR multiplex. Forensic Sci Int 120(3):177–188
Prinz M, Sansone M (2001) Y chromosome-specific short tandem repeats in forensic casework. Croat Med J 42(3):288–291
Walker JA, Garber RK, Hedges DJ, Kilroy GE, Xing J, Batzer MA (2004) Resolution of mixed human DNA samples using mitochondrial DNA sequence variants. Anal Biochem 325(1):171–173
Kristinsson R (2011) Mitochondrial DNA analysis by denaturing high-performance liquid chromatography for the characterization and separation of mixtures in forensic samples. Dissertations, University of Denver, US
Pfeifer CM, Klein-Unseld R, Klintschar M, Wiegand P (2012) Comparison of different interpretation strategies for low template DNA mixtures. Forensic Sci Int Genet 6(6):716–722
Benschop C, Haned H, Sijen T (2013) Consensus and pool profiles to assist in the analysis and interpretation of complex low template DNA mixtures. Int J Legal Med 127(1):11–23
Gill P, Haned H, Bleka O, Hansson O, Dørum G, Egeland T (2015) Genotyping and interpretation of STR-DNA: low-template, mixtures and database matches—twenty years of research and development. Forensic Sci Int Genet 18:100–117
Mcdonald A, Jones E, Lewis J, O’Rourke P (2015) Y-STR analysis of digital and/or penile penetration cases with no detected spermatozoa. Forensic Sci Int Genet 15(2):84–89
Purps J, Geppert M, Nagy M, Roewer L (2015) Validation of a combined autosomal/Y-chromosomal STR approach for analyzing typical biological stains in sexual-assault cases. Forensic Sci Int Genet 19:238–242. https://doi.org/10.1016/j.fsigen.2015.08.002
De lPM, Phillips C, Fondevila M, Gelabertbesada M, Carracedo Á, Lareu MV (2017) A forensic multiplex of nine novel pentameric-repeat STRs. Forensic Sci Int Genet 29:154–164
Haas C, Hanson E, Anjos MJ, Bar W, Banemann R, Berti A, Borges E, Bouakaze C, Carracedo A, Carvalho M, Castella V, Choma A, De Cock G, Dotsch M, Hoff-Olsen P, Johansen P, Kohlmeier F, Lindenbergh PA, Ludes B, Maronas O, Moore D, Morerod ML, Morling N, Niederstatter H, Noel F, Parson W, Patel G, Popielarz C, Salata E, Schneider PM, Sijen T, Sviezena B, Turanska M, Zatkalikova L, Ballantyne J (2012) RNA/DNA co-analysis from blood stains—results of a second collaborative EDNAP exercise. Forensic Sci Int Genet 6(1):70–80. https://doi.org/10.1016/j.fsigen.2011.02.004
Haas C, Hanson E, Anjos MJ, Banemann R, Berti A, Borges E, Carracedo A, Carvalho M, Courts C, De Cock G, Dotsch M, Flynn S, Gomes I, Hollard C, Hjort B, Hoff-Olsen P, Hribikova K, Lindenbergh A, Ludes B, Maronas O, McCallum N, Moore D, Morling N, Niederstatter H, Noel F, Parson W, Popielarz C, Rapone C, Roeder AD, Ruiz Y, Sauer E, Schneider PM, Sijen T, Court DS, Sviezena B, Turanska M, Vidaki A, Zatkalikova L, Ballantyne J (2013) RNA/DNA co-analysis from human saliva and semen stains-results of a third collaborative EDNAP exercise. Forensic Sci Int Genet 7(2):230–239. https://doi.org/10.1016/j.fsigen.2012.10.011
Haas C, Hanson E, Anjos MJ, Ballantyne KN, Banemann R, Bhoelai B, Borges E, Carvalho M, Courts C, De Cock G, Drobnic K, Dotsch M, Fleming R, Franchi C, Gomes I, Hadzic G, Harbison SA, Harteveld J, Hjort B, Hollard C, Hoff-Olsen P, Huls C, Keyser C, Maronas O, McCallum N, Moore D, Morling N, Niederstatter H, Noel F, Parson W, Phillips C, Popielarz C, Roeder AD, Salvaderi L, Sauer E, Schneider PM, Shanthan G, Court DS, Turanska M, van Oorschot RA, Vennemann M, Vidaki A, Zatkalikova L, Ballantyne J (2014) RNA/DNA co-analysis from human menstrual blood and vaginal secretion stains: results of a fourth and fifth collaborative EDNAP exercise. Forensic Sci Int Genet 8(1):203–212. https://doi.org/10.1016/j.fsigen.2013.09.009
Haas C, Hanson E, Banemann R, Bento AM, Berti A, Carracedo A, Courts C, Cock G, Drobnic K, Fleming R, Franchi C, Gomes I, Hadzic G, Harbison SA, Hjort B, Hollard C, Hoff-Olsen P, Keyser C, Kondili A, Maronas O, McCallum N, Miniati P, Morling N, Niederstatter H, Noel F, Parson W, Porto MJ, Roeder AD, Sauer E, Schneider PM, Shanthan G, Sijen T, Syndercombe Court D, Turanska M, van den Berge M, Vennemann M, Vidaki A, Zatkalikova L, Ballantyne J (2015) RNA/DNA co-analysis from human skin and contact traces-results of a sixth collaborative EDNAP exercise. Forensic Sci Int Genet 16:139–147. https://doi.org/10.1016/j.fsigen.2015.01.002
Uchimoto ML, Beasley E, Coult N, Omelia EJ, World D, Williams G (2013) Considering the effect of stem-loop reverse transcription and real-time PCR analysis of blood and saliva specific microRNA markers upon mixed body fluid stains. Forensic Sci Int Genet 7(4):418–421. https://doi.org/10.1016/j.fsigen.2013.04.006
Voskoboinik L, Darvasi A (2011) Forensic identification of an individual in complex DNA mixtures. Forensic Sci Int Genet 5(5):428–435
Alonso A, Salas A, Albarrán C, Arroyo E, Castro A, Crespillo M, Di LA, Lareu MV, Cubría CL, Soto ML (2002) Results of the 1999-2000 collaborative exercise and proficiency testing program on mitochondrial DNA of the GEP-ISFG: an inter-laboratory study of the observed variability in the heteroplasmy level of hair from the same donor. Forensic Sci Int 125:1):1–1):7
Prieto L, Montesino M, Salas A, Alonso A, Albarrã nC, Alvarez S, Crespillo M, Di LA, Doutremepuich C, Fernã n-FI (2003) The 2000-2001 GEP-ISFG Collaborative Exercise on mtDNA: assessing the cause of unsuccessful mtDNA PCR amplification of hair shaft samples. Forensic Sci Int 134(1):46–53
Salas A, Prieto L, Montesino M, Albarrán C, Arroyo E, Paredesherrera MR, Di LA, Doutremepuich C, Fernándezfernández I, Ag DLV (2005) Mitochondrial DNA error prophylaxis: assessing the causes of errors in the GEP'02-03 proficiency testing trial. Forensic Sci Int 148(2–3):191–198
Crespillo M, Paredes MR, Prieto L, Montesino M, Salas A, Albarran C, Alvareziglesias V, Amorin A, Bernielllee G, Brehm A (2006) Results of the 2003-2004 GEP-ISFG collaborative study on mitochondrial DNA: focus on the mtDNA profile of a mixed semen-saliva stain. Forensic Sci Int 160(2–3):157–167
Montesino M, Salas A, Crespillo M, Albarrán C, Alonso A, Alvarez-Iglesias V, Cano JA, Carvalho M, Corach D, Cruz C (2007) Analysis of body fluid mixtures by mtDNA sequencing: an inter-laboratory study of the GEP-ISFG working group. Forensic Sci Int 168(1):42–56
Montesino M, Picornell A, Brehm A (2008) 2006 GEP-ISFG collaborative exercise on mtDNA: reflections about interpretation, artefacts, and DNA mixtures. Forensic Sci Int Genet 2(2):126–133
Zhang L, Ding M, Pang H, Xing J, Xuan J, Wang C, Lin Z, Han S, Liang K, Li C (2016) Mitochondrial DNA typing of laser-captured single sperm cells to differentiate individuals in a mixed semen stain. Electrophoresis 37(15–16):2273–2277
Castella V, Gervaix J, Hall D (2013) DIP–STR: highly sensitive markers for the analysis of unbalanced genomic mixtures. Hum Mutat 34(4):644–654
Cereda G, Biedermanna A (2014) An investigation of the potential of DIP-STR markers for DNA mixture analyses. Forensic Sci Int Genet 11(4):229–240
Oldoni F, Castella V, Hall D (2015) A novel set of DIP-STR markers for improved analysis of challenging DNA mixtures. Forensic Sci Int Genet 19:156–164
Tan Y, Wang L, Wang H, Tian H, Li Z, Wang Q, Jian H, Cao S, Liang W, Zhang L (2017) An investigation of a set of DIP-STR markers to detect unbalanced DNA mixtures among the southwest Chinese Han population. Forensic Sci Int Genet 31:34–39. https://doi.org/10.1016/j.fsigen.2017.08.014
Oldoni F, Castella V, Hall D (2017) Application of DIP-STRs to sexual/physical assault investigations: eight case reports. Forensic Sci Int Genet 30:106–113. https://doi.org/10.1016/j.fsigen.2017.06.010
Pakstis AJ, Fang R, Furtado MR, Kidd JR, Kidd KK (2012) Mini-haplotypes as lineage informative SNPs and ancestry inference SNPs. Eur J Hum Genet 20(11):1148–1154
Kidd KK, Pakstis AJ, Speed WC, Lagace R, Chang J, Wootton S, Ihuegbu N (2013) Microhaplotype loci are a powerful new type of forensic marker. Forensic Sci Int Genet Suppl Ser 4(1):e123–e124. https://doi.org/10.1016/j.fsigss.2013.10.063
Kidd KK, Pakstis AJ, Speed WC, Lagacé R, Chang J, Wootton S, Haigh E, Kidd JR (2014) Current sequencing technology makes microhaplotypes a powerful new type of genetic marker for forensics. Forensic Sci Int Genet 12(9):215–224
Kidd KK, Speed WC (2015) Criteria for selecting microhaplotypes: mixture detection and deconvolution. Investig Genet 6(1):1
Kidd KK (2016) Proposed nomenclature for microhaplotypes. Hum Genomics 10(1):16
Zhu J, Zhou N, Jiang Y, Wang L, He W, Peng D, Su Q, Mao J, Chen D, Liang W, Zhang L (2015) FLfinder: a novel software for the microhaplotype marker. Forensic Sci Int Genet Suppl Ser 5:e622–e624. https://doi.org/10.1016/j.fsigss.2015.10.002
Bill M, Gill P, Curran J, Clayton T, Pinchin R, Healy M, Buckleton J (2005) PENDULUM-a guideline-based approach to the interpretation of STR mixtures. Forensic Sci Int 148(2–3):181–189. https://doi.org/10.1016/j.forsciint.2004.06.037
Buckleton JS, Jo-Anne B, Duncan T (2016) Forensic DNA evidence interpretation, 2nd edn. CRC Press
Kelly H, Bright JA, Buckleton JS, Curran JM (2014) A comparison of statistical models for the analysis of complex forensic DNA profiles. Sci Justice 54(1):66–70
Haned H, Slooten K, Gill P (2012) Exploratory data analysis for the interpretation of low template DNA mixtures. Forensic Sci Int Genet 6(6):762–774
Slooten K (2015) Distinguishing between donors and their relatives in complex DNA mixtures with binary models. Forensic Sci Int Genet 21(1–2):95–109
Gill P, Kirkham A, Curran J (2007) LoComatioN: a software tool for the analysis of low copy number DNA profiles. Forensic Sci Int 166(2):128–138. https://doi.org/10.1016/j.forsciint.2006.04.016
Puch-Solis R, Rodgers L, Mazumder A, Pope S, Evett I, Curran J, Balding D (2013) Evaluating forensic DNA profiles using peak heights, allowing for multiple donors, allelic dropout and stutters. Forensic Sci Int Genet 7(5):555–563. https://doi.org/10.1016/j.fsigen.2013.05.009
Taylor D, Bright JA, Buckleton J (2013) The interpretation of single source and mixed DNA profiles. Forensic Sci Int Genet 7(5):516–528
Bille TW, Weitz SM, Coble MD, Buckleton J, Bright JA (2015) Comparison of the performance of different models for the interpretation of low level mixed DNA profiles. Electrophoresis 35(21–22):3125–3133
Bleka Ø, Benschop CC, Storvik G, Gill P (2016) A comparative study of qualitative and quantitative models used to interpret complex STR DNA profiles. Forensic Sci Int Genet 25:85–96
Biedermann A, Taroni F (2012) Bayesian networks for evaluating forensic DNA profiling evidence: a review and guide to literature. Forensic Sci Int Genet 6(2):147–157
Pascali VL, Merigioli S (2012) Joint Bayesian analysis of forensic mixtures. Forensic Sci Int Genet 6(6):735–748
Cereda G, Biedermann A, Hall D, Taroni F (2014) Object-oriented Bayesian networks for evaluating DIP-STR profiling results from unbalanced DNA mixtures. Forensic Sci Int Genet 8(1):159–169
Curran JM (2008) A MCMC method for resolving two person mixtures. Sci Justice 48(4):168–177
Gill P, Brenner CH, Buckleton JS, Carracedo A, Krawczak M, Mayr WR, Morling N, Prinz M, Schneider PM, Weir BS (2006) DNA commission of the International Society of Forensic Genetics: recommendations on the interpretation of mixtures. Forensic Sci Int 160(2):90–101. https://doi.org/10.1016/j.forsciint.2006.04.009
Gill P, Gusmão L, Haned H, Mayr WR, Morling N, Parson W, Prieto L, Prinz M, Schneider H, Schneider PM, Weir BS (2012) DNA commission of the International Society of Forensic Genetics: recommendations on the evaluation of STR typing results that may include drop-out and/or drop-in using probabilistic methods. Forensic Sci Int Genet 6(6):679–688. https://doi.org/10.1016/j.fsigen.2012.06.002
Morling N, Bastisch I, Gill P, Schneider PM (2007) Interpretation of DNA mixtures—European consensus on principles. Forensic Sci Int Genet 1(3–4):291–292
Stringer P, Scheffer JW, Scott P, Lee J, Goetz R, Ientile V, Eckhoff C, Turbett G, Carroll D, Harbison SA (2009) Interpretation of DNA mixtures—Australian and New Zealand consensus on principles. Forensic Sci Int Genet 3(2):144–145
Dørum G, Bleka Ø, Gill P, Haned H, Snipen L, Sæbø S, Egeland T (2014) Exact computation of the distribution of likelihood ratios with forensic applications. Forensic Sci Int Genet 9(3):93–101
Taylor D, Buckleton J, Evett I (2015) Testing likelihood ratios produced from complex DNA profiles. Forensic Sci Int Genet 16:165–171
Marsden CD, Rudin N, Inman K, Lohmueller KE (2016) An assessment of the information content of likelihood ratios derived from complex mixtures. Forensic Sci Int Genet 22:64–72. https://doi.org/10.1016/j.fsigen.2016.01.008
Benschop CCG, Haned H, Jeurissen L, Gill PD, Sijen T (2015) The effect of varying the number of contributors on likelihood ratios for complex DNA mixtures. Forensic Sci Int Genet 19:92–99. https://doi.org/10.1016/j.fsigen.2015.07.003
Slooten K (2017) Accurate assessment of the weight of evidence for DNA mixtures by integrating the likelihood ratio. Forensic Sci Int Genet 27:1–16. https://doi.org/10.1016/j.fsigen.2016.11.001
Haned H, Benschop CCG, Gill PD, Sijen T (2015) Complex DNA mixture analysis in a forensic context: evaluating the probative value using a likelihood ratio model. Forensic Sci Int Genet 16:17–25. https://doi.org/10.1016/j.fsigen.2014.11.014
Benschop CCG, van de Merwe L, de Jong J, Vanvooren V, Kempenaers M, Kees van der Beek CP, Barni F, Reyes EL, Moulin L, Pene L, Haned H, Sijen T (2017) Validation of SmartRank: a likelihood ratio software for searching national DNA databases with complex DNA profiles. Forensic Sci Int Genet 29:145–153
Buckleton J, Curran J (2008) A discussion of the merits of random man not excluded and likelihood ratios. Forensic Sci Int Genet 2(4):343–348. https://doi.org/10.1016/j.fsigen.2008.05.005
Van Nieuwerburgh F, Goetghebeur E, Vandewoestyne M, Deforce D (2009) Impact of allelic dropout on evidential value of forensic DNA profiles using RMNE. Bioinformatics 25(2):225–229. https://doi.org/10.1093/bioinformatics/btn608
Fung TWK, Chung YK (2013) Evaluation of forensic evidence in DNA mixture using RMNE. In: The 59th World Statistics Congress (WSC) of the International Statistical Institute (ISI), Hong Kong, China
Butler JM (2015) Chapter 1—data interpretation overview. In: Advanced topics in forensic DNA typing: interpretation. Academic Press, San Diego, pp 3–24. https://doi.org/10.1016/B978-0-12-405213-0.00001-4
Slooten KJ, Egeland T (2016) Exclusion probabilities and likelihood ratios with applications to mixtures. Int J Legal Med 130(1):39–57
Bille T, Bright JA, Buckleton J (2013) Application of random match probability calculations to mixed STR profiles. J Forensic Sci 58(2):474–485
Carracedo A, Schneider PM, Butler J, Prinz M (2012) Focus issue—analysis and biostatistical interpretation of complex and low template DNA samples. Forensic Sci Int Genet 6(6):677–678
Curran JM, Gill P, Bill MR (2005) Interpretation of repeat measurement DNA evidence allowing for multiple contributors and population substructure. Forensic Sci Int 148(1):47–53
Haned H (2011) Forensim: an open-source initiative for the evaluation of statistical methods in forensic genetics. Forensic Sci Int Genet 5(4):265–268
Haned H, Gill P (2011) Analysis of complex DNA mixtures using the Forensim package. Forensic Sci Int Genet Suppl Ser 3(1):e79–e80
Haned H, Pène L, Lobry JR, Dufour AB, Pontier D (2011) Estimating the number of contributors to forensic DNA mixtures: does maximum likelihood perform better than maximum allele count? J Forensic Sci 56(1):23–28
Gill P, Haned H (2013) A new methodological framework to interpret complex DNA profiles using likelihood ratios. Forensic Sci Int Genet 7(2):251–263. https://doi.org/10.1016/j.fsigen.2012.11.002
Gill P, Haned H, Eduardoff M, Santos C, Phillips C, Parson W (2015) The open-source software LRmix can be used to analyse SNP mixtures. Forensic Sci Int Genet Suppl Ser 5:e50–e51. https://doi.org/10.1016/j.fsigss.2015.09.020
Bleka Ø, Eduardoff M, Santos C, Phillips C, Parson W, Gill P (2017) Open source software EuroForMix can be used to analyse complex SNP mixtures. Forensic Sci Int Genet 31:105–110. https://doi.org/10.1016/j.fsigen.2017.08.001
Cowell RG, Graversen T, Lauritzen SL, Mortera J (2015) Analysis of forensic DNA mixtures with artefacts. J Roy Stat Soc C-APP 64(1):1–48
Bleka Ø, Storvik G, Gill P (2016) EuroForMix: an open source software based on a continuous model to evaluate STR DNA profiles from a mixture of contributors with artefacts. Forensic Sci Int Genet 21:35–44. https://doi.org/10.1016/j.fsigen.2015.11.008
Perlin MW, Legler MM, Spencer CE, Smith JL, Allan WP, Belrose JL, Duceman BW (2011) Validating TrueAllele® DNA mixture interpretation. J Forensic Sci 56(6):1430–1447
Hansson O, Gill P (2011) Evaluation of GeneMapper®; ID-X mixture analysis tool. Forensic Sci Int Genet Suppl Ser 3(1):e11–e12
Balding DJ (2013) Evaluation of mixed-source, low-template DNA profiles in forensic science. Proc Natl Acad Sci USA 110(30):12241–12246
Inman K, Rudin N, Cheng K, Robinson C, Kirschner A, Inman-Semerau L, Lohmueller KE (2015) Lab Retriever: a software tool for calculating likelihood ratios incorporating a probability of drop-out for forensic DNA profiles. BMC Bioinformatics 16(1):298
Moretti TR, Just RS, Kehl SC, Willis LE, Buckleton JS, Bright JA, Taylor DA, Onorato AJ (2017) Internal validation of STRmix for the interpretation of single source and mixed DNA profiles. Forensic Sci Int Genet 29:126–144. https://doi.org/10.1016/j.fsigen.2017.04.004
Coble MD, Buckleton J, Butler JM, Egeland T, Fimmers R, Gill P, Gusmão L, Guttman B, Krawczak M, Morling N, Parson W, Pinto N, Schneider PM, Sherry ST, Willuweit S, Prinz M (2016) DNA Commission of the International Society for Forensic Genetics: recommendations on the validation of software programs performing biostatistical calculations for forensic genetics applications. Forensic Sci Int Genet 25:191–197
Guo F, Yu J, Zhang L, Li J (2017) Massively parallel sequencing of forensic STRs and SNPs using the Illumina((R)) ForenSeq DNA Signature Prep Kit on the MiSeq FGx Forensic Genomics System. Forensic Sci Int Genet 31:135–148. https://doi.org/10.1016/j.fsigen.2017.09.003
Funding
This study was supported by grants from the National Key R&D Program of China (No. 2016YFC0800703), the Standard Program of Shanghai Municipality (No. 16DZ0501600, No. 16DZ1205500), the Public Interest Research Grant Programs of National Research Institutes (No. GY2017D-2), and the National Natural Science Foundation (No. 81 625013, No. 81772028). The funders had no role in study design, data analysis, publishing decisions, or manuscript preparation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Tao, R., Wang, S., Zhang, J. et al. Separation/extraction, detection, and interpretation of DNA mixtures in forensic science (review). Int J Legal Med 132, 1247–1261 (2018). https://doi.org/10.1007/s00414-018-1862-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-018-1862-0