Abstract
Caveolin-1 (Cav-1) and Actin-Related Protein 2/3 Complex, Subunit 1B (ARPC1B) have been implicated in various human cancers, yet its role in tumorigenesis remains controversial. Therefore, this study aims to determine the protein expression of these two genes in oral squamous cell carcinomas (OSCCs) and to evaluate the clinical and prognostic impact of these genes in OSCC. Protein expressions of these two genes were determined by immunohistochemistry technique. The association between Cav-1 and ARPC1B with clinico-pathological parameters was evaluated by Chi-square test (or Fisher exact test where appropriate). Correlation between the protein expressions of these 2 genes with survival was analyzed using Kaplan–Meier and Cox regression models. Cav-1 and ARPC1B were found to be significantly over-expressed in OSCC compared to normal oral mucosa (p = 0.002 and p = 0.033, respectively). Low level of ARPC1B protein expression showed a significant correlation with lymph node metastasis (LNM) (p = 0.010) and advanced tumor staging (p = 0.003). Kaplan–Meier survival analyses demonstrated that patients with over-expression of Cav-1 protein were associated with poor prognosis (p = 0.030). Adjusted multivariate Cox regression model revealed that over-expression of Cav-1 remained as an independent significant prognostic factor for OSCC (HRR = 2.700, 95 % CI 1.013–7.198, p = 0.047). This study demonstrated that low-expression of ARPC1B is significantly associated with LNM and advanced tumor staging whereas high expression of Cav-1 can be a prognostic indicator for poor prognosis in OSCC patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Globally, oral cancer is ranked as one of the tenth most common cancers in the world [1, 2]. According to Global Burden of Cancer (GLOBOCAN), it was estimated that 263,900 oral cancer cases (including lip cancer) occur annually, whereby 128,000 oral cancers result in death [1]. Histologically, more than 90 % of cases were reported as epithelial neoplasms with majority of them being oral squamous cell carcinomas (OSCCs) [3]. Despite the fact that OSCC can be controlled by surgical excision and radiotherapy, the 5 year survival rate still remains dismal [4] due to delay in diagnosis and lack of specific molecular markers that can predict tumor progression and prognosis [5]. Therefore, the need for identification of new molecular markers to predict the clinical behavior and prognosis of OSCC cannot be overemphasized. Moreover, an extended knowledge of these predictive markers would facilitate better characterization of these tumors through an understanding of the various molecular pathways of carcinogenesis. This in turn will help to design individual specific molecular targeted therapies that are now considered to be a promising strategy [5, 6].
A recent review paper by Hunt et al. [7] suggested that molecular markers like EGFR, RAS, CCND1, BRAF, PIK3CA, p53, CDKN2A, and NOTCH play fundamental roles in the pathogenesis of HNSCC [7]. These molecular markers enable to classify the tumor subtype and aid as prognostic markers for better patient management [6].
To date, clinical-pathological parameters such as tumor size, lymph node metastasis, and tumor stage have been considered to be the most reliable determinant for prognosis and optimal patient management [5]. Emerging high throughput technologies such as microarray has identified new molecular markers that potentially could be employed as predictive or prognostic as well as therapeutic targets for the treatment of oral cancer [5]. Based on our previous microarray study, we identified two candidate genes that were found to be differentially expressed in OSCC samples, namely Caveolin 1 (Cav-1) and Actin-Related Protein 2/3 Complex, Subunit 1B (ARPC1B) [8]. It has been indicated in the literature that these two genes are highly involved in tumorigenesis [9–11]. As there is a lack of understanding of their role in oral carcinogenesis, therefore, they were selected for the current study.
Caveolin-1 (Cav-1), a 21–24 kDa protein, is the main component of caveolae of plasma membranes and acts as a regulator in vesicle trafficking, intracellular cholesterol homeostasis, and lipoprotein metabolism that give impact to the actin cytoskeleton and cell polarity remodeling [12]. The implication of Cav-1 expression in various cancers, including oral cancer is still controversial due to the observations of both low and high expression of this gene. Previous studies have demonstrated low expression of Cav-1 in ovarian, breast, and lung cancers, whereas high expression of this gene in bladder cancer, esophageal squamous cell carcinoma (ESCC), and nasopharyngeal carcinoma (NPC) was associated with metastasis and poor prognosis [13]. To date, there are only two studies that have been carried out to determine the role of Cav-1 in oral carcinogenesis. Cav-1 has been reported to be over-expressed in OSCC and was found to be associated with lymph node metastasis which is a major determinant of prognosis for OSCC [14]. A recent study by Nohata et al. [15] concluded that Cav-1 acts as an oncogene involved in cell migration and invasion thus implicating its role in oncogenic cell transformation and metastasis [9, 13]. However, in contrast, Zhang et al. (2008) [16] showed that over-expression of Cav-1 inhibited cancer cell proliferation and invasion, and increased apoptosis suggesting that the role of this gene in oral carcinogenesis is still not fully elucidated.
Actin-related protein 2/3 complex, Subunit 1B (ARPC1B) is a component of the Arp2/3 complex which is involved in the regulation of actin polymerization that stabilizes the actin filaments and forms microspikes in lamellipodia to promote cell movement [17]. A recent study has demonstrated that over-expression of ARPC1B leads to increased tumorigenicity by centrosome amplification in breast cancer [18]. Besides breast cancer, ARPC1B was also shown to be over-expressed in radiation resistant intraocular choroidal melanoma cells [10]. However, Kaneda et al. [11] found that reduced expression of this gene was associated with dysplastic morphology. To the best of our knowledge, there has been no study reporting on the relationship between ARPC1B expression and OSCC.
Since the role of these 2 genes in oral carcinogenesis, whether as an oncogene or tumor suppressor gene is still undetermined in the reported literature, we hoped to assess the utility of these genes as potential prognostic indicators for tumor progression. Hence, this current study aimed to determine the expression levels of Cav-1 and ARPC1B in OSCCs, and to determine their clinical and prognostic significance among OSCC patients.
Methodology
Specimen and data collection
In this study, 77 archived formalin-fixed, paraffin-embedded (FFPE) OSCC specimens were obtained from the Oral Pathology Diagnostic and Research Laboratory at the University of Malaya for the assessment of Cav-1 and ARPC1B protein expression using immunohistochemistry technique. The OSCC tissue specimens were derived from the tongue (excluding the base of the tongue), buccal mucosa, gum, palate, floor of mouth, and lip (C00-06). Normal oral mucosal tissues included alveolar mucosal tissues obtained from minor surgical procedure of impacted wisdom teeth. Socio-demographic, clinico-pathological, and survival data of all OSCC samples were obtained from the Malaysian Oral Cancer Database and Tissue Bank System (MOCDTBS) coordinated by the Oral Cancer Research and Coordinating Centre, University of Malaya (OCRCC-UM) [19]. The American Joint Committee on cancer staging criteria was used for tumor staging [20]. This study was approved by Medical Ethics Committee, Faculty of Dentistry, University of Malaya [MEC no: DFOP1108/0083(L)].
Tissue microarray construction
Tissue microarrays (TMAs) were constructed from the 77 OSCC FFPE tissues according to procedures described previously [21]. Briefly, 3 cores in 1-mm sizes from the selected areas of donor blocks were transferred to the recipient paraffin blocks and were incubated overnight at 37 °C. 4-μm-thick sections were then sectioned on poly-l-lysine slides for the immunohistochemistry study.
Immunohistochemistry and scoring system
Immunohistochemistry (IHC) was performed on 4-µm-thick FFPE TMA sections using the Envision technique, DAKO REAL EnVision Detection System and Peroxidase/DAB+ (Dako Corporation, Carpinteria, CA, USA) according to the manufacturer’s protocol. FFPE sections were de-paraffinized in Xylene and gradually rehydrated in descending grades of ethanol. Antigen retrieval was carried out using an electric pressure cooker (110 °C, 20 min) in 10 mM citrate buffer (pH 6.0). The sections were immersed in blocking solution (Dako Corporation, Carpinteria, CA, USA) for 10 min at room temperature followed by washing with Phosphate-buffered saline (pH 7.4) and 0.1 % Tween 20 (PBST) to block the endogenous peroxidase activity. The sections were then incubated with anti-Arp2/3 Complex 41 Kd Subunit (ARPC1B) (N-Term), antibody (10645-1-AP, 1:1000 dilution, Protein Tech Group, Chicago, IL), and anti-Caveolin 1 (Cav-1) antibody (Abcam, Cambridge, MA, ab18199, 1:100) for 1 h at room temperature, followed by incubation with peroxidase labeled secondary antibody from the Envision kit (Dako Corporation, Carpinteria, CA, USA) for 1 h for immunoreactivity performances. Sections were then stained with 3′3 diaminobenzidine substrate chromogen (Dako Corporation, Carpinteria, CA, USA) for 5 min for colorization, followed by counterstaining with Mayer’s hematoxylin, dehydration, and mounted. For negative procedural control, the Cav-1 and ARPC1B antibodies were replaced by phosphate-buffered saline (PBS) whereas for positive control, esophageal squamous cell carcinoma and renal carcinoma tissues were used for Cav-1 and ARPC1B protein expression, respectively.
Digitalized immuno-stained TMA spots were analyzed and scored based on a semi-quantitative scoring system using the TMA software module 1.15.2 (3DHISTECH, Budapest, Hungary) as described previously [21]. Intensity scores were quantified using the following criteria: negative = 0; weak = 1; moderate = 2; and strong = 3. The proportion of immunopositive cells was quantified as follows: 0 = negative; 1 = <10 %; 2 = 10–50 %; 3 = 51–80 %; and 4 = ≥80 % of positive cells. The final immuno-reactive score was determined by multiplying the intensity and the proportion scores of the stained cells to obtain an immuno-reactive score ranging from 0 to 12. All the scoring was done by two oral pathologists (TG and AR). Cores with discrepant scores were discussed by both pathologists to achieve a consensus to derive the final score. The mean of consolidated immuno-reactive scores for each case was recorded. A cut-off point of ≥5 and ≥6 was used as high level of ARPC1B and Cav-1 expression, respectively, while the counterpart <5 and <6 were used as low level of expression for ARPC1B and Cav-1 which was derived from the 25th percentile of the respective mean immunoscores of Cav-1 and ARPC1B. The immunostaining of Cav-1 was considered as positive when there was strong and diffuse membrane and granular cytoplasmic staining in the tumor cells, while ARPC1B immunostaining was considered as positive when there was strong and diffuse cytoplasmic staining in tumor cells as demonstrated in positive controls. Staining was regarded as negative in cases of absent, focal, and weak staining within the tumor cells.
Statistical analysis
The association between the protein expression of ARPC1B and Cav-1 and the selected clinico-pathological parameters was analyzed using Chi-square test (or Fisher exact test where appropriate). Survival curves were plotted and compared by the log rank tests using the Kaplan–Meier analysis. In addition, Cox regression analysis was conducted to evaluate the utility of protein expression of these genes as an independent prognostic factor. All statistical analyses were performed using the SPSS statistical package (SPSS version 12.0, Chicago, IL, USA), with p values <0.05 considered as statistically significant.
Results
Cav-1 and ARPC1B protein expression and its correlation with clinico-pathological parameters
For IHC analysis of Cav-1 protein, the epithelial cells of normal oral mucosal tissues showed a strong staining at the basement membrane but weak to negative cytoplasmic staining toward the spinous and keratinized layer (Fig. 1). The endothelial cells and fat cells also stained positive and were regarded as internal controls (Fig. 1). More than 80 % of OSCCs exhibited a strong membrane and granular cytoplasmic staining in the epithelial tumor cells (Fig. 1). The high expression of Cav-1 protein was statistically different between OSCC and normal oral mucosal tissues (p = 0.033), but it did not show significant association with any of the clinico-pathologic parameters studied.
In IHC analysis of ARPC1B protein, the epithelial cells of normal oral mucosal tissues showed a strong cytoplasmic staining within the spinous cell layer of the epithelium and weak to negative staining of the basal and parabasal layers of the epithelium. Muscles bundles stained positive and were regarded as internal controls. More than 75 % of OSCCs displayed a strong cytoplasmic staining of epithelial tumor cells. The low expression of ARPC1B protein was statistically different between OSCC and normal oral mucosal tissues (p = 0.002). Low expression of ARPC1B significantly correlated with lymph node metastasis (p = 0.010) and advanced tumor stages (0.003) (Table 1).
Protein expression of ARPC1B and Cav-1 in correlation with survival status
The follow-up time for patients included in our study ranged from 3 to 93 months (mean 31.6 months, median 21.0 months)
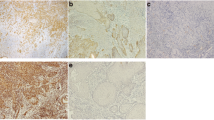
Immunohistochemistry of ARPC1B. Hyperplastic oral mucosa a H&E stain (magnification ×400 and ×1600); b anti-ARPC1B antibody immunostaining showed positive cytoplasmic staining within the spinous and superficial cell layers of the epithelium and weak to negative staining of the para basal and basal layers of the epithelium in hyperplastic oral mucosa (magnification ×400 and ×1600). OSCC c and e H&E stained (magnification ×800 and ×1600); d anti-ARPC1B antibody immunostaining showed negative expression and f strong expression in the cytoplasm of the epithelial tumor cells (magnification ×800 and ×1600). g anti-ARPC1B antibody immunostaining showed strong expression in the cytoplasm of renal cell carcinoma cells that acted as positive tissue control (magnification ×800 and ×1600). h–n Immunohistochemistry of Cav-1. Hyperplastic oral mucosa (H) H&E stain (magnification 400x and 1600x); i anti-Cav-1 antibody immunostaining showed strong positive staining of the basal layer but weak to negative staining of the spinous and superficial layers of hyperplastic oral mucosa (magnification ×400 and ×1600). OSCC j and l H&E stained (magnification ×800 and ×1600); k anti-Cav-1 antibody immunostaining showed low expression and (M) strong membrane and granular cytoplasmic staining in the epithelial tumor cells (magnification ×800 and ×1600). n anti-Cav-1 antibody immunostaining showed strong membrane and granular cytoplasmic staining in the esophagus squamous cell carcinoma that acted as positive tissue control (magnification ×800 and ×1600)
The 5 year survival rates for low and high protein expression of Cav-1 were 46.43 and 27.51 %, respectively. Results of the overall survival rate analysis demonstrated a significant association between high Cav-1 protein expression and poor prognosis (p = 0.030) (Fig. 2). After adjustment for selected socio-demographic (age, gender and risk habits) and clinico-pathological parameters (tumor size, lymph node metastasis, tumor staging, and pattern of invasion), high expression of Cav-1 remained a significant prognostic factor for overall survival of OSCC (HRR = 2.700, 95 % CI 1.013–7.198, p = 0.047, Table 2).
The 5 year survival rate for low and high expression of ARPC1B protein was 18.05 and 34.01 %, respectively. Results from Kaplan–Meier analysis showed no significant association between ARPC1B protein expression and prognosis (p = 0.651) (Fig. 2). After adjustment for selected socio-demographic (age, gender and risk habits) and clinico-pathological parameters (tumor size, lymph node metastasis, tumor staging and pattern of invasion), the multivariate cox regression analysis showed that low expression of ARPC1B was not a significant prognostic factor for the overall survival of OSCC (HRR = 1.319, 95 % CI 0.577–3.014, p = 0.511, Table 2).
Discussion
Oral cancer is distinguished by its uncontrollable proliferative characteristics as well as migration and invasive capabilities. Dissemination of tumor cells into cervical lymph nodes and distant sites is a major determinant of prognosis. [22] Thus, active genes involved in tumor metastasis can be considered as potential prognostic and therapeutic targets for OSCC.
There has always been a great interest in predicting tumor progression using different molecular markers in OSCC. Galactin-7 has been suggested to be one such potential clinical marker [23]. Similarly, over-expression of stefin A gene could be utilized to identify OSCC patients at increased risk for disease relapse [24]. A recent study by Choi et al. [25] revealed that over-expression of HIF-1α or HSP70 which are hypoxia signaling markers was associated with lymph node metastasis and poor prognosis in head and neck SCC.
In the current study, the difference in expression of ARPC1B and Cav-1 between OSCC and normal oral epithelium indicated the involvement of these genes in tumorigenesis. Furthermore, we demonstrated that low expression of ARPC1B correlated with lymph node metastasis and advanced tumor stages in OSCC patients, suggesting that this gene may have clinical relevance in the aggressive progression of OSCC. In addition, high expression of Cav-1 was found to be associated with poor survival, suggesting that this gene may have a prognostic influence for OSCC. The failure of detection of lymph node metastasis significantly affects the survival outcomes of OSCC patients. It has been estimated that approximately one third of the patients clinically diagnosed as having negative cervical lymph nodes (N0) may actually have positive cervical LNs [26]. Therefore, the current findings suggest that the evaluation of these genes in OSCC biopsies could contribute to a higher accuracy to determine specific resection procedures in OSCC patients in order to improve patient prognosis.
Researchers have tried to establish the clinical value of Cav-1 in OSCC and head and neck SCC; however, none of the studies have reported its prognostic significance [14, 16, 27, 28]. In our study, despite lack of significant association between expression of Cav-1 with clinico-pathologic parameters (tumor size, lymph node metastasis, tumor staging, and pattern of invasion), over-expression of Cav-1 protein was found to be associated with poor prognosis. This observation reflects the role of Cav-1 as a tumor regulator involved in promoting tumor cell proliferation, cell migration and metastasis, resistance to apoptotic signals, chemotherapeutic drug resistance, and angiogenesis [29] However, in contrast to our study, Xue et al. [27] in their study on oral tongue SCC found a significant correlation between Cav-1 with clinical stage and histologic grade but did not observe any association with poor survival in OSCC using Quantum dots immunofluorescence histochemistry (QDs-IHC) method. A difference in findings between our study with Xue et al. [27] may be explained due to the following reasons: (a) different methods for assessment of protein expression (immunohistochemistry vs QD-IHCs method); (b) samples for this study were derived from different subsites such as tongue, buccal mucosa, floor of the mouth, and gum while Xue et al. [27] focused only on tongue OSCCs; (c) different scoring criteria for immuno-evaluation. Apart from that, the failure of obtaining significant association of Cav-1 with clinical-pathological parameters might be due to the controversial roles of Cav-1 in oral carcinogenesis. To date, several studies [14, 27] have demonstrated that over-expression of Cav-1 gene is important in oral carcinogenesis. This is in contrast to studies by Han et al. [28] and Zhang et al. [16] who demonstrated that Cav-1 plays inhibitory roles in oral carcinogenesis. Therefore, whether Cav-1 acts as an oncogene or tumor suppressor gene in oral carcinogenesis warrants further insight.
To date, only few studies have investigated ARPC1B expression and its clinical applications in various cancers, such as in breast carcinomas [18] and intraocular choroidal melanomas [10]. ARPC1B has been shown to play an important role in regulating actin polymerization and allowing the dynamic assembly and disassembly of actin filament for cell migration [17]. In the current study, under-expression of ARPC1B protein showed significant association with advanced disease stage and lymph node metastasis. This could be explained by the findings from a recent study which demonstrated that low expression of ARPC1B gene would promote the activities of Arp2/3-nucleating core in focal adhesions sites involved in cell migration and adhesion [30]. Down regulation of the Arp2/3 complex may also be attributed to methylation of its subunit p41-Arc, leading to loss of expression and development of dysplastic morphology. In addition, Zucchini et al. [31] suggested that the down regulation of ARPC1B in osteosarcoma might be one of the causal effects that leads to inhibition of metastasis by decreasing dynamic actin disassembly that is crucial for cancer cell migration. These observations reflect the migration activity of ARPC1B in regulation of the focal adhesions and actin filaments which promotes tumor cell metastasis [30]. Although loss of ARPC1B has been reported in various cancers, the mechanism of this gene in OSCC remains unclear, and further downstream analysis should be performed to clarify its role in oral carcinogenesis.
There were some limitations in the current study. Firstly, the tumor sample size was relatively small. This might have attributed to our failure to detect association between ARPC1B expressions with poor survival. Secondly, the precise role of Cav-1 either as a tumor suppressor or oncogene in OSCC remains elusive, and this might have contributed to the failure of obtaining an association of Cav-1 with various clinical parameters. Further validations using larger number of tumor samples should be performed.
In conclusion, this is the first study to identify that over-expression of Cav-1 is an independent prognostic marker for OSCC. Additionally, increased expression of ARPC1B protein in OSCC and its correlation with advanced stage and lymph node metastasis provide further evidence to its role in genesis and progression of OSCC.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2):69–90. doi:10.3322/caac.20107
Warnakulasuriya S (2009) Global epidemiology of oral and oropharyngeal cancer. Oral Oncol 45(4–5):309–316. doi:10.1016/j.oraloncology.2008.06.002
Neville BW, Day TA (2002) Oral cancer and precancerous lesions. CA Cancer J Clin 52(4):195–215
Silverman S Jr (2001) Demographics and occurrence of oral and pharyngeal cancers. The outcomes, the trends, the challenge. J Am Dent Assoc 132(Suppl):7S–11S
Mahfouz ME, Rodrigo JP, Takes RP, Elsheikh MN, Rinaldo A, Brakenhoff RH, Ferlito A (2010) Current potential and limitations of molecular diagnostic methods in head and neck cancer. Eur Arch Otorhinolaryngol 267(6):851–860. doi:10.1007/s00405-009-1177-3
Grimminger CM, Danenberg PV (2011) Update of prognostic and predictive biomarkers in oropharyngeal squamous cell carcinoma: a review. Eur Arch Otorhinolaryngol 268(1):5–16. doi:10.1007/s00405-010-1369-x
Hunt JL, Barnes L, Lewis JS Jr, Mahfouz ME, Slootweg PJ, Thompson LD, Cardesa A, Devaney KO, Gnepp DR, Westra WH, Rodrigo JP, Woolgar JA, Rinaldo A, Triantafyllou A, Takes RP, Ferlito A (2014) Molecular diagnostic alterations in squamous cell carcinoma of the head and neck and potential diagnostic applications. Eur Arch Otorhinolaryngol 271(2):211–223. doi:10.1007/s00405-013-2400-9
Cheong SC, Chandramouli GV, Saleh A, Zain RB, Lau SH, Sivakumaren S, Pathmanathan R, Prime SS, Teo SH, Patel V, Gutkind JS (2009) Gene expression in human oral squamous cell carcinoma is influenced by risk factor exposure. Oral Oncol 45(8):712–719. doi:10.1016/j.oraloncology.2008.11.002
Routray S (2014) Caveolin-1 in oral squamous cell carcinoma microenvironment: an overview. Tumour biol J Int Soc Oncodevelop Biol Med. doi:10.1007/s13277-014-2482-z
Kumagai K, Nimura Y, Mizota A, Miyahara N, Aoki M, Furusawa Y, Takiguchi M, Yamamoto S, Seki N (2006) Arpc1b gene is a candidate prediction marker for choroidal malignant melanomas sensitive to radiotherapy. Invest Ophthalmol Vis Sci 47(6):2300–2304. doi:10.1167/iovs.05-0810
Kaneda A, Kaminishi M, Nakanishi Y, Sugimura T, Ushijima T (2002) Reduced expression of the insulin-induced protein 1 and p41 Arp2/3 complex genes in human gastric cancers. Int J Cancer J Int du Cancer 100(1):57–62. doi:10.1002/ijc.10464
Staubach S, Hanisch FG (2011) Lipid rafts: signaling and sorting platforms of cells and their roles in cancer. Expert Rev Prot 8(2):263–277. doi:10.1586/epr.11.2
Williams TM, Lisanti MP (2005) Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am J Physiol Cell Physiol 288(3):C494–C506. doi:10.1152/ajpcell.00458.2004
Hung KF, Lin SC, Liu CJ, Chang CS, Chang KW, Kao SY (2003) The biphasic differential expression of the cellular membrane protein, caveolin-1, in oral carcinogenesis. J Oral Pathol Med Off Publ Int Assoc Oral Pathol Am Acad Oral Pathol 32(8):461–467
Nohata N, Hanazawa T, Kikkawa N, Mutallip M, Fujimura L, Yoshino H, Kawakami K, Chiyomaru T, Enokida H, Nakagawa M, Okamoto Y, Seki N (2011) Caveolin-1 mediates tumor cell migration and invasion and its regulation by miR-133a in head and neck squamous cell carcinoma. Int J Oncol 38(1):209–217
Zhang H, Su L, Muller S, Tighiouart M, Xu Z, Zhang X, Shin HJ, Hunt J, Sun SY, Shin DM, Chen ZG (2008) Restoration of caveolin-1 expression suppresses growth and metastasis of head and neck squamous cell carcinoma. Br J Cancer 99(10):1684–1694. doi:10.1038/sj.bjc.6604735
Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ (1997) The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol 138(2):375–384
Molli PR, Li DQ, Bagheri-Yarmand R, Pakala SB, Katayama H, Sen S, Iyer J, Chernoff J, Tsai MY, Nair SS, Kumar R (2010) Arpc1b, a centrosomal protein, is both an activator and substrate of Aurora A. J Cell Biol 190(1):101–114. doi:10.1083/jcb.200908050
Zain RB, Athirajan V, Ghani WM, Razak IA, Raja Latifah RJ, Ismail SM, Sallam AA, Bustam AZ, Rahman ZA, Hussien A, Talib N, Cheong SC, Jallaludin A (2013) An oral cancer biobank initiative: a platform for multidisciplinary research in a developing country. Cell Tissue Banking 14(1):45–52. doi:10.1007/s10561-012-9298-0
Sobin LH, Gospodarowicz MK, Wittekind C (2011) TNM classification of malignant tumours. Wiley
Vincent-Chong VK, Salahshourifar I, Karen-Ng LP, Siow MY, Kallarakkal TG, Ramanathan A, Yang Y-H, Khor GH, Rahman ZAA, Ismail SM, Prepageran N, Wan Mustafa WM, Abraham MT, Tay KK, Cheong SC, Zain RB (2014) Overexpression of MMP13 Is Associated with Clinical Outcomes and Poor Prognosis in Oral Squamous Cell Carcinoma. Sci World J 2014:12. doi:10.1155/2014/897523
Kunzel J, Psychogios G, Mantsopoulos K, Grundtner P, Waldfahrer F, Iro H (2014) Lymph node ratio as a predictor of outcome in patients with oropharyngeal cancer. Eur Arch Otorhinolaryngol 271(5):1171–1180. doi:10.1007/s00405-013-2513-1
Mesquita JA, Queiroz LM, Silveira EJ, Gordon-Nunez MA, Godoy GP, Nonaka CF, Alves PM (2015) Association of immunoexpression of the galectins-3 and -7 with histopathological and clinical parameters in oral squamous cell carcinoma in young patients. Eur Arch Otorhinolaryngol. doi:10.1007/s00405-014-3439-y
Anicin A, Gale N, Smid L, Kos J, Strojan P (2013) Expression of stefin A is of prognostic significance in squamous cell carcinoma of the head and neck. Eur Arch Otorhinolaryngol 270(12):3143–3151. doi:10.1007/s00405-013-2465-5
Choi HG, Kim JS, Kim KH, Sung MW, Choe JY, Kim JE, Jung YH (2015) Expression of hypoxic signaling markers in head and neck squamous cell carcinoma and its clinical significance. Eur Arch Otorhinolaryngol 272(1):219–228. doi:10.1007/s00405-014-2954-1
Zanaruddin SN, Saleh A, Yang YH, Hamid S, Mustafa WM, Khairul Bariah AA, Zain RB, Lau SH, Cheong SC (2013) Four-protein signature accurately predicts lymph node metastasis and survival in oral squamous cell carcinoma. Hum Pathol 44(3):417–426. doi:10.1016/j.humpath.2012.06.007
Xue J, Chen H, Diao L, Chen X, Xia D (2010) Expression of caveolin-1 in tongue squamous cell carcinoma by quantum dots. Euro J Histochem EJH 54(2):e20
Han SE, Park KH, Lee G, Huh YJ, Min BM (2004) Mutation and aberrant expression of Caveolin-1 in human oral squamous cell carcinomas and oral cancer cell lines. Int J Oncol 24(2):435–440
Goetz JG, Lajoie P, Wiseman SM, Nabi IR (2008) Caveolin-1 in tumor progression: the good, the bad and the ugly. Cancer Metast Rev 27(4):715–735. doi:10.1007/s10555-008-9160-9
Chorev DS, Moscovitz O, Geiger B, Sharon M (2014) Regulation of focal adhesion formation by a vinculin-Arp2/3 hybrid complex. Nature Comm 5:3758. doi:10.1038/ncomms4758
Zucchini C, Rocchi A, Manara MC, De Sanctis P, Capanni C, Bianchini M, Carinci P, Scotlandi K, Valvassori L (2008) Apoptotic genes as potential markers of metastatic phenotype in human osteosarcoma cell lines. Int J Oncol 32(1):17–31
Acknowledgement
This study was supported by University of Malaya Research Grant (RP002E-13HTM) and University of Malaya Postgraduate Research Fund (PPPC/C1-2012/DEGC/24). The authors acknowledged the Oral Cancer Research and Coordinating Centre (OCRCC), University of Malaya (UM) for providing tissue and data from the Malaysian Oral Cancer Database and Tissue Bank System (MOCDTBS). The authors also thanked the clinicians and pathologists from Ministry of Health Malaysia for their clinical and pathologic expertise.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest for this research.
Rights and permissions
About this article
Cite this article
Auzair, L.B.M., Vincent-Chong, V.K., Ghani, W.M.N. et al. Caveolin 1 (Cav-1) and actin-related protein 2/3 complex, subunit 1B (ARPC1B) expressions as prognostic indicators for oral squamous cell carcinoma (OSCC). Eur Arch Otorhinolaryngol 273, 1885–1893 (2016). https://doi.org/10.1007/s00405-015-3703-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00405-015-3703-9