Abstract
Purpose
The current study aimed to investigate the role of long intergenic noncoding 01433 (LINC01433) in the proliferation, migration and invasion of nasopharyngeal carcinoma (NPC).
Methods
Real-time quantitative PCR (RT-qPCR) was performed to determine the expressions of LINC01433 and miR-506-3p in NPC samples and cell lines. The effects of LINC01433 on cell proliferation, migration and invasion were measured by CCK-8, wound healing assay and Transwell, respectively. In addition, Pearson correlation analysis, starBase, RNA immunoprecipitation, luciferase assay, Western blot and functional experiments were conducted to detect and confirm the relationship between LINC01433 and miR-506-3p.
Results
LINC01433 level was noticeably elevated in NPC tissues and cell lines. As the expression of LINC01433 in 5-8F cells was the highest in NPC cell lines and the expression of LINC01433 in SUNE1 cells was the lowest, 5-8F and SUNE1 cells were therefore selected as the target cells for following experiments. Furthermore, miR-506-3p was predicted as the target of LINC01433, and the two were negatively correlated with each other. Interestingly, overexpression of LINC01433 promoted proliferation, migration and invasion of NPC cells, while miR-506-3p reversed such effects of LINC01433. Moreover, LINC01433 silencing had the opposite effects to LINC01433 overexpression. Furthermore, miR-506-3p overexpression inhibited the expressions of MMP2, N-cadherin, p-PI3K and p-Akt, and promoted the expressions of E-cadherin and TIMP-2, and partially reversed the role of LINC01433 in promoting cancer development.
Conclusion
The current findings reveal that LINC01433 regulates NPC cell biological progress through miR-506-3p.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nasopharyngeal carcinoma (NPC), which is a malignant tumor that occurs in the head and neck, is characterized by early metastasis and recurrence [1]. Though the disease has a low incidence in the world, it has a high incidence in Southern China [2]. Many currently known factors such as genetics, environment, virus and diet are involved in the pathogenesis of NPC involves [1, 2], but its specific molecular mechanism is still unclear. Therefore, it is highly necessary to study the molecular mechanism of NPC.
Long-chain non-coding RNAs (lncRNAs) located in the “dark region” of a gene, moreover, lncRNAs are mainly transcribed by RNA polymerase II [3]. At present, evidence increasingly showed that lncRNA plays a key regulatory role in tumorigenesis [4,5,6,7]. For example, Geng et al. found that lncRNA HOX transcript antisense RNA is significantly higher in hepatocellular carcinoma than in normal tissues, and that its expression level is closely related to lymph node metastasis [8]. Zhang et al. showed that overexpression of lncRNA MALAT-1 promotes cell biological function in non-small cell lung cancer (NSCLC) [9]. In recent years, the role of lncRNA in NPC has also been reported [10], for example, Sun et al. found that the expression of lncRNA (LET) in tumors was significantly down-regulated in NPC group, noticeably, LET was closely related to tumor volume and tumor lymph nodes [11]. Yang et al. [12] screened 856 lncRNAs at whole-genome level for abnormal expression in primary NPC and chronic nasopharyngitis by high-throughput microarray method. Nie et al. [13] found that the expressions of HOTAIR in NPC were markedly upper than that in non-cancerous nasopharyngeal tissues.
Long intergenic noncoding 01433 (LINC01433) is an lncRNA located on chromosome 20p13 and contains 840 nucleotides [14]. Research found that LINC01433 could promote cancer development such as in NSCLC and breast cancer [15, 16]. LINC01433 is significantly increased in NSCLC tissues, and upregulation of LINC01433 promotes the proliferation, migratory and invasive ability of lung cancer cells. Recent findings indicated that the many lncRNAs in the cytoplasms widely participate in transcription regulation through interact on miRNAs or mRNAs [17,18,19]. It has been found that LINC01433 expression is up-regulated in hepatocellular carcinoma cells compared with normal cells, and that to a certain extent, LINC01433 promotes the proliferation of hepatocellular carcinoma through regulating the miR-1301/STAT3 signal pathway [20]. Therefore, we speculated that LINC01433 also functions in NPC cells through regulating certain miRNA.
LncRNAs clinically relevant to NPC have been reported, however, the role and mechanism of LINC01433 in the disease has not been explored before. Therefore, the current study intended to further explore the level of LINC01433 in NPC tissues and cell lines, and explored the mechanism of action of LINC01433 and its targeted miRNAs in the regulation of NPC development.
Materials and methods
NPC tissues, cell lines and culture
From March 2018 to September 2019, NPC tissues were obtained from 45 patients diagnosed with the disease in Sir Run Run Shaw Hospital, Zhejiang University School of Medicine. All patients had signed informed consent, and the study was approved by the Ethics Committee of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine (ZUE201801022). Human nasopharyngeal epithelial cell line (NP69, Chinese Academy of Medical Sciences & Peking Unlon dediol Colilaga) and NPC cell lines (5-8F, 6-10B, SUNE1 and C666-1, Jennio-bio, Guangzhou, China) were used in this study.
The cells were cultured in RPMI-1640 medium (11879020, Gibco, USA) containing 10% FBS (30067334, ThermoFisher, USA) and containing 4% of penicillin and streptomycin (SV30010, Hyclone, USA) in a Herocell C1 carbon dioxide incubator (37 °C, 5% CO2). One day after the cells were cultured, the original medium was replaced and the cells were continued to be cultured.
Transfection
The cells were cultured for 12 to 16 h (h). The cells were transfected using liposome method. PcDNA™3.1(+)-control (V79020), LINC01433, were purchased from ThermoFisher, sh-control, and sh-LINC01433 plasmids were ordered by GENESEED company (Guangzhou, China), while miR-506-3p mimic (miR10002878-1-5), mimic negative control (miRB160401025525-2-1), and miR-506-3p inhibitor (miR20002878-1-5) and inhibitor negative control (miR2N0000001-1-5) were purchased from RIBOBIO Company (https://www.ribobio.com/). Cell transfection was performed in accordance with the protocol of Lipofectamine 3000 Transfection Reagent (L3000015, ThermoFisher, USA).
Bioinformatics prediction
The target gene of LINC01433 was analyzed by starBase (http://starbase.sysu.edu.cn/index.php). The target genes of miR-506-3p were predicted by starBase, miRDB (http://mirdb.org/), TargetScan7 (http://www.targetscan.org/vert_72/) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) websites. The Cancer Genome Atlas (TCGA) website (https://www.cancer.gov/about-cancer) was used to analyze the expressions of target genes in NPC.
RNA immunoprecipitation (RIP)
The binding of LINC01433 to argonaute 2 (Ago2) proteins was performed on the basis of the RIP kit instructions (RIP-12RXN, Merck Millipore, USA). Briefly, cell extracts were incubated with antibodies and magnetic beads at 4 °C overnight. The above complex was resuspended with RIP-wash buffer. After separation by proteinase K, RNA was extracted and then detected by RT-qPCR. RIP antibodies: Ago2 (ab32381, 1:50, Abcam, UK) and IgG (1: 100, ab109489).
Dual luciferase activity assay
The specific sequence of LINC01433-wt and LINC01433-mut was cloned into pmirGLO luciferase Vectors (E1330; Promega, CA, USA). To study the relationship of LINC01433 and miRNA, SUNE1 and 5-8F cells were transfected with miR-506-3p mimic, while those transfected with blank served as control. The luciferase activity was measured according to Dual-Luciferase Reporter Assay System (Catalog number E1910, Promega, USA). The firefly luciferase activity of the reporter gene and Renilla luciferase reporter gene were measured in a GloMax 20/20 luminescence meter (Promega,, USA).
Real-time quantitative polymerase chain reaction (RT-qPCR)
The cells and tissues in each group were lysed to extract total RNAs at 4 °C using Trizol method [21]. The purity and concentration of RNAs were detected by NanoDrop 2000 UV–Vis (ND-LITE-PR, ThermoFisher, USA). The steps of RNA reverse transcription into cDNA follow the operation of MicroRNA Reverse Transcription Kit (4366597, ThermoFisher, USA) and PrimeScript RT reagent Kit (RR047A, TaKaRa, Japan). RT-qPCR was conducted in the Thermo Fisher Scientific real-time PCR system (QuantStudio 3, USA). The relative expressions were analyzed using the 2−ΔΔCt method. GAPDH was employed as control for mRNA and lncRNA, while U6 employed for miRNA. The sequences of the primers were listed below (5′-3′): LINC01433-F: TGCCTTTGCTGCTGTATGA, LINC01433-R: CTCCAAAGGACAGGCATGA; miR-506-3p-F: GAGTAGAGTCGTATCCAGTGCAA, miR-506-3p-R: CGTATCCAGTGCGTGTCGT; GAPDH-F: GTCAGCCGCATCTTCTTTTG, GAPDH-R: GCGCCCAATACGACCAAATC; U6-F: CTCGCTTCGGCAGCACA, U6-R: AACGCTTCACGAATTTGCGT.
CCK-8 assay
The concentration of NPC cells was determined to be 1 × 104 /mL, and cells were cultured for 24 h. After 24, 48, 72 h of transfection treatment, 10 μL CCK8 solution (HY-K0301, MedChemExpress, USA) was added to the cells. After culture for 2–4 h at 37 °C, cell viability was quantified by OD value at 450 nm through the SpectraMax iD3 multifunctional microplate reader (Molecular Devices, USA).
Cell migration analysis
The NPC cells were seeded at 1 × 105 for 24 h and scratched using a sterile sampler tip, and the floating cells were removed by PBS and then cultured for 24 h. Cell migration images were observed under a microscope (BZ-8100, Keyence, Japan). Image-pro Plus 4.1 analysis software (Media Cybernetics Company, USA) was used to measure the cell migration and take images.
Transwell
Invasion ability of the NPC cells was measured by 24-well Transwell (CLS3398, Sigma, Germany). The cells were seeded at 1 × 105 cells/well in the upper chamber pre-coated by 200 mg/mL Matrigel (354230, BD, USA). The supernatant chamber was supplemented with serum-free medium, while the lower chamber was added to 20% serum medium. After incubation at 37 °C for 24 h, the cells across the membrane were fixed by 4% paraformaldehyde at ambient temperature for 10 min and stained by 0.1% crystal violet at ambient temperature for 20 min. Cell number was calculated from four fields of view.
Western blot
An RIPA lysate (P0013B-100 mL, Beyotime, China) was used to isolate total proteins from each cell lines, and the protein concentration was detected by the bicinchoninic acid (BCA) kit (PC0020-50T, Solarbio LIFE SCIENCES, China). 30 μg proteins from each cell was separated by 10% SDS-PAGE and transferred to a methanol-treated nitrocellulose filter membrane (PVDF, Millipore, and USA). The membrane was incubated with primary antibodies overnight at 4 °C, the specific antibodies information were as follows: MMP2 (72 kDa; 1:1000; ab215986; Abcam, UK), TIMP-2 (24 kDa; 1:1000; ab180630), E-cadherin (97 kDa; 1:50; ab1416), N-cadherin (130 kDa; 1 µg/ml; ab18203), GAPDH (36 kDa;1:5000; ab8245), p-PI3K (ab182651, 1:1000; 84 kDa), PI3K (ab191606, 1:1000; 85 kDa), p-Akt (ab38449, 1:1000; 56 kDa), and Akt (ab8805, 1:500, 55 kDa). The target bands were incubated by corresponding secondary antibody (Anti-rabbit IgG, 1:5000, #7074, CST, USA; Anti-mouse IgG, 1:5000, #7076, CST, USA) for 2 h at room temperature. Finally, ECL ultrasensitive luminous solution (SL1350-100 ml, Coolaber, China) and American Bio-Rad GelDoc XR + gel imaging system (USA) was used to detect the signals, and analyze and count gray value of the bands.
Data analysis
The data were shown as the mean ± SD, performed by SPSS 20.0 software (USA). Statistics significances between groups were determined with one-way ANOVA and student’s t test. Comparison of gene expression between adjacent tissues and cancer tissues were analyzed using paired t-test. The linear relationship between LINC01433 and miR-506-3p in NPC was analyzed by Pearson correlation analysis. P < 0.05 had statistically significant.
Results
LINC01433 was high-expressed in NPC tissues and cell lines
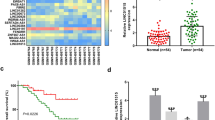
RT-qPCR was used to detect expression levels of LINC01433 in NPC and adjacent tissues. As shown in Fig. 1a, the results showed that LINC01433 expression in NPC was significantly higher than that in adjacent tissues (P < 0.001). In addition, the expression level of LINC01433 in NP69 and NPC cell lines was detected, and showed that the level of LINC01433 in NPC cell lines was significantly higher than that in NP69 (P < 0.001, Fig. 1b). Among them, the expression of LINC01433 in 5-8F cells was the highest in the four NPC cell lines, while the expression of LINC01433 in SUNE1 cells was the lowest. Therefore, 5-8F and SUNE1 cells were used in the following study.
Expression levels of long intergenic noncoding 01433 (LINC01433) in NPC clinical specimens and cell lines. a LINC01433 was up-regulated in cancer tissues compared with adjacent tissues from 45 NPC patients. LINC01433 levels were determined by RT-qPCR. GAPDH served as housekeeping-gene controls. b LINC01433 expressions of NPC cell lines. The expression level of LINC01433 was detected by RT-qPCR and found significantly increased in NPC cell lines compared with NP69. The expression levels are presented as the means ± standard deviation (RT-qPCR: quantitative real-time polymerase chain reaction; ***P < 0.001 vs. Adjacent tissue; ###P < 0.001 vs. NP69)
LINC01433 regulated proliferation, migration and invasion of NPC cells
The level of LINC01433 was significantly increased after transfection of LINC01433 overexpressed plasmid in SUNE1 cells (P < 0.001, Fig. 2a). Moreover, sh-LINC01433 was transfected into 5-8F cells, and we found that LINC01433 level were significantly reduced (P < 0.001, Fig. 2b). Overexpression of LINC01433 increased the proliferation of SUNE1 cells (P < 0.05, Fig. 2c), while down-regulation of LINC01433 inhibited the proliferation of 5-8F cells (P < 0.05, Fig. 2d). Wound healing experiments revealed that wound healing of cells transfected with LINC01433 overexpressed plasmid was faster than the SUNE1 cells transfected with NC-control, and the migration rate of 5-8F cells transfected with sh-LINC01433 were significantly lower than those transfected with sh-control (P < 0.01, Fig. 2e–g). At the same time, Transwell analysis revealed that overexpression of LINC01433 promoted cell invasion, while the invasion ability of sh-LINC01433 transfected cells was reduced (P < 0.001, Fig. 2h–j).
LINC01433 regulated proliferation, migration and invasion of NPC cells. a The level of LINC01433 transfection with control or NCc-control or LINC01433 in SUNE1 cells was analyzed by RT-qPCR. b The level of LINC01433 transfection with control or sh-control or sh-LINC01433 in 5-8F cells was analyzed by RT-qPCR. c The proliferation of SUNE1 cells transfected with control or NC-control or LINC01433 was performed by CCK-8 assay. d The proliferation ability of 5-8F cells transfected with control or sh-control or sh-LINC01433 was performed by CCK-8 assay. e–g Wound scratch assay in control or NC-control or LINC01433 transfected SUNE1 cells and in control or sh-control or sh-LINC01433 transfected 5-8F cells was shown. Scale bars = 200 μm. h–j Transwell invasion assay in control or NC-control or LINC01433 transfected SUNE1 cells and in control or sh-control or sh-LINC01433 transfected 5-8F cells was shown. Scale bars = 50 μm. Each experiment was repeated three times (NPC, nasopharyngeal carcinoma; RT-qPCR, quantitative real-time polymerase chain reaction; CCK-8, cell counting kit-8; *P < 0.05, **P < 0.01, ***P < 0.001 vs. NC-control; #P < 0.05, ##P < 0.01, ###P < 0.001 vs. sh-control)
LINC01433 was negatively correlated with the target gene miR-506-3p
To further study the regulatory mechanism of LINC01433 in NPC, the potential miRNA binding site of LINC01433was predicted by starBase, and the results showed that there are seven possible target genes for LINC01433 (including miR-506-3p, miR-4428, miR-1301-3p, miR-5047, miR-3605-5p, miR-2116-3p, miR-124-3p). Then we further examined the levels of these seven miRNAs in NPC samples, and found that miR-506-3p was most abnormally expressed in NPC tissues (P < 0.05, Fig. 3a). StarBase predicted that there was a binding site between LINC01433 and miR-506-3p (Fig. 3b), and then conducted RIP and dual luciferase reporter gene experiments to further validate the prediction that miR-506-3p directly targets LINC01433 in SUNE1 and 5-8F cell lines (Fig. 3c–f). Pearson correlation analysis showed a negative correlation between LINC01433 and miR-506-3p in NPC (r = − 0.308, P = 0.040, Fig. 3g). At the same time, the results showed that overexpression of LINC01433 reduce the miR-506-3p expression in SUNE1 cells, while silencing LINC01433 could increase the miR-506-3p expression in 5-8F cells. (P < 0.001, Fig. 3h–i).
LINC01433 was negatively correlated with the target gene miR-506-3p. a RT-qPCR was used to determine the levels of seven target miRNAs in NPC samples. ▲P < 0.05, ▲▲P < 0.01, ▲▲▲P < 0.001 vs. Adjacent tissue. b StarBase, c, d RNA immunoprecipitation and dual luciferase activity (e, f) were used to identify miR-506-3p binding sites in LINC01433 (^^^P < 0.001 vs. blank, △△△P < 0.001 vs. IgG). g Pearson’s correlation curve revealed the negative relevance between LINC01433 and miR-506-3p levels (n = 45, r = − 0.308, P = 0.040). h The miR-506-3p expression of SUNE1 cells transfected with control, NC-control or LINC01433 was performed by RT-qPCR. i The miR-506-3p expression of 5-8F cells transfected with control, sh-control or sh-LINC01433 was performed by RT-qPCR. RT-qPCR: quantitative real-time polymerase chain reaction; ***P < 0.001 vs. NC-control; ###P < 0.001 vs. sh-control
MiR-506-3p partly reversed the regulatory effects of LINC01433 NPC cells
Rescue experiments were performed to determine the regulatory relationship between LINC01433 and miR-506-3p (Fig. 4a, b). The expression of miR-506-3p in miR-506-3p mimic + LINC01433 group was evidently lower than that of mimic group (P < 0.001), and the miR-506-3p expression in miR-506-3p inhibitor + sh-LINC01433 group was evidently higher than that of inhibitor group (P < 0.05, Fig. 4a, b). MiR-506-3p mimic inhibited the proliferation of SUNE1 cells, whereas down-regulating miR-506-3p promoted the proliferation of 5-8F cells (P < 0.05, Fig. 4c, d). Wound healing experiments showed that miR-506-3p mimic inhibited SUNE1 cell migration (P < 0.001), miR-506-3p inhibitor promoted 5-8F cell migration (P < 0.05), and miR-506-3p reversed the role of LINC01433 on cell migration (P < 0.001, Fig. 4e–g). Furthermore, Transwell analysis indicated that miR-506-3p mimic inhibited cell invasion (P < 0.001), and that cell invasion of mimic + LINC01433 group was significantly higher than that of mimic group (P < 0.001). Silencing of miR-506-3p promoted cell invasion (P < 0.05, Fig. 4h–i), and the inhibitory ability of inhibitor + sh-LINC01433 group was markedly reduced compared with the sh-LINC01433 group (P < 0.001, Fig. 4h–j).
MiR-506-3p partly reversed the regulation of LINC01433 on the proliferation, migration and invasion of nasopharyngeal carcinoma cells. a, b The expression of miR-506-3p in miR-506-3p mimic + LINC01433 group was significantly lower than that of mimic group, and the expression of miR-506-3p in miR-506-3p inhibitor + sh-LINC01433 group was significantly higher than that of inhibitor group. c The proliferation ability of SUNE1 cells transfected with control, mimic control, mimic, LINC01433 + mimic or LINC01433 was performed by CCK-8 assay. d The proliferative ability of 5-8F cells transfected with control, inhibitor control, inhibitor, sh-LINC01433 + inhibitor or sh-LINC01433 was performed by CCK-8 assay. e–g Wound healing assay in control or mimic control or mimic or LINC01433 + mimic or LINC01433 transfected SUNE1 cells and in control or inhibitor control or inhibitor or sh-LINC01433 + inhibitor or sh-LINC01433 transfected 5-8F cells was shown. Scale bars = 200 μm. h–j Transwell invasion assay in control or mimic control or mimic or LINC01433 + mimic or LINC01433 transfected SUNE1 cells and in control or inhibitor control or inhibitor or sh-LINC01433 + inhibitor or sh-LINC01433 transfected 5-8F cells was shown. Scale bars = 50 μm. Each experiment was repeated three times (RT-qPCR: quantitative real-time polymerase chain reaction; CCK-8: cell counting kit-8; *P < 0.05, **P < 0.01, ***P < 0.001 vs. control; ###P < 0.001 vs. mimic control; ^^P < 0.01, ^^^P < 0.001 vs. mimic; △P < 0.05, △△△P < 0.001 vs. LINC01433 + mimic; ▲P < 0.05, ▲▲▲P < 0.001 vs. inhibitor control; ∇P < 0.05 vs. inhibitor; ‡P < 0.05, ‡‡‡P < 0.001 vs. sh-LINC01433 + inhibitor)
LINC01433 targeted miR-506-3p to regulate the expressions of EMT- and PI3K/Akt-related proteins in NPC
In the SUNE1 cell line, as shown in Fig. 5a, b, up-regulation of miR-506-3p inhibited the expressions of MMP2 and N-cadherin and promoted the expressions of TIMP-2 and E-cadherin (P < 0.001). Moreover, the expressions of MMP2 and N-cadherin were increased and the expressions of TIMP-2 and E-cadherin were reduced in mimic + LINC01433 group compared with mimic group (P < 0.05, Fig. 5a, b). In contrast, in the 5-8F cell line, down-regulation of miR-506-3p promoted MMP2 and N-cadherin expressions and inhibited TIMP-2 and E-cadherin expressions (P < 0.05, Fig. 5c, d). Moreover, the levels of MMP2 and N-cadherin were reduced and the expressions of TIMP-2 and E-cadherin were increased in cells transfected with sh-LINC01433 (P < 0.05, Fig. 5c, d). As shown in Fig. 5e–g, miR-506-3p down-regulated the level of p-PI3K and p-Akt, while LINC01433 could reverse the action of miR-506-3p mimic and promoted PI3K and Akt activation (P < 0.05). Down-regulation of miR-506-3p promoted the level of p-PI3K and p-Akt, while sh-LINC01433 could reverse the action of miR-506-3p inhibitor and inhibited PI3K and Akt activation (P < 0.01, Fig. 5h–j).
LINC01433 targeted miR-506-3p to regulate the expressions of EMT- and PI3K/Akt- related proteins in NPC. a, b The levels of MMP2, TIMP-2, E-cadherin, and N-cadherin in SUNE1 cells transfected control, mimic control, mimic, LINC01433 + mimic or LINC01433 were detected by Western blot. c, d The levels of MMP2, TIMP-2, E-cadherin, N-cadherin in 5-8F cells transfected control, inhibitor control, inhibitor, sh-LINC01433 + inhibitor or sh-LINC01433 were detected by Western blot. e–g The levels of p-PI3K, PI3K, p-Akt, and Akt in SUNE1 cells transfected control, mimic control, mimic, LINC01433 + mimic or LINC01433 were detected by Western blot. h–j The levels of p-PI3K, PI3K, p-Akt, and Akt in 5-8F cells transfected control, inhibitor control, inhibitor, sh-LINC01433 + inhibitor or sh-LINC01433 were detected by Western blot (***P < 0.001 vs. control; ###P < 0.001 vs. mimic control; ^P < 0.05, ^^^P < 0.001 vs. mimic; △P < 0.05, △△P < 0.01, △△△P < 0.001 vs. LINC01433 + mimic; ▲▲▲P < 0.001 vs. inhibitor control; ∇∇P < 0.01, ∇∇∇P < 0.001 vs. inhibitor; ‡‡‡P < 0.001 vs. sh-LINC01433 + inhibitor)
MiR-506-3p may alter biological characteristics of NPC cells via target genes
We used different databases for bioinformatics analysis, and found 462 genes that were predicted to be commonly targeted by miR-506-3p in different databases (Fig. 6a). Subsequently, these 462 target mRNAs were intersected with the NPC differentially expressed genes analyzed in TCGA, and 63 mRNAs were obtained (Fig. 6b, Supplemental Table1).
Prediction of downstream target genes of miR-506-3p. a The starBase (http://starbase.sysu.edu.cn/index.php), miRDB (http://mirdb.org/), TargetScan7 (http://www.targetscan.org/vert_72/) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) websites were used to predict the target mRNAs of miR-506-3p. b 63 mRNAs were targeted by miR-506-3p and abnormally expressed in NPC, as visualized by Venn diagram
Discussion
LncRNAs were initially defined as by-products of RNA polymerase II transcription and has no biological functions [8, 13, 22]. However, studies have increasingly shown that lncRNA is involved in tumorigenesis, development, invasion, metastasis and other biological processes [22,23,24]. Therefore, the role of LINC01433 in NPC was investigated in current study.
Recently, efforts have been made on determining the relationship between lncRNA and NPC cell proliferation, invasion and metastasis [12, 22, 23, 25,26,27,28]. For example, Zhang et al. compared some abnormal expressions of lncRNAs in primary NPC and metastatic NPC, and found that HOTAIR and H19 were up-regulated in metastatic NPC, and that the expressions of HOTAIR and H19 may be associated with metastasis of NPC [25]. LINC01433 is a tumor-associated lncRNA, and study observed that LINC01433 is involved in regulation of cancer development [15, 16, 20]. Study observed that LINC01433 promotes the biological function and chemotherapy resistance of gastric cancer cells, and its mechanism may stabilize the YAP to play a carcinogenic role by enhancing the interaction between USP9X and YAP [29]. The present study showed that the mRNA level of LINC01433 in NPC tissues was noticeably higher, similarly, the expression level in NPC cells was also noticeably increased than that in human nasopharyngeal epithelial cell lines. Moreover, up-regulation of endogenously high-expressed LINC01433 greatly enhanced biological function of SUNE1 cells. Our results may demonstrate that the role of LINC01433 in NPC cell lines may be similar to that in gastric cancer cells [29].
In recent years, miRNAs play important roles in maintaining normal cell functions, while dysregulation of expressions of miRNAs will lead to tumor progression and cancer incidence [30, 31]. Evidence indicated that miR-506 is abnormally expressed in various tumors [32,33,34,35]. The expression of miR-506 in cervical cancer cell line was observably lower than that in normal tissues [36]. Liu et al. [37] demonstrated that miR-506 regulated cell proliferation and senescence through CDK4 and CDK6. Sun et al., showed that miR-506 was down-regulated in human breast malignant and breast cancer cell lines. In addition, Zhang et al. [38] found that miR-506 was down-regulated in NPC cell lines and tissues, which is consistent with our findings. Though the expression pattern of miR-506 in pancreatic cancer, gastric cancer, ovarian cancer and other cancers has been previously reported, but its role is different [32, 39]. To our knowledge, this is the first discovery that miR-506-3p targeted LINC01433 and up-regulation of miR-506-3p attenuated NPC cell biological function, and partially reversed the cancer-promoting effects of LINC01433. Besides, miRNA can regulate cell biology characteristics by target mRNA. For instance, Liang et al. [40] revealed that miR-506 attenuated tumor growth and metastasis though down-regulating LHX2. We identified that 63 mRNAs were targeted by miR-506-3p and abnormally expressed in NPC. What has caught our attention is that among the 63 downstream targets of miR-506-3p, DEPDC1 is overexpressed in nasopharyngeal carcinoma and is associated with cancer progression [41]. Therefore, our next study will try to verify whether LINC01433 can up-regulate DEPDC1 by inhibiting the expression of miR-506-3p, and ultimately lead to the development of NPC.
EMT, MMP, and MMP tissue inhibitors (TIMP) are known to play key roles in tumor cell invasion and metastasis [42,43,44,45]. E-cadherin and N-cadherin are two protein markers indicative of EMT [46]. Scholars have found that the expression of E-cadherin is down-regulated to different extent in malignant tumors, and up-regulation of N-cadherin can promote the transformation of tumor epithelial cells into stromal cells with an invasive phenotype and is easy to metastasize. The findings suggested that E-cadherin and N-cadherin expressions and the degree of tumor stage, metastasis, aggressiveness and prognosis are closely related [47,48,49]. MMP and metalloproteinase inhibitor (TMP) function together to regulate the homeostasis of ECM and integrity of the basement membrane [45]. Furthermore, TIMP-2 could inhibit soluble MMP and membrane type MMP [50]. MMP2 and TIMP-2 are key factors for tumor metastasis and invasion [43, 45], specifically, MMP2 promotes tumor metastasis, while TIMP-2 inhibits tumor metastasis [51]. Four tumor-associated markers, namely, MMP2, TIMP-2, E-cadherin, and N-cadherin, were studied in the present study, and the data showed that overexpression of miR-506-3p inhibited tumor cells migration and invasion, partially reversed the promotion of tumor growth by LINC01433, and the results are consistent with previous studies [15]. In addition, LINC01433 also showed high expression in breast cancer cells, and more interestingly, down-regulation of LINC01433 inhibited the proliferation, migration and EMT of breast cancer cells [16]. This experiment proved that LINC01433 is a tumor-promoting gene. However, scholars have discovered that the mechanism of LINC01433 in breast cancer cells is activated through the miR-2116-3p/MYC axis [16], which also shows that LINC01433 may play a role through multiple regulatory pathways. The PI3K/Akt pathway plays a vital role in regulating various cell functions, and has become a major focus of academic attention [52]. Thus, we also tested the PI3K/Akt pathway protein and found that LINC01433 activated the PI3K / Akt pathway by down-regulating miR-506-3p. Of course, whether LINC01433/miR-506-3p acts on PI3K/Akt pathway through target gene DEPDC1 needs further study.
Conclusion
In conclusion, LINC01433 regulates the biological characteristics of NPC cells through miR-506-3p, indicating that LINC01433 has a pro-carcinogenic effect on NPC progression. These findings provide better understandings on the molecular mechanism of NPC.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
References
Paul P, Deka H, Malakar AK, Halder B, Chakraborty S (2018) Nasopharyngeal carcinoma: understanding its molecular biology at a fine scale. Eur J Cancer Prevent 27(1):33–41. https://doi.org/10.1097/cej.0000000000000314
Chua MLK, Wee JTS, Hui EP, Chan ATC (2016) Nasopharyngeal carcinoma. Lancet (Lond, Engl) 387(10022):1012–1024. https://doi.org/10.1016/s0140-6736(15)00055-0
Renganathan A, Felley-Bosco E (2017) Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol 1008:199–222. https://doi.org/10.1007/978-981-10-5203-3_7
Cai Z, Xu K, Li Y, Lv Y, Bao J, Qiao L (2017) Long noncoding RNA in liver cancer stem cells. Discov Med 24(131):87–93
Li J, Li Z, Zheng W, Li X, Wang Z, Cui Y, Jiang X (2017) LncRNA-ATB: An indispensable cancer-related long noncoding RNA. Cell Prolif 50:6. https://doi.org/10.1111/cpr.12381
Wang J, Su Z, Lu S, Fu W, Liu Z, Jiang X, Tai S (2018) LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta 485:229–233. https://doi.org/10.1016/j.cca.2018.07.004
Huang HW, Xie H, Ma X, Zhao F, Gao Y (2017) Upregulation of LncRNA PANDAR predicts poor prognosis and promotes cell proliferation in cervical cancer. Eur Rev Med Pharmacol Sci 21(20):4529–4535
Geng YJ, Xie SL, Li Q, Ma J, Wang GY (2011) Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res 39(6):2119–2128. https://doi.org/10.1177/147323001103900608
Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, Wang Y, Ming H (2017) Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun 490(2):406–414. https://doi.org/10.1016/j.bbrc.2017.06.055
He R, Hu Z, Wang Q, Luo W, Li J, Duan L, Zhu YS, Luo DX (2017) The role of long non-coding RNAs in nasopharyngeal carcinoma: as systemic review. Oncotarget 8(9):16075–16083. https://doi.org/10.18632/oncotarget.14211
Sun Q, Liu H, Li L, Zhang S, Liu K, Liu Y, Yang C (2015) Long noncoding RNA-LET, which is repressed by EZH2, inhibits cell proliferation and induces apoptosis of nasopharyngeal carcinoma cell. Med Oncol (Northwood, Lond, Engl) 32(9):226. https://doi.org/10.1007/s12032-015-0673-0
Yang QQ, Deng YF (2015) Genome-wide analysis of long non-coding RNA in primary nasopharyngeal carcinoma by microarray. Histopathology 66(7):1022–1030. https://doi.org/10.1111/his.12616
Nie Y, Liu X, Qu S, Song E, Zou H, Gong C (2013) Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci 104(4):458–464. https://doi.org/10.1111/cas.12092
Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X (2001) The sequence of the human genome. Science 291(5507):1304–1351. https://doi.org/10.1126/science.1058040
Qian B, Wang X, Mao C, Jiang Y, Shi Y, Chen L, Liu S, Wang B, Pan S, Tao Y, Shi H (2018) Long non-coding RNA linc01433 promotes migration and invasion in non-small cell lung cancer. Thorac Cancer 9(5):589–597. https://doi.org/10.1111/1759-7714.12623
Wu M, Wu W, Ding J, Yang J (2019) LINC01433/miR-2116-3p/MYC feedback loop promotes cell proliferation, migration, and the epithelial-mesenchymal transition in breast cancer. Cancer Biotherapy Radiopharmaceut 34(6):388–397. https://doi.org/10.1089/cbr.2019.2772
Zhang A, Xu M, Mo YY (2014) Role of the lncRNA-p53 regulatory network in cancer. J Mol Cell Biol 6(3):181–191. https://doi.org/10.1093/jmcb/mju013
Ballantyne MD, McDonald RA, Baker AH (2016) lncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol Ther 99(5):494–501. https://doi.org/10.1002/cpt.355
Kong YG, Cui M, Chen SM, Xu Y, Xu Y, Tao ZZ (2018) LncRNA-LINC00460 facilitates nasopharyngeal carcinoma tumorigenesis through sponging miR-149-5p to up-regulate IL6. Gene 639:77–84. https://doi.org/10.1016/j.gene.2017.10.006
Huang H, Bu YZ, Zhang XY, Liu J, Zhu LY, Fang Y (2019) LINC01433 promotes hepatocellular carcinoma progression via modulating the miR-1301/STAT3 axis. J Cell Physiol 234(5):6116–6124. https://doi.org/10.1002/jcp.27366
Rio DC, Ares M Jr, Hannon GJ (2010) Nilsen TW (2010) Purification of RNA using TRIzol (TRI reagent). Cold Spring Harbor Protocols 6:5439. https://doi.org/10.1101/pdb.prot5439
Xie L, Hu Z, Wang X, Li Z (2013) Expression of long noncoding RNA MALAT1 gene in human nasopharyngeal carcinoma cell lines and its biological significance. J Southern Med Univ 33(5):692–697
Bo H, Gong Z, Zhang W, Li X, Zeng Y, Liao Q, Chen P, Shi L, Lian Y, Jing Y, Tang K, Li Z, Zhou Y, Zhou M, Xiang B, Li X, Yang J, Xiong W, Li G, Zeng Z (2015) Upregulated long non-coding RNA AFAP1-AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget 6(24):20404–20418. https://doi.org/10.18632/oncotarget.4057
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, Waye MM (2015) The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 6(26):22513–22525. https://doi.org/10.18632/oncotarget.4154
Zhang W, Wang L, Zheng F, Zou R, Xie C, Guo Q, Hu Q, Chen J, Yang X, Yao H, Song E, Xiang Y (2015) Long noncoding RNA expression signatures of metastatic nasopharyngeal carcinoma and their prognostic value. Biomed Res Int 2015:618924. https://doi.org/10.1155/2015/618924
Zhang S, Zhong G, He W, Yu H, Huang J, Lin T (2016) lncRNA up-regulated in nonmuscle invasive bladder cancer facilitates tumor growth and acts as a negative prognostic factor of recurrence. J Urol 196(4):1270–1278. https://doi.org/10.1016/j.juro.2016.05.107
Nie GH, Li Z, Duan HF, Luo L, Hu HY, Yang WQ, Nie LP, Zhu RF, Chen XF, Zhang W (2017) lncRNA C22orf32-1 contributes to the tumorigenesis of nasopharyngeal carcinoma. Oncol Lett 13(6):4487–4492. https://doi.org/10.3892/ol.2017.6021
Ma DD, Yuan LL, Lin LQ (2017) LncRNA HOTAIR contributes to the tumorigenesis of nasopharyngeal carcinoma via up-regulating FASN. Eur Rev Med Pharmacol Sci 21(22):5143–5152. https://doi.org/10.26355/eurrev_201711_13831
Zhang C, Qian H, Liu K, Zhao W, Wang L (2019) A Feedback loop regulation of LINC01433 and YAP promotes malignant behavior in gastric cancer cells. OncoTargets Therapy 12:7949–7962. https://doi.org/10.2147/OTT.S222903
Qadir MI, Faheem A (2017) miRNA: a diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr 27(3):197–204. https://doi.org/10.1615/CritRevEukaryotGeneExpr.2017019494
Puik JR, Meijer LL, Le Large TY, Prado MM, Frampton AE, Kazemier G, Giovannetti E (2017) miRNA profiling for diagnosis, prognosis and stratification of cancer treatment in cholangiocarcinoma. Pharmacogenomics 18(14):1343–1358. https://doi.org/10.2217/pgs-2017-0010
Yu F, Lv M, Li D, Cai H, Ma L, Luo Q, Yuan X, Lv Z (2015) MiR-506 over-expression inhibits proliferation and metastasis of breast cancer cells. Med Sci Monitor 21:1687–1692. https://doi.org/10.12659/msm.893522
Yao J, Qin L, Miao S, Wang X, Wu X (2016) Overexpression of miR-506 suppresses proliferation and promotes apoptosis of osteosarcoma cells by targeting astrocyte elevated gene-1. Oncol Lett 12(3):1840–1848. https://doi.org/10.3892/ol.2016.4827
Luo Y, Sun R, Zhang J, Sun T, Liu X, Yang B (2015) miR-506 inhibits the proliferation and invasion by targeting IGF2BP1 in glioblastoma. Am J Transl Res 7(10):2007–2014
Erice O, Munoz-Garrido P, Vaquero J, Perugorria MJ, Fernandez-Barrena MG, Saez E, Santos-Laso A, Arbelaiz A, Jimenez-Aguero R, Fernandez-Irigoyen J, Santamaria E, Torrano V, Carracedo A, Ananthanarayanan M, Marzioni M, Prieto J, Beuers U, Oude Elferink RP, LaRusso NF, Bujanda L, Marin JJG, Banales JM (2018) MicroRNA-506 promotes primary biliary cholangitis-like features in cholangiocytes and immune activation. Hepatol (Baltim) 67(4):1420–1440. https://doi.org/10.1002/hep.29533
Gong M, Chen C, Zhao H, Sun M, Song M (2018) miR-506 suppresses cervical cancer cell proliferation both in vitro and in vivo. Neoplasma 65(3):331–338. https://doi.org/10.4149/neo_2018_170112N25
Liu G, Sun Y, Ji P, Li X, Cogdell D, Yang D, Parker Kerrigan BC, Shmulevich I, Chen K, Sood AK, Xue F, Zhang W (2014) MiR-506 suppresses proliferation and induces senescence by directly targeting the CDK4/6-FOXM1 axis in ovarian cancer. J Pathol 233(3):308–318. https://doi.org/10.1002/path.4348
Zhang Z, Ma J, Luan G, Kang L, Su Y, He Y, Luan F (2015) MiR-506 suppresses tumor proliferation and invasion by targeting FOXQ1 in nasopharyngeal carcinoma. PLoS ONE 10(4):e0122851. https://doi.org/10.1371/journal.pone.0122851
Li J, Ju J, Ni B, Wang H (2016) The emerging role of miR-506 in cancer. Oncotarget 7(38):62778–62788. https://doi.org/10.18632/oncotarget.11294
Liang TS, Zheng YJ, Wang J, Zhao JY, Yang DK, Liu ZS (2019) MicroRNA-506 inhibits tumor growth and metastasis in nasopharyngeal carcinoma through the inactivation of the Wnt/beta-catenin signaling pathway by down-regulating LHX2. J Exp Clin Cancer Res 38(1):97. https://doi.org/10.1186/s13046-019-1023-4
Feng X, Zhang C, Zhu L, Zhang L, Li H, He L, Mi Y, Wang Y, Zhu J, Bu Y (2017) DEPDC1 is required for cell cycle progression and motility in nasopharyngeal carcinoma. Oncotarget 8(38):63605–63619. https://doi.org/10.18632/oncotarget.18868
Lawicki S, Zajkowska M, Glazewska EK, Bedkowska GE, Szmitkowski M (2017) Plasma levels and diagnostic utility of VEGF, MMP-2 and TIMP-2 in the diagnostics of breast cancer patients. Biomarkers 22(2):157–164. https://doi.org/10.1080/1354750x.2016.1252955
Shrestha B, Bajracharya D, Byatnal AA, Kamath A, Radhakrishnan R (2017) May High MMP-2 and TIMP-2 expressions increase or decrease the aggressivity of oral cancer? Pathol Oncol Res 23(1):197–206. https://doi.org/10.1007/s12253-016-0149-3
Yi GZ, Feng WY, Zhou Q, Liu YW, Qi ST (2017) The impact of MMP-2 and its specific inhibitor TIMP-2 expression on the WHO grade and prognosis of gliomas in Chinese population: a meta-analysis. Mol Neurobiol 54(1):22–30. https://doi.org/10.1007/s12035-015-9539-x
Wang T, Hou J, Jian S, Luo Q, Wei J, Li Z, Wang X, Bai P, Duan B, Xing J, Cai J (2018) miR-29b negatively regulates MMP2 to impact gastric cancer development by suppress gastric cancer cell migration and tumor growth. J Cancer 9(20):3776–3786. https://doi.org/10.7150/jca.26263
Lei H, Gao Y, Xu X (2017) LncRNA TUG1 influences papillary thyroid cancer cell proliferation, migration and EMT formation through targeting miR-145. Acta Biochim Biophys Sin 49(7):588–597. https://doi.org/10.1093/abbs/gmx047
Derycke LD, Bracke ME (2004) N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int J Develop Biol 48(5–6):463–476. https://doi.org/10.1387/ijdb.041793ld
Blaschuk OW (2015) N-cadherin antagonists as oncology therapeutics. Philos Trans R Soc Lond B Biol Sci 370(1661):20140039. https://doi.org/10.1098/rstb.2014.0039
Petrova YI, Schecterson L, Gumbiner BM (2016) Roles for E-cadherin cell surface regulation in cancer. Mol Biol Cell 27(21):3233–3244. https://doi.org/10.1091/mbc.E16-01-0058
Zhu Y, Yan L, Zhu W, Song X, Yang G, Wang S (2019) MMP2/3 promote the growth and migration of laryngeal squamous cell carcinoma via PI3K/Akt-NF-kappaB-mediated epithelial-mesenchymal transformation. J Cell Physiol. https://doi.org/10.1002/jcp.28242
Kolude B, Adisa AO, Lawal AO, Adeyemi BF, Akinyamoju AO (2015) Stoichiometric expression of MMP-2/TIMP-2 in benign and malignant tumours of the salivary gland. Tumour Biol 36(4):2351–2357. https://doi.org/10.1007/s13277-014-2842-8
Xia P, Xu XY (2015) PI3K/Akt/mTOR signaling pathway in cancer stem cells: from basic research to clinical application. Am J Cancer Res 5(5):1602–1609
Funding
This work was supported by the Scientific Research Project of Zhejiang Educational Committee [Grant no. Y201636243].
Author information
Authors and Affiliations
Contributions
Substantial contributions to conception and design: MZ. Data acquisition, data analysis and interpretation: ZD, SH, MX. Drafting the article or critically revising it for important intellectual content: MZ. Final approval of the version to be published: All authors. Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of the work are appropriately investigated and resolved: All authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, M., Dong, Z., Hu, S. et al. LINC01433 targets miR-506-3p to promote the biological progress of nasopharyngeal carcinoma cells. Eur Arch Otorhinolaryngol 278, 3363–3374 (2021). https://doi.org/10.1007/s00405-021-06607-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-021-06607-w