Abstract
GEMIN5 is essential for core assembly of small nuclear Ribonucleoproteins (snRNPs), the building blocks of spliceosome formation. Loss-of-function mutations in GEMIN5 lead to a neurodevelopmental syndrome among patients presenting with developmental delay, motor dysfunction, and cerebellar atrophy by perturbing SMN complex protein expression and assembly. Currently, molecular determinants of GEMIN5-mediated disease have yet to be explored. Here, we identified SMN as a genetic suppressor of GEMIN5-mediated neurodegeneration in vivo. We discovered that an increase in SMN expression by either SMN gene therapy replacement or the antisense oligonucleotide (ASO), Nusinersen, significantly upregulated the endogenous levels of GEMIN5 in mammalian cells and mutant GEMIN5-derived iPSC neurons. Further, we identified a strong functional association between the expression patterns of SMN and GEMIN5 in patient Spinal Muscular Atrophy (SMA)-derived motor neurons harboring loss-of-function mutations in the SMN gene. Interestingly, SMN binds to the C-terminus of GEMIN5 and requires the Tudor domain for GEMIN5 binding and expression regulation. Finally, we show that SMN upregulation ameliorates defective snRNP biogenesis and alternative splicing defects caused by loss of GEMIN5 in iPSC neurons and in vivo. Collectively, these studies indicate that SMN acts as a regulator of GEMIN5 expression and neuropathologies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mutations in RNA-binding protein (RBP)-encoded genes have surfaced as significant factors contributing to numerous neurological disease [9, 27, 41]. RBPs are key contributors to numerous aspects of RNA processing and defects in the physiological functions of RBPs can lead to the neurological diseases amyotrophic lateral sclerosis (ALS) and spinal muscular atrophy (SMA) [9]. SMA is characterized by degeneration and loss of anterior horn cells in the spinal cord that leads to muscle weakness and atrophy resulting from the loss of Survival Motor Neuron (SMN) protein [6, 32, 33]. SMN interacts with GEMIN5, other GEMIN proteins, and Smith-core (Sm) proteins to form the SMN Complex [3, 73]. The SMN complex is responsible for the biogenesis of small nuclear ribonucleoproteins (snRNPs), which are essential components of splicing machinery [3, 72, 73].
We, as well as others, previously identified biallelic variants in the gene GEMIN5 that give rise to a neurological syndrome which features developmental delay, cerebellar atrophy, and predominant motor dysfunction along with hypotonia [28, 48, 50]. GEMIN5 is a highly conserved multifunctional protein involved in numerous aspects of RNA processing that includes the biogenesis of snRNPs through the SMN complex [4, 23, 72, 74], as well as the ability to regulate aspects of translation at a global level [12, 45]. GEMIN5 interacts with numerous SMN complex members but is directly associated with GEMIN2 and SMN [17, 42, 71]. The initiator of the SMN complex is GEMIN5, as it specifically recognizes each unique snRNP code to begin the process of snRNP-core assembly [4, 23, 72, 74]. The majority of GEMIN5 variants appear to cause loss of function by reducing GEMIN5 protein expression and disrupting snRNP-core complex dynamics [28, 48]. In patients with SMA, the disease severity is associated with the amount of functional SMN protein production which correlates to the degree of snRNP assembly defects [6, 32]. Recent advancements have shown remarkable success in treating SMA patient symptoms through restoration of functional SMN expression using either antisense oligonucleotides (ASOs; Nusinersen), small molecules (Risdiplam), or SMN gene replacement therapies (Onasemnogene abeparvovec-xioi) [37, 68, 70]. However, the implications of SMN-upregulating therapies to ameliorate snRNP defects in other neuroglial diseases beyond SMA have yet to be explored.
Here, we identified SMN as a genetic modifier of GEMIN5-mediated neurodegeneration in vivo and as a regulator of GEMIN5 expression in mammalian cells and iPSC neurons. Specifically, genetic upregulation of SMN via ectopic expression and administration of the SMN2 FDA-approved antisense oligonucleotide, Nusinersen, significantly increased the physiological levels of GEMIN5 in mammalian cells. Furthermore, a drastic reduction in SMN expression in SMA motor neurons revealed a notable decrease in GEMIN5 protein. Upregulation of SMN via lentiviral expression and Nusinersen administration significantly increased GEMIN5 expression in loss-of-function GEMIN5 patient iPSCs. Importantly, we found the Tudor domain of SMN is required for GEMIN5 interaction and regulation. Finally, ectopic expression of SMN rescued the snRNP biogenesis defects exhibited in mutant GEMIN5 iPSC-derived neurons as well as loss-of-function GEMIN5 in vivo. We also show that lentiviral SMN expression significantly rescued the inclusion of alternatively spliced isoforms identified in GEMIN5 iPSC neurons. Taken together, these observations provide further insights into the biological mechanisms of SMN and suggest SMN as a positive regulator of GEMIN5-mediated neurodegeneration and GEMIN5 expression.
Materials and methods
Drosophila lines
The fly lines used for this study and motor function assay can be found in Sup table 1. All Drosophila stocks were cultured on standard media on a 12-h light/dark cycle.
Genetic crosses
The Gal4/UAS system was used for targeted misexpression of the inducible transgene (Brand & Perrimon, 1993). All Gal4/UAS crosses were maintained at 25 °C and 29 °C to sample different induction levels of transgenes [47, 55]. UAS-dicer/CyO; tub-Gal4/TM3 and UAS-dicer/CyO; GMR-Gal4/Tb stocks were generated to enhance inducible RNAi potency [10] in UAS-rigKK RNAi, or UAS-SMN alone (FLAG tagged) or both. The GMR-Gal4 drives expression in the differentiating retinal precursor cells [39], whereas tub-GAL4 drives expression ubiquitously. The adults were assessed for phenotypes and frequency analysis was performed. Images of the left eyes of F1 generation, adult female Drosophila were taken at day 1 (or as indicated for ageing experiments) using a Leica M205C dissection microscope equipped with a Leica DFC450 camera. External eye degeneration was quantified using a previously published scoring system [31]. Briefly, eyes were examined for the presence or absence of the following features: supernumerary inter-ommatidial bristles, abnormal bristle orientation, ommatidial fusion, ommatidial pitting, disorganization of ommatidial array, and retinal collapse. If the following features were present, one point was given. Two points were added if the affected area involved more than 5% of the eye and four points were added if the affected area was involved more than 50% of the eye. Comparisons between genotypes were made using Student’s t test assuming equal variances. Eye size was measured using ImageJ software (NIH). Adult fly images were captured at 5X by placing the flies sideways on a clean slide. The images were taken on an MrC5 color camera mounted on an Axioimager.Z1 Zeiss Apotome using a Z-sectioning function of Axiovision software 4.6.3 [21]. The final images were prepared using Adobe Photoshop CS6 software. A detailed description of the fly crosses can be found in Sup table 2.
Larval preparations and immunohistochemistry
Third instar larvae were dissected, fixed, and immunostained as previously described [1]. Briefly, animals were dissected in ice-cold phosphate-buffered saline (PBS) (Lonza, #17–516 F), fixed in 4% formaldehyde, washed three times in PBS, incubated in 5% Triton-X-100/PBS for 20 min, washed three times in 0.1% PBST (0.1% Triton-X-100/PBS), and incubated overnight with primary antibodies: Drosophila coilin (gift from Dr. Joseph Gall, Carnegie Institution for Science, Baltimore, MD; 1:100) and Drosophila SmB (Y12, Novusbio, 1:100). Larvae were washed three times in 0.1% PBST and incubated with secondary antibodies: anti-rabbit Alexa Flour 568, 1:100 (Invitrogen, #651727); anti-mouse Alexa Flour 647, 1:100 (Invitrogen, #28181). Stained larvae were mounted using DAPI Fluoroshield (Sigma-Aldrich, #F6182). Images were collected on a Nikon A1 eclipse Ti confocal microscope.
Plasmids
The lentiviral vectors used to express EGFP (pLV[Exp]-CMV > EGFP, VB900088-2229upx) and SMN1 (pLV[Exp]-EGFP-CMV > hSMN1, VB201023-1249tdf) in mammalian cells were constructed and packaged by VectorBuilder. The following plasmids were constructed and packaged by VectorBuilder: HA-GEMIN5-WT (VB210409-1232kze), HA-WD40 (VB210819-1201kfk), HA-∆RBS (VB210819-1225rn), HA-∆WD40 (VB211122-1218exw), HA-SMN (VB220512-1296yke), HA-SMN Ex1-5 (VB220512-1302gqd), HA-SMN ∆Ex3 (VB220512-1299vvx), and HA-SMN ∆Ex5 (VB220512-1301jan).
Cell culture and differentiation of iPSCs into neuronal cells
SMA patient motor neurons: HUVEC iPSC line was a generous gift from the Kurian Lab [43]. WTC11 line was generated by the Conklin Lab and purchased from Erasmus MC iPS Core Facility. CS13iSMA was purchased from Cedars Sinai iPSC Core Facility. HGK1, HGK4a, HGK13, HGK22, and HGK27 were generated by Dr. Ludwig Heesen and iPIERIAN [14, 20]. A description of the SMA patient lines can be found in Sup table 3. Motor neuron differentiation was performed as described before, with some modifications [18]. To generate the clumps for embryoid body formation, similar size squares were scratched on a ~ 90% confluent well, treated with collagenase type IV, and resuspended in Essential™6 medium (A1516401, Gibco™) on ultra-low attachment flask (Corning). The first 2 days of the differentiation, neuronal basic medium (DMEM/F12 and Neurobasal supplemented with N2 and B27 without vitamin A) were supplemented with 3 µM CHIR99021 (4423, Tocris Bioscience), 0.2 µM LDN-193189 (S2618, Selleckchem), 40 µg SB431542 (1614, Tocris Bioscience), and 5 µM Y-27632 (S1049, Selleckchem). From day 3 on, neuronal media was supplemented with 0.1 µM retinoic acid (Sigma) and 500 nM SAG (Merck Millipore). From day 8 on until the end of the differentiation, BDNF (10 ng/ml, Peprotech) and GDNF (10 ng/ml, Peprotech) were added to the media. From day 9 to 16, neuronal media was supplemented with 20 µM DAPT (2634, Tocris Bioscience). On day 11, clumps with motor neuron progenitors were dissociated into single cells for plating on 20 µg/ml laminin-coated wells. From day 17 on, maturation media containing 10 ng/ml of BDNF, GDNF and CNTF (Peprotech) was added. Every other day media was changed by replacing half of the medium.
GEMIN5 iPSC-derived neurons: We used two different mutant GEMIN5 iPSC lines harboring L1068P and H913R variants characterized in [28]. In short, we reprogrammed peripheral blood mononuclear cells (PBMCs) from a homozygous GEMIN5 L1068P/L1068P patient (referred to as GEMIN5L1068P) and an unaffected parent (L1068P/ + , referred to as control) into induced pluripotent stem cell lines (iPSC). In addition, we used CRISPR/Cas9 to engineer an additional mutant GEMIN5 cell line, GEMIN5 H913R (H913R/ + , referred to as control) and homozygous variant GEMIN5 H913R/H913R (referred to as GEMIN5H913R) in a healthy isogenic iPSC line. The iPSCs were cultured and maintained in mTeSR media (STEMCELL technologies) on Matrigel-coated plates. For neuronal differentiation, ~ 0.6 million cells were plated and let to grow for up to 80–90% confluency in mTeSR 1 for 2 days. For the first phase of differentiation, the confluent iPSC cells were grown for 6 days in N2B27 Neurobasal/DMEM-F12 medium (1:1 v/v) containing 1% N2 (Gibco, 17,502–048), 2% B27 (Gibco, 17,054–044), 1% Glutamax (Gibco), and non-essential amino acids (NEAA) (Gibco, 11,140,050) along with 10 µM SB431542 (STEMCELL technologies), 0.1 µM LDN (Sigma SML0559), 1 µM retinoic acid (RA) (Sigma R2625), and 1 µM smoothened agonist (SAG, Cayman chemicals 11,914). For days 7 to 14, cells were grown in N2B27 media supplemented with 1 µM RA, 1 µM SAG, 10 µM DAPT (Cayman, 13,197), and 16 µM SU5406 (Cayman, 131,825). On day 14, cells were dissociated using TrypLE/ DNase I (Invitrogen) and cultured on poly-ornithine and laminin-coated coverslips or plates in neuronal media containing neurobasal medium, N2, B27, 0.4 mg/ml ascorbic acid (Sigma, A4403), 10 µg/ml human brain-derived neurotrophic factor (BDNF) (Peprotech, 45,002), 10 µg/ml glial cell derived neurotrophic factor (GDNF) (Peprotech, 45,010), 10 µg/ml ciliary neurotrophic factor (CNTF) (Peprotech, 45,013), 1% Glutamax, and NEAA.
Lentiviral production and transduction
Lentiviral transfer vectors encoding EGFP and SMN were co-transfected with Lenti packaging plasmids (OriGene) into HEK cells using the Turbofectin transfection reagent (OriGene) according to the manufacturer’s instructions. Following an initial media change, lentiviral supernatant was collected at 24 and 48 h post-transfection prior to filtration and overnight incubation at 4 °C with 1X Lenti Concentrator Solution (OriGene). The Lenti Particles were then centrifuged at 4000 × g for 120 min at 4 °C. The resulting pellet was recentrifuged at 4000 × g for 5 min prior to re-suspension in ice-cold, sterile PBS. Pellets were then allowed to dissolve for 1–2 days at 4 °C. Resuspended lentiviral particles were then aliquoted. Neuron transductions were performed by diluting lentiviral particles at an MOI of 5 in neuronal differentiation media. Media changes were performed after 48 h of incubation and all experiments were initiated at 120 + hours post-transduction.
Transfections and ASOs
HEK293T cells were transiently transfected with plasmids using Lipofectamine 3000 (Invitrogen L3000001) and used 48 h after transfection. All ASOs used in this study were obtained from Dharmacon (Lafayette, CO, USA), and encompassed phosphorothioate backbone and 2′OMe modifications throughout the entire sequence. HEK293T cells were transiently transfected with control 10’mer and ASO N1 ASO’s at 25, 50, and 100 nM for 48 h. iPSC neurons were transiently transfected with control 10’mer and ASO N1 (Nusinersen) ASO’s at 500 nM for 72 h. The ASO N1 has the same sequence of Nusinersen and was constructed with 2'-Omethyl RNA Bases and Phosphorothioate backbone modifications. ASO sequences are as follows: ASO N1: 5′-AUUCACUUUCAUAAUGCUGG-3′. Control: 5′-UUGCCUUUCU-3′.
Immunofluorescence
The HEK293T cells and neurons were fixed in 4% paraformaldehyde (PFA) for 15 min and blocked in 0.1% Triton-X in PBS and 5% normal goat serum for 1 h. The cells were treated overnight with the following antibodies: rabbit anti-GEMIN5 (Millipore Sigma HPA037393, 1:1,000), mouse anti-Smb (Y12) (NovusBio, 1:500), mouse anti-SMN (BD transduction 610,646, 1:1,000), mouse anti-coilin (Santa Cruz sc-55594, 1:500), chicken anti- beta-III Tubulin (NOVUS Biologicals NB100-1612- 1:1,000), and goat anti-MAP2 (Synaptic System-188 004, 1:1,000). Alexa fluor-488, -568, and -647 secondary antibodies were used from Invitrogen. The cells were mounted using fluoroshield™ with DAPI (Sigma) and images were taken at 60 X using Nikon A1-T216.3 confocal microscope.
Quantification of immunofluorescent staining: Images used for quantification were acquired at matched exposure times or laser settings and processed using identical settings. Quantification of the confocal immunostaining was performed in a blinded fashion and maximum intensity images were used for analysis in Fiji ImageJ software. Regions of interest (ROI) were drawn around the nuclear and cytoplasmic regions and referred to as “total intensity”. The raw Integrated Density Values were detected for the total intensity of each cell and normalized to the area of each cell for the final total intensity value. The nuclear region was identified by DAPI signal and the cytoplasmic region according to the plasmid fluorescent reporter construct [16, 49].
Western blot (WB) analysis
Fly heads were collected from each cross and snap-frozen on dry ice. Heads were crushed on dry ice and incubated in RIPA buffer (150 mM NaCl, 1% NP40, 0.1% SDS, 1% sodium deoxycholate, 50 mM NaF, 2 mM EDTA, 2 mM DTT, 0.2 mM Na orthovanadate, and 1 X Roche protease inhibitor #11,836,170,001). Lysates were sonicated and centrifuged to remove debris. Supernatants were boiled in Laemmli buffer (Boston Bioproducts, #BP-111R) for 5 min and loaded onto 4%–12% Nupage Bis–Tris gels (Novex/Life Technologies). Proteins were transferred using the iBlot2 (Life Technologies, #13,120,134) onto nitrocellulose (iBlot 2 NC regular Stacks, Invitrogen, #IB23001). Western blots were blocked with 2.5% milk solution (BLOT-QuickBlocker reagent, EMB Millipore, #WB57-175GM) and incubated with primary antibody overnight: rabbit Drosophila Rig, 1–1:000 (developed by Bio Boster Biological Technology Co. Ltd, Wuhan, China), rabbit Drosophila Smn (developed by Bio Boster Biological Technology Co. Ltd, Wuhan, China), and mouse anti-tubulin, 1:10,000 (66,031, Protein Tech). Blots were washed and incubated in secondary antibody for 1 h and imaged on a Licor imager (Odyssey CLx). All western blots were run in triplicate using biological replicates. Protein levels were quantified using Image J software, and statistical analysis was performed with GraphPad Prism six software.
HEK293T cells and differentiated neurons were dissociated in TrypLE/DNase and cells were pelleted down at 250xg at room temperature. The cells were washed with PBS and lysed in RIPA buffer containing 150 mM NaCl, 50 mM NaF, 2 mM EDTA, 0.2 mM Na orthovanadate, 1% sodium deoxycholate, 2 mM DTT, 1% NP40, 0.1% SDS, and protease inhibitor (Roche 11,836,170,001). The lysates were sonicated and centrifuged at 10,000 xg for 15 min at 4 °C. The concentrations of proteins in the supernatant were measured by Pierce™ BCA protein assay kit (Thermo Scientific 23,227). Equal concentration of supernatant was boiled with 1X Laemmli buffer and the proteins were separated using 4–12% NuPage bis–Tris gel (Novex/Life Technologies). Proteins were transferred onto nitrocellulose (Invitrogen IB23001) using the iBlot2 (Life Technologies 13,120,134). The blots were blocked in 2.5% QuickBlocker reagent (EMB Millipore WB57-175GM) and probed overnight with the following antibodies: mouse anti-tubulin (SIGMA, 1:10,000), anti-beta-actin HRP-conjugated (1:10,000, HRP-60008, Proteintech), anti-GEMIN5 (Protein Tech p-7210531, 1:1,000), mouse anti-GEMIN2 [2E17] (1:2,000), mouse anti-GEMIN4 [F-2] (Santa Cruz sc-166418, 1:1,000), and mouse anti-SMN (1:3000, 610,646, BD Biosciences).
Secondary antibodies used were anti-rabbit DYLight 800 and anti-mouse 680 (Invitrogen, 1:10,000). The blots were imaged using Licor imager (Odyssey CLx). All the blots were run in triplicates and the integrated band densities were calculated using image studio software (Licor).
RT-PCR and qRT-PCR
Total RNA was isolated using the PureLinkTM RNA mini kit (Invitrogen). RNA quantity and purity (260/280 and 260/230 ratios) were determined using a NanoDrop ND-1000 spectrophotometer. The iScript Select cDNA Synthesis Kit (Bio-Rad) was then used to produce cDNA from 500 ng of RNA from each sample. For PCR reactions, the cDNA was synthesized in 20uL reactions using PCR supermix (Invitrogen) following the manufacturer’s instructions. PCR products were separated on 6% TBE gels (Invitrogen) and visualized by ethidium bromide staining. Analysis and quantifications of SMN2 splice products were performed using ImageJ software. The primers used in the PCR reaction are outlined in Sup table 4.
For qRT-PCR, all cDNA samples were run on 96-well plates (Applied Biosystems; 4,306,737) on a 7300 Real-Time PCR System (Applied Biosystems) using the Bio-Rad iQ Supermix (170–8862). Cycle threshold (CT) values were recorded and analyzed following the comparative (CT) method using Prism 6 (GraphPad Software) for statistical analyses. All primers for qPCR were designed PrimerQuest primer design tool (Integrated DNA Technologies). The primers were designed with the PrimerTime qPCR Assay tool (Integrated DNA Technologies). IDT PrimeTime qPCR Assays were used as the primer/probe solutions. The primers and probes used for qPCR assays are listed in Sup table 4.
Simplex real-time PCR SMN copy number
Simplex real-time PCR analysis was performed as previously described with some modifications. Human serum albumin (ALB) was employed as an endogenous reference gene. Primers and TaqMan MGB 50-labeled VIC probes for the SMN1 and SMN2 exon 7 locus used in this study were as previously described [2, 44]. Primers and TaqMan MGB 50-labeled FAM probe for the reference gene Albumin map are in exon 12 and have been designed using Integrated DNA technology software. All primers and probes for the copy-number assay are in Sup table 4. Sample DNA was diluted to the final theoretical concentration of 5.0 ng/mL. Simplex PCR was carried out in quadruplicate for each sample. Each reaction was performed in a total volume of 20 µL containing 10.0 ng of genomic DNA, 10 µL iQ multiplex powermix (Bio-Rad), 300 nM of the gene-specific primers, and 100 nM of the gene-specific MGB probe. PCR conditions were 3 min 95 °C and 40 cycles consisting of 15 s 95 °C, and 1 min 60 °C. The copy number of SMN1 or SMN2 was calculated using the following equation: SMN1 (SMN2) copy number = raw SMN1 (SMN2) CT value/(raw albumin CT value/2).
In vitro snRNP assembly
Cytoplasmic extracts from the differentiated neurons were prepared using NE-PER nuclear and cytoplasmic extraction kit (Thermo Scientific 78,835) and the protein concentration was measured by Pierce™ BCA protein assay kit. U1snRNA were transcribed from gel-eluted and linearized DNA template by in vitro transcription using T7 RNA polymerase and m7G cap analogue. pCp-Cy3 (Cytidine-5'-phosphate-3'-(6-aminohexyl) phosphate) (Jena Bioscience) was transferred to the 3′-hydroxyl group on U1snRNA by T4 RNA ligase (ThermoFisher). The snRNP assembly reaction was carried out as previously described [28]. In short, 5 µg of pCp-Cy3 labeled U1snRNAs were incubated with 50 µg of cytoplasmic extract, 10 µM tRNA, and 2.5 mM ATP at 30 degrees C for 90 min. The reaction samples were loaded onto a native 6% TBE polyacrylamide gel (Novex/Life Technologies). The gel was run at 150 V at 4 degrees and was imaged using Licor imager [28].
RNA sequencing and alternative splicing (AS) analysis
RNA was isolated from iPSC-derived differentiated neurons from control, mutant GEMIN5H913R iPSC neurons with EGFP, and EGFP-SMN lentiviral expression using the PureLink™ RNA mini kit (Invitrogen). RNA from all samples was extracted, assessed for quality control, and sequenced via paired-end 2 X 150 bases at approximately 60 million paired-end reads per sample on an Illumina HiSeq platform. RNA library preparations and RNA sequencing reactions were conducted at GENEWIZ, LLC. For consistency, we first performed gene expression analysis for all samples by mapping to the genome (Ensembl Version GRCh38.96) using the STAR aligner. Principal Components Analysis (PCA) confirmed that the samples were sequentially ordered in gene expression space, as expected. Alternative splicing analysis was conducted using rMATS (V4.1.1) with Ensembl gene models [53]. Initially, the categories of alternative splicing events, such as skipped exon (SE), mutually exclusive exon, alternative 5’ splice site, alternative 3’ splice site, and retained intron, were identified and the schematics were plotted. Further, mapped BAM files were used as an input to detect differences in specific regions of transcripts from a particular gene based on the percentage of splicing inclusion and detect the differential splicing between controls and mutant GEMIN5H913R neurons with and without SMN expression. Output from the analysis included both the reads mapped to splice junctions as well as reads mapped to both splice junctions and exon body. To gain a systematic view of the alternative splicing, we retrieved the PSI values for these events in and created sashimi plots. Sashimi plots of selecting representative genes as determined by rMATS analysis were plotted using rmats2sashimiplot wrapper by calculating the average inclusion level, average read depth, and the average number of junction-spanning reads of each group.
Statistical analysis
Statistical analyses were done on GraphPad Prism using one-way ANOVA (analysis of variance) followed by a Bonferroni or Tukey post hoc test for comparison between two or more groups.
Results
SMN is a genetic modifier of GEMIN5-mediated neurodegeneration in vivo
To investigate if the interacting partners of GEMIN5 could compensate for the neurodegeneration associated with the loss of GEMIN5 (Rigor mortis, the Drosophila homologue of human GEMIN5) in vivo, we conducted a screen of Drosophila SMN complex members and assessed their impact on the effects of Rigor mortis (Rig) knock down (KD) (Fig. 1a). Utilizing the glass multiple reporter (GMR), Rig RNAi-expressing animals were crossed with control, Luciferase (LUC) overexpression (OE) (positive control to rule out gal4 dilution), Smn RNAi, Smn OE, and Gemin2 OE-mediated fly lines to drive targeted expression of the transgenes to Drosophila eyes. UAS-dicer was used to enhance the KD of Rig over 50% and resulted in severe external eye degeneration and ommatidial disorganization phenotypes as compared to control animals (Fig. 1a, Sup Fig. 1a–c). Overexpression of Luciferase and KD of Smn had no effect on the external eye degeneration of Rig KD animals (Fig. 1c-d). Surprisingly, ectopic expression of Drosophila Smn significantly suppressed the external eye degeneration phenotypes in Rig RNAi-expressing animals (Fig. 1e). Contrary to Smn over expression, we found that ectopic expression of Drosophila Gemin2 significantly exacerbated eye toxicity in Rig RNAi-expressing animals as compared to controls (Sup Fig. 1d-e). In addition, ectopic expression of Smn significantly restored the decreased eye size of Rig RNAi expressing animals (Fig. 1f). Further, we found that Smn overexpression continued to suppress Rig RNAi induced external eye degeneration over time (Sup Fig. 1f). We observed a similar suppression in the loss of Rig phenotype among both male and female animals.
Smn upregulation suppresses loss of Gemin5 neurodegeneration in vivo. a Schematic representing the experimental to investigating the genetic manipulation of the Drosophila snRNP complex proteins in the background of loss of Gemin5 (Rig) animals in vivo. Drosophila expressing UAS-Rig RNAi were crossed with the GMR-gal4 driver for targeted expression to the Drosophila eye and Tubulin-gal4 driver for ubiquitous expression. b Representative images of female Drosophila eyes expressing GMR-gal4 and Rig RNAi crossed with control (W1118), Luciferase OE, Smn RNAi, and Smn OE. c, d, e Quantification of eye degeneration of control and Rig RNAi flies combined with (c) Luciferase OE, (d) Smn RNAi, and (e) Smn OE (n = 20 Drosophila per group, one-way ANOVA w/ Tukey’s multiple comparisons). Error bars indicate S.E.M. ****p < 0.0001. NS, not significant. f Quantification of relative eye sizes of control and Rig RNAi flies combined with Smn OE (n = 5 Drosophila per group, one-way ANOVA w/ Tukey’s multiple comparisons). Error bars indicate S.E.M. *p < 0.05; ****p < 0.001. NS, not significant. g, h qPCR analysis from (n = 4 biological replicates of 6 Drosophila heads per group) confirms significant knockdown of endogenous (g) Rig in the Rig KD groups and significant overexpression of (h) Smn in the Smn OE crosses (one-way ANOVA w/ Tukey’s multiple comparisons). Error bars indicate S.E.M. *p < 0.05; ****p < 0.0001; NS, not significant. i Representative male images of eclosed Drosophila adults expressing Control, Rig RNAi (pupal lethality), Rig RNAi in combination with over expression of Smn, and Smn OE. j The percentage of eclosed adults for control and Rig RNAi animals combined with Smn OE (n = 100 Drosophila per replicate, one-way ANOVA w/ Tukey’s multiple comparisons). Error bars indicate S.E.M. ****p < 0.0001
These results prompted us to examine the expression levels of Rig and Smn in the Rig RNAi-expressing animals via qPCR and Western Blot (WB). We found that the endogenous levels of Rig mRNA (decreased by ~ 65–70% with dicer) and protein (Fig. 1g, Sup Fig. 1g-h) were significantly reduced upon KD of Rig, as compared to control. In addition, Smn mRNA (decreased by ~ 50%) and protein were also significantly reduced in Rig KD animals (Fig. 1h, Sup Fig. 1i). WB and qPCR confirmed ectopic expression of Smn in Rig KD animals and had no effect on the efficacy of silencing expression of Rig via RNA interference (Fig. 1g-h, Sup Fig. 1g-i). More importantly, GEMIN5 appears to be essential for motor function, as patients with homozygous mutations manifest neurological symptoms at very early stages [28, 48, 50]. Thus, we sought to determine if Smn had any effect on the motor function phenotypes observed in Rig KD animals. We used Tubulin-GAL4 and UAS-Dicer (to enhance KD of Rig) to drive ubiquitous expression of Rig RNAi in flies ectopically expressing Smn (Fig. 1i). We observed complete pupal lethality of Rig KD animals compared to controls, suggesting severe developmental defects associated with loss of Rig. Interestingly, upregulation of Smn significantly ameliorated the developmental defects observed in Rig KD animals, with up to almost 100% of animals eclosing to the adult stage (Fig. 1j). Together, these results indicate that upregulation of Smn in Rig KD animals rescues morphological defects and motor dysfunction associated with the loss of Gemin5 in vivo.
SMN regulates the expression of GEMIN5 in mammalian cells
SMN protein-upregulating therapies have demonstrated remarkable success in suppressing SMA patient symptoms caused by loss-of-function mutations in the SMN1 gene [37, 68, 70]. However, these therapies have been limited to only SMA patients and little is known about how SMN-upregulating therapies directly affect GEMIN5. To further understand these effects, we transfected HEK293T cells with EGFP control and EGFP-SMN (dual promoter lentiviral construct resulting in coexpression of SMN and EGFP separately) in a dose-dependent manner and measured the protein expression of GEMIN5 and SMN via WB (Fig. 2a). We found that the protein levels of endogenous GEMIN5 were significantly increased upon ectopic expression of SMN (Fig. 2b). In addition, we found that EGFP-SMN also notably increased the fluorescent intensity of both SMN and GEMIN5 in HEK293T cells (Sup Fig. 2a). Similarly, we asked if ectopic expression of other SMN complex members had any effect on the levels of GEMIN5. We ectopically expressed HA-GEMIN2 and HA-GEMIN4 in HEK293T cells and observed no significant changes in GEMIN5 expression compared to controls (Sup Fig. 3a-e).
SMN expression increases the levels of GEMIN5 in HEK293T cells. a WB analysis showing the levels of GEMIN5 and SMN proteins after ectopic overexpression of EGFP-SMN at 0 ug (untransfected), 1 µg, and 2 µg in HEK293T cells (EGFP alone and untransfected HEK293T cells were used as control). b Quantification of proteins from panel A showing a significant increase in GEMIN5 SMN after overexpression of SMN (n = 3 blots per condition, unpaired Student’s t test between 0 µg and each condition). Error bars indicate S.E.M. *p < 0.05; **p < 0.01; ****p < 0.0001. NS, not significant. c WB analysis showing the levels of GEMIN5 and SMN after dose-dependent administration of ASO Control (Ctrl) and the SMN2 oligonucleotide ASO N1. d, e Quantification of GEMIN5 and SMN protein levels after administration of 0 (untransfected), 25, 50, & 100 nm of (d) control ASO and (e) ASO N1 (n = 3 blots per condition, unpaired Student’s t test between 0 nm and each condition). Error bars indicate S.E.M. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. NS, not significant. f, g Quantification of the mRNA levels showing a significant increase in (f) GEMIN5 and (g) SMN after EGFP-SMN expression (n = 3, one-way ANOVA w/ Bonferroni test). Error bars indicate S.E.M. ***p < 0.001; ****p < 0.0001. h, i Quantification of GEMIN5 and SMN2-FL mRNA levels after administration of 0 (untransfected), 25, 50, & 100 nm of (h) control ASO and (i) ASO N1 (n = 4, unpaired Student’s t test between 0 nm and each condition). Error bars indicate S.E.M. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. NS, not significant
GEMIN5 is significantly reduced in motor neurons derived from SMA patient iPSC lines. a WB analysis showing the levels of GEMIN5 and SMN in motor neurons differentiated from iPSCs from two healthy controls and 6 SMA patients (type 1, type 2 & type 3). b, c Quantification of (b) GEMIN5 and (c) SMN showing a significant decrease in both proteins in SMA type 1 motor neurons compared to two healthy controls (n = 3 biological replicates from three independent differentiations, one-way ANOVA w/ Tukey’s multiple comparisons). Error bars indicate S.E.M. **p < 0.01; ***p < 0.001. d, e Quantification of (e) GEMIN5 and (e) SMN showing a decrease in both proteins in SMA type 2 motor neurons compared to two healthy controls (n = 3 biological replicates from three independent differentiations, one-way ANOVA w/ Tukey’s multiple comparisons). Error bars indicate S.E.M. *p < 0.05. NS, not significant. f, g Quantification of (f) GEMIN5 and (g) SMN showing a significant decrease in only SMN protein in SMA type 3 motor neurons compared to two healthy controls (n = 3 biological replicates from 3 independent differentiations, one-way ANOVA w/ Tukey’s multiple comparisons). Error bars indicate S.E.M. ****p < 0.0001. NS, not significant. h Pearson correlation of SMN and GEMIN5 levels in the healthy controls and SMA type 1 motor neurons exhibits a strong positive correlation between the levels of both proteins across all samples (R = 0.93) (n = average of three biological replications, Pearson correlation coefficient). i Pearson correlation of SMN and GEMIN5 levels in the healthy controls and 2 SMA type 2 motor neurons exhibits a strong positive correlation between the levels of both proteins across all samples (R = 0.95) (n = average of three biological replicates, Pearson correlation coefficient). j Pearson correlation of SMN and GEMIN5 levels in the healthy controls and 3 SMA type 3 motor neurons exhibits a moderate positive correlation between the levels of both proteins across all samples (R = 0.5) (n = average of three biological replicates, Pearson correlation coefficient)
Based on these results, we asked if upregulation of SMN2 via the FDA-approved Nusinersen (referred to as ASO N1), antisense oligonucleotide, had any effects on GEMIN5 expression. We treated HEK293T cells with ASO N1 and a scrambled control ASO (ASO Ctrl) at 3 different concentrations (25 nM, 50 nM, 100 nM) and assessed the protein levels of GEMIN5 and SMN via WB (Fig. 2c). Treatment with ASO N1 resulted in a significant increase in GEMIN5 and SMN (Fig. 2e), compared to cells treated with the control ASO (Fig. 2d). In addition, we found that 25 nm of Cy3 ASO N1 notably increased the fluorescent intensity of both SMN and GEMIN5 in HEK293T cells (Fig. S2b). To determine the effectiveness of the N1-targeting ASO, we performed PCR analysis to assess the percentage of SMN2 exon 7 inclusion in HEK293T cells after being treated with 0, 25, 50, and 100 nM of the control and ASO N1 ASOs (Sup Fig. 4a). Treatment with ASO N1 produced an increase in the inclusion of SMN2 exon 7 with the highest inclusion percentage being ~ 80% at 100 nM for ASO N1 compared to ~ 45% for the control (Sup Fig. 4b).
Upregulation of SMN rescues the levels of GEMIN5 in mutant GEMIN5 iPSC-derived neurons. a Representative blots showing the levels of GEMIN5 and SMN after lentiviral transduction of EGFP and EGFP-SMN in unaffected control (H913R/ +) and patient (H913R/H913R) GEMIN5H913R iPSC neurons. b, c Quantification of (b) GEMIN5 and (c) SMN proteins from GEMIN5H913R iPSC neurons in panel A. (n = 4 blots per condition, one-way ANOVA w/ Tukey’s multiple comparisons). Error bars indicate S.E.M. *p < 0.05; **p < 0.01; ****p < 0.0001; NS, not significant. d Representative blots showing the levels of GEMIN5 and SMN after lentiviral transduction of EGFP and EGFP-SMN in mutant GEMIN5L1068P iPSC-derived neurons. e, f Quantification of (e) GEMIN5 and (f) SMN proteins from mutant-derived GEMIN5L1068P iPSC neurons in panel d. (n = 8 blots per condition, unpaired Student’s t test). Error bars indicate S.E.M. **p < 0.01; ****p < 0.0001. g Representative confocal images of mutant GEMIN5L1068P-derived iPSC neurons transduced with EGFP control and lentiviral EGFP-SMN from the same set of neuronal differentiations. The neurons were probed for EGFP, endogenous SMN (to assess the level of SMN OE), and endogenous GEMIN5. The cell nuclei were stained with DAPI. Scale bar = 10 µm and 5 µm for zoom. h, i Quantitative analysis displaying the total intensity of (h) SMN and (i) GEMIN5 from panel G measured as integrated density values (IDV) divided by cell area (n = 170 neurons, unpaired Student’s t test). Error bars indicate S.E.M. ****p < 0.0001. j Representative blots showing the protein levels of GEMIN5 and SMN in mutant GEMIN5L1068P neurons after administration of ASO Control and ASO N1 at 500 nM. k, l. Quantitative analysis of (k) GEMIN5 and (l) SMN proteins displaying an increase in expression after 500 nM of ASO Control or ASO N1 administration (n = 6 blots, unpaired Student’s t test). Error bars indicate S.E.M. **p < 0.01; ***p < 0.001. m Representative confocal images of mutant GEMIN5L1068P-derived iPSC neurons transfected with 500 nm of Cy3 ASO Control and Cy3 ASO N1 from the same set of neuronal differentiations. The neurons were probed for, endogenous SMN (to assess the level of SMN OE), Cy3, and endogenous GEMIN5. The cell nuclei were stained with DAPI. Scale bar = 10 µm and 5 µm for zoom. n, o Quantitative analysis displaying the total intensity of (n) SMN and (o) GEMIN5 from panel G measured as integrated density values (IDV) divided by cell area (n = 60 neurons, unpaired Student’s t test). Error bars indicate S.E.M. ****p < 0.0001. p Representative blots showing the protein levels of GEMIN5 and SMN in GEMIN5L1068P neurons after 0, 4, 8, 12, and 24 h of cycloheximide (CHX) treatment. q, r Quantitative analysis of the rate of degradation of (q) GEMIN5 and (r) SMN proteins after CHX treatment at 0, 4, 8, 12, and 24 h. (n = 3 blots, nonlinear regression-one phase decay)
In addition, to assess if upregulation of GEMIN5 was restricted to the protein level or also had an effect at the mRNA level in response to SMN expression, we measured the mRNA levels of GEMIN5 and SMN via qPCR. We found that the mRNA levels of GEMIN5 (Fig. 2f) were significantly increased in response to EGFP-SMN (Fig. 2g). Similarly, we assessed the mRNA expression of GEMIN5 and SMN2 after ASO N1 treatment in HEK293T cells and observed a significant increase in both GEMIN5 and SMN2 (Fig. 2i) mRNA compared to controls (Fig. 2h). These observations suggest that upregulation of both SMN transcripts (SMN1 & SMN2) in HEK293T cells significantly increases the overall expression of GEMIN5.
Since we identified increased SMN expression as a regulator of GEMIN5, we asked how a reduction in SMN expression influenced GEMIN5 levels. To investigate this, we measured the levels of GEMIN5 and SMN in 6 SMA patient iPSC samples harboring LOF mutations in the SMN gene. We differentiated 2 healthy control iPSC lines and 6 SMA patient iPSC lines (across 3 types of SMA) into motor neurons and conducted WB analysis (Fig. 3a). The SMA type 1 neurons displayed a significant decrease in the levels of both GEMIN5 (~ 50% reduction) (Fig. 3b) and SMN (~ 85% reduction) (Fig. 3c), as compared to the 2 control motor neuron samples. In addition, GEMIN5 (~ 30% reduction) (Fig. 3d) and SMN (~ 80% reduction) (Fig. 3e) were also notably reduced in both SMA type 2 motor neurons, as compared to the controls. Lastly, all SMA type 3 motor neuron samples did not result in a significant decrease in GEMIN5 expression (Fig. 3f), while displaying a drastic reduction in SMN (~ 60% reduction) (Fig. 3g). This suggests the effect of reduced GEMIN5 expression in SMA motor neurons to be dosage-dependent, as over a 75% decrease in SMN expression is needed to significantly influence GEMIN5 expression. Furthermore, to confirm these findings, we assessed for the levels of GEMIN2 protein in the SMA type 1 and type 2 motor neurons. GEMIN2 was also significantly reduced (over 50%) in both SMA type 1 and 2 motor neuron samples (Sup Fig. 5a-b). Importantly, we observed a strong positive correlation between the levels of SMN and GEMIN5 in the SMA type 1 and type 2 motor neurons (Fig. 3h-i), while only exhibiting a moderate positive correlation in the SMA type 3 motor neurons. These data suggest a strong association between SMN and GEMIN5 expression.
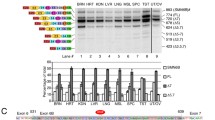
The RNA-binding sites of GEMIN5 and the Tudor domain of SMN are required for the GEMIN5-SMN interaction. a Schematic illustration of constructed HA-GEMIN5 constructs. HA-GEMIN5-WT harbors full-length GEMIN5 protein. HA-WD40 harbors only the n-terminus of GEMIN5 containing 13 tryptophan aspartic acid repeat domains (WD40). HA-∆RBS lacks the C-terminus of GEMIN5 and harbors the N-terminus and the tetratricopeptide repeat (TPR)-like domains of GEMIN5. HA-∆WD40 lacks the N-terminus of GEMIN5 and harbors the TPR-like region and the c-terminus containing the RNA-binding sites of GEMIN5. b Representative blots of co-immunoprecipitation from HEK cells expressing HA-GEMIN5 constructs and EGFP-SMN. Immunoprecipitation with HA antibody showed SMN in the input samples and pulled down with HA-GEMIN5-WT and HA-∆WD40, but absent from HA-WD40 and HA-∆RBS. Immunoprecipitation with SMN showed HA-GEMIN5 in the input samples and pulled down with HA-GEMIN5-WT and ∆WD40, but absent from HA-WD40 and HA-∆RBS; * and arrows indicate bands of interest. c Representative blot of Immunoprecipitated HA-GEMIN5-WT and HA-∆WD40 HEK cell lysates with HA antibody and treated with RNase A. Treatment with RNase A slightly decreases the interaction between SMN and HA-GEMIN5-WT and HA-∆WD40; * and arrows indicate bands of interest. d Schematic illustration of constructed HA-SMN constructs. HA-SMN-WT harbors full-length SMN protein. HA-SMN Ex1-5 harbors exons 1–5 and lacks exons 6–8 of SMN. HA-SMN ∆Ex3 lacks exon 3 of SMN which harbors the Tudor domain. HA-SMN ∆Ex5 lacks exon 5 which harbors the proline-rich region of SMN. e Representative blots of immunoprecipitation from HEK cells expressing HA-SMN constructs. Immunoprecipitation with HA antibody showed endogenous GEMIN5 in the input samples and pulled down with HA-SMN-WT and HA-SMN ∆Ex5, but absent from HA-SMN Ex1-5 and HA-SMN ∆Ex3. f Quantitative analysis of immunoprecipitated endogenous GEMIN5 pulled down with the HA-SMN constructs (n = 3 blots, one-way ANOVA w/ Tukey’s multiple comparisons). Error bars indicate S.E.M. ***p < 0.001. NS, not significant. g Representative blots of HEK cells transfected with HA-SMN-WT, HA-SMN ∆Ex5, and HA-SMN ∆Ex5 showing the levels endogenous GEMIN5 and SMN. h Quantitative analysis shows upregulation of GEMIN5 and SMN protein in HA-SMN-WT and HA- SMN ∆Ex5 only (n = 4 blots, one-way ANOVA w/ Tukey’s multiple comparisons). Error bars indicate S.E.M. **p < 0.01; ***p < 0.001
SMN upregulation restores the loss of GEMIN5 in mutant GEMIN5 iPSC neurons
We previously reported that patients with biallelic mutations in GEMIN5 exhibited a significant reduction in GEMIN5 expression levels, resulting in defective SMN complex formation [28]. We asked if upregulation of SMN had any effect on the expression levels of GEMIN5 in mutant GEMIN5 iPSC-derived neurons. Utilizing a lentiviral expression system, we expressed EGFP and EGFP-SMN in mutant GEMIN5H913R iPSC-derived neuronal cells and assessed the expression levels of GEMIN5 and SMN via WB. In GEMIN5H913R-derived neurons expressing EGFP control, we found the expression levels of GEMIN5 to be drastically reduced up to ̴70–80% compared to the unaffected controls (Fig. 4a). Further, we also observed a reduction in the protein levels of SMN in mutant GEMIN5H913R neurons expressing EGFP as compared to controls. Surprisingly, lentiviral expression of SMN (increased by ~ 40%) (Fig. 4c) in mutant GEMIN5H913R neurons significantly increased the protein levels of GEMIN5 (Fig. 4b), as compared to GEMIN5H913R neurons expressing EGFP control. Next, we asked if lentiviral SMN expression had a similar effect on GEMIN5 in GEMIN5L1068P iPSC-derived neurons (Fig. 4d). Lentiviral expression of SMN (increased by ~ 30%) (Fig. 4f) in mutant GEMIN5L1068P neurons significantly increased the protein levels of mutant GEMIN5 (Fig. 4e), as compared to GEMIN5L1068P neurons expressing EGFP control. To gain further insights into mechanisms behind SMN’s influence on mutant GEMIN5, we assessed if SMN expression had any effect on the localization pattern of GEMIN5 in mutant GEMIN5L1068P iPSC-derived neuronal cells (Fig. 4g). We previously reported that biallelic mutations in GEMIN5 result in a loss of cytoplasmic GEMIN5 expression [28]. We found a significant increase in total SMN (Fig. 4h) and GEMIN5 (Fig. 4i) immunoreactivity in mutant GEMIN5L1068P neuronal cells expressing lentiviral EGFP-SMN, as compared to mutant GEMIN5L1068P neuronal cells expressing EGFP control. Furthermore, we asked if lentiviral SMN expression had any effect on GEMIN5 mRNA in control and mutant GEMIN5 iPSC neurons. In neurons harboring GEMIN5H913R, we found that the levels of GEMIN5 mRNA were significantly increased in response to lentiviral EGFP-SMN in both control and mutant GEMIN5 neuronal cells (Sup Fig. 6a-b). Thus, the effect of SMN upregulation on GEMIN5 appears to be at the transcriptional and translational level.
SMN expression alleviates reduced snRNP biogenesis in GEMIN5-mediated disease. a Confocal images of control and mutant GEMIN5 L1068P neurons transduced with EGFP and lentiviral EGFP-SMN from the same set of neuronal differentiations. The neurons were probed for the snRNP marker Y12 (Sm), GFP, and the neuronal marker microtubule-associated protein 2 (MAP2). The cell nuclei were stained with DAPI. Scale bar = 10 µm. b Quantitative analysis observing a significant decrease in nuclear snRNP expression in L1068P neurons after lentiviral transduction of EGFP. Lentiviral transduction of EGFP-SMN significantly rescues the levels of nuclear snRNP expression in L1068P neurons (n = 75–100 neurons, one-way ANOVA w/ Tukey’s multiple comparisons). Error bars indicate S.E.M. *p < 0.05; **p < 0.01; ****p < 0.0001. NS, not significant. c Confocal images of control and mutant GEMIN5 H913R neurons transduced with EGFP and lentiviral EGFP-SMN from the same set of neuronal differentiations. The neurons were probed for the small nuclear ribonucleoprotein interacting partner, Coilin (cajal bodies, CBs), GFP, and GEMIN5. The cell nuclei were stained with DAPI. Scale bar = 10 µm. d Quantitative analysis observing the average number of CBs per cell in control and mutant H913R neuronal cells with EGFP and EGFP-SMN lentiviral expression. Only CBs above 0.2 microns were quantified (n = 60–80 neurons, one-way ANOVA w/ Tukey’s multiple comparisons). Error bars indicate S.E.M. **p < 0.01; NS, not significant. e The percentage of cells with CBs in control and mutant H913R neuronal cells with EGFP and EGFP-SMN expression (n = 60–80 neurons, one-way ANOVA w/ Bonferroni test). Error bars indicate S.E.M. **p < 0.01; NS, not significant. f Representative gel displaying complete in vitro snRNP assembly formation using 3′ Cy3-biotin-labeled U1 snRNA and the cytoplasmic extract from control and L1068P neurons with lentirviral SMN and ASO N1 administration. Untransfected HEK293T cells and HEK293T transfected with GEMIN5 shRNA were used as a positive control for assembly formation. g Representative quantification of 3 biological replicate sample displaying the intensity of the snRNP assembly between groups (n = 3, one-way ANOVA w/ Tukey’s multiple comparisons). Error bars indicate S.E.M. *p < 0.05; **p < 0.01. NS, not significant. h Confocal images of the ventral nerve cord (VNC) of control and ubiquitously expressing GEMIN5 KD Drosophila larvae with and without Smn over expression stained with the snRNP marker Sm (Y12) and nuclei (DAPI). Loss of GEMIN5 in vivo results in a reduction of nuclear Sm which is rescued by upregulation of Smn. Scale bar = 10 µM. i. Confocal images of the ventral nerve cord (VNC) of control and ubiquitously expressing GEMIN5 KD Drosophila larvae with and without Smn over expression stained with the small nuclear ribonucleoprotein interacting partner, dCoilin (CBs) and nuclei (DAPI). Loss of GEMIN5 in vivo results in a drastic reduction of CB formation which is rescued by upregulation of Smn. Scale bar = 10 µM
Additionally, we asked if Nusinersen ASO (ASO N1) exerted similar effects as to lentiviral SMN on the increase of GEMIN5 and SMN expression in mutant GEMIN5L1068P neurons (Fig. 4j). To investigate this, we treated GEMIN5L1068P neurons with 500 nM of ASO N1 and scrambled control ASO and found a significant increase in the expression levels of both GEMIN5 (Fig. 4k) and SMN (Fig. 4l). To assess the efficacy of ASO N1 administration, we performed PCR analysis on the GEMIN5L1068P neurons and found that ASO N1 administration at 500 nM reduced exon 7 SMN2 skipping from 50 to 25% (Sup Fig. 6c-d). This resulted in about a 25% increase in SMN2 mRNA (Sup Fig. 6e), which also resulted in a 25% increase in GEMIN5 mRNA (Sup Fig. 6f). In addition, we assessed the impact of ASO N1 administration on the localization patterns of endogenous SMN and GEMIN5 in mutant GEMIN5L1068P iPSC-derived neuronal cells (Fig. 4m). Interestingly, administration of Cy3 ASO N1 significantly increased the fluorescent intensity of both SMN (Fig. 4n) and GEMIN5 (Fig. 4o) in mutant GEMIN5L1068P iPSC-derived neuronal cells compared to the scrambled Cy3 ASO control. Thus, ASO N1 exerts similar effects as lentiviral SMN in mutant GEMIN5 iPSC-derived neurons and increases the expression of both SMN and GEMIN5 at the mRNA and protein level.
In addition, to examine other possible underlying mechanisms responsible for the increased protein levels of GEMIN5 in response to SMN expression, we compared GEMIN5’s protein stability between mutant GEMIN5L1068P neurons expressing EGFP and EGFP-SMN (Fig. 4p). We assessed the protein expression of GEMIN5 and SMN after 0, 4, 8, 12, and 24 h of cycloheximide (CHX) treatment. We identified a steady decrease in GEMIN5 expression in neurons expressing EGFP control after 2 h of CHX treatment, whereas we observed a higher expression of GEMIN5 up to 8 h after CHX treatment in response to lentiviral SMN compared to controls (Fig. 4q). Likewise, SMN protein levels showed a steady reduction after 2 h of CHX treatment in EGFP expressing controls, whereas a higher expression of SMN was observed up to 8 h after CHX treatment in neurons expressing lentiviral SMN (Fig. 4r).
In patients with SMA, the number of innate copies of SMN2 is considered the most important phenotypic modifier of the disease, as an increase in SMN2 copy number results in a higher expression of functional SMN protein [7, 11, 46, 69]. Our current findings indicate that the levels of endogenous GEMIN5 and SMN present in patients with mutations in GEMIN5 may play a role in the regulation of other SMN complex components and possibly contribute to GEMIN5-mediated neurological phenotypes. To assess the copy numbers of both SMN1 and SMN2 in the unaffected parent and the GEMIN5L1068P patient, we extracted DNA from the patient iPSCs and conducted simplex Real-Time PCR SMN copy-number quantification [44, 61]. Interestingly, we found that both the unaffected parent and mutant GEMIN5L1068P individual harbor 2 copies of both SMN1 and SMN2 (Sup Fig. 7a-b), suggesting the reduction in SMN protein levels in the GEMIN5L1068P individual to be caused by the loss-of-function biallelic mutations in GEMIN5. Overall, these findings indicate that upregulation of SMN influences the levels of GEMIN5 in mutant GEMIN5 iPSCs and may demonstrate that the expression levels of these two SMN complex components are inter-dependent in neuronal cell types.
Lentiviral SMN expression significantly impacts the alternative splicing profile in mutant GEMIN5H913R neurons. a Representative schematic of the five-alternative splicing (AS) event types reported by rMATS. The types of AS events include retained intron (RI), alternative 5’ splice site (A5’SS), alternative 3’ splice site (A3’SS), mutually exclusive exons (MXE), and exon skipping (SE). b The number of significant splicing events with an increased or decreased inclusion per AS category in GEMIN5H913R neurons expressing EGFP control and EGFP-SMN compared to control (FDR < 0.05, ΔΨ > 5%). GEMIN5H913R neurons expressing EGFP-SMN result in a higher number of increased inclusions and reduce the number of decreased inclusions in GEMIN5H913R neurons expressing EGFP control. c Percentage of splicing events per category that resulted in an increased inclusion event between mutant GEMIN5H913R neurons with EGFP and SMN OE compared to control. Mutant GEMIN5H913R neurons with SMN saw a rescue in decreased inclusions and an increased percentage in included AS events (SE, A5SS, A3SS, and RI) compared to mutant GEMIN5H913R neurons with EGFP. d Analysis showing the number of genes that exhibited a significantly improved AS event (increased inclusion) per category between mutant GEMIN5H913R neurons with and without lentiviral SMN expression (FDR < 0.05, ΔΨ > 5%). e Gene ontology (GO)-based annotation of the genes with a significantly improved splicing event after SMN transduction via DAVID (v6.7). The y-axis represents GO annotations of the up-key words. The x-axis represents the Fold Enrichment Score for each GO annotation (FDR < 0.05). f, g, h Sashimi plots of the improved exon skipping events after SMN transduction for the genes (f) FNBP1, (g) EHBP1, and (h) SPTSSB in GEMIN5H913R neurons. The axes represent the counts of RNA-seq reads that span the junctions in each genomic region. Red indicates the GEMIN5H913R neurons with EGFP, while orange indicates the GEMIN5H913R neurons with SMN overexpression. The estimated Ψ values are shown on the right
The Tudor of SMN is required for GEMIN5 interaction and regulation
To identify the domains responsible for GEMIN5 and SMN interaction, we co-transfected HEK293T cells with full-length HA-GEMIN5-WT, HA-WD40 (n-terminus only), HA-∆RBS (deleted c-terminus), HA-∆WD40 (deleted n-terminus), and EGFP-SMN, and tested via co-immunoprecipitation (Fig. 5a). Interestingly, disruption of the c-terminus of GEMIN5 which harbors two RNA-binding sites abolished GEMIN5’s interaction with SMN, suggesting that the functional RNA-binding sites of GEMIN5 are required for efficient interaction with SMN protein (Fig. 5b). To assess if this interaction was RNA dependent, we treated HA-GEMIN5-WT and HA-∆WD40 immunoprecipitated lysates with RNase A. We found that RNase A slightly decreased the interaction between GEMIN5 and SMN, suggesting their interaction to be partially regulated through RNA (Fig. 5c). Likewise, to identify the SMN protein domains required for the interaction with GEMIN5, HA-SMN-WT (full length), HA-SMN-Ex1-5 (lacking exon 7—main product of SMN2 gene), HA-SMN-∆Ex3 (deletion of the SMN Tudor domain), and HA-SMN-∆Ex5 (deletion of the proline-rich region of SMN) were transfected in HEK293T cells and tested by immunoprecipitation (Fig. 5d). Surprisingly, immunoprecipitation with the SMN2 product (HA-SMN-Ex1-5 which lacks E7 of SMN) and deletion of the SMN Tudor domain (HA-SMN-∆Ex3) completely abolished the interaction with GEMIN5 (Fig. 5e-f). Of note, deletion of the SMN Tudor domain may result in expression of non-functional SMN protein, which in turn does not interact with endogenous GEMIN5. To assess if the Tudor domain was also required for SMN regulating GEMIN5 expression, we transfected HEK cells with HA-SMN-WT, HA-SMN-∆Ex5, and HA-SMN-∆Ex3, and analyzed the protein expression of both GEMIN5 and SMN (Fig. 5g). HA-SMN-WT and HA-SMN-∆Ex5 both increased the levels of endogenous SMN, which resulted in an increase in GEMIN5 expression (Fig. 5g). These results suggest that the Tudor domain of SMN is required for efficient GEMIN5 binding and regulation of expression.
SMN expression rescues the loss of splicing machinery and improves alternative splicing defects in GEMIN5-mediated disease
GEMIN5 is an essential component of the SMN complex that is required for biogenesis of the spliceosome snRNP core. Upon final completion of snRNP-core assembly in the cytoplasm, the snRNPs are imported into the nucleus and transiently localize in nuclear body structures termed Cajal Bodies (CBs) [8, 38, 59]. More importantly, disruption of snRNP-core assembly resulting from the loss of SMN complex proteins leads to a destabilized SMN complex and significant disruptions in CB formation [5, 54]. These findings prompted us to ask if mutations in GEMIN5 disrupt snRNP biogenesis and import into CBs. First, we examined the presence of nuclear U snRNPs in GEMIN5L1068P neuronal cells by immunostaining for SmB (Y12) (Fig. 6a). We observed a drastic decrease in nuclear U snRNP expression in GEMIN5L1068P neurons as compared to control. These findings prompted us to ask if lentiviral SMN had any effect on U snRNP expression in the GEMIN5L1068P neurons. Interestingly, we observed a significant increase in nuclear U snRNP in both control and mutant GEMIN5L1068P neurons expressing SMN (Fig. 6b). Since we saw a decrease in imported U snRNPs in mutant GEMIN5 neurons, we also observed for the presence of CBs in mutant GEMIN5H913R neuronal cells (Fig. 6c). GEMIN5H913R neuronal cells expressing EGFP displayed a significant reduction in the average number of CBs per cell (Fig. 6d) and the percentage of neuronal cells that harbored CBs (Fig. 6e) compared to control. Surprisingly, lentiviral expression of SMN restored the loss of CB formation and increased the percentage of neuronal cells harboring CBs in mutant GEMIN5H913R neuronal cells. There appeared to be no obvious localization of GEMIN5 into the CB structures. Next, we wanted to assess if SMN upregulation was sufficient to rescue mature snRNP assembly in GEMIN5L1068P neurons. To investigate this, we reconstituted the snRNP core in vitro using transcribed 3′Cy3-biotinylated-U1snRNA and cytoplasmic extracts from control and mutant GEMIN5L1068P neurons transduced with EGFP-SMN and treated with 500 nM of scramble control and ASO N1 ASO (Fig. 6f). We also used cytoplasmic extract from HEK293T cells transfected with scramble shRNA and GEMIN5 shRNA as a positive control [28]. Assembly formation was drastically reduced in GEMIN5L1068P neurons with the scrambled control ASO compared to control neurons. We observed a significant rescue in assembly formation in GEMIN5L1068P neurons transduced with lentiviral SMN and treated with the ASO N1 (Fig. 6g).
To determine if there are also snRNP-core assembly phenotypes associated with loss of Gemin5 in vivo, we utilized the UAS/gal4 system and used Tubulin-GS-gal4 (GeneSwitch) induced by a mild dose of RU486 to induce ubiquitous expression of Rig KD throughout larval development. First, we dissected Drosophila larval brains of control, Rig KD, and Rig KD Smn overexpressing animals in the ventral nerve cord (VNC) and immunostained for Drosophila U snRNP (dSmb) and cell nuclei (DAPI) (Fig. 6h). Similarly to the mutant GEMIN5 iPSC-derived neurons, we found that dSmB was drastically reduced in Rig KD animals compared to control. We therefore investigated if ectopic expression of Smn rescued dSmB expression in Drosophila brains of Rig KD animals and observed a noticeable rescue in dSmB expression (Fig. 6h). Next, we investigated the expression of Drosophila Coilin in the brains of control, Rig KD, and Rig KD Smn over expressing larvae. We observed a drastic reduction in dCoilin expression and CB formation in Rig KD animals compared to control. Surprisingly, upregulation of Smn restored dCoilin expression in Drosophila brains of Rig KD animals (Fig. 6i). These results suggest that mutations and loss of Gemin5 in vivo disrupt snRNP assembly and upregulation of SMN partially rescues snRNP phenotypes associated with GEMIN5-mediated disease.
The results above suggest that SMN upregulation rescues snRNP defects exhibited in GEMIN5-mediated disease. Since snRNP biogenesis is a key driver of RNA splicing events, we investigated the alternative splicing (AS) profile of mutant GEMIN5H913R neurons with lentiviral expression of EGFP and EGFP-SMN compared to control neurons using rMATS. rMATS calculates the inclusion of a given differentially expressed exon as percent spliced-in (PSI or Ψ) and assesses the fraction of a gene’s mRNA across five main alternative splicing patterns, retained intron (RI), alternative 5' splice site (A5SS), 3' alternative splice site (A3SS), mutually exclusive exon (MXE), and exon skipping (SE) (Fig. 7a, Sup Dataset 1). Using an FDR of < 0.05 and a ΔΨ > 5%, we identified a total of 2,950 significant splicing events (1,758 with a significantly increased Ψ and 1,192 with a significantly decreased Ψ in mutant GEMIN5H913R neurons. Exon skipping represented the largest number of AS events between the two groups with a total of 2,009/2,950 (68.1%) (Fig. 7b). Importantly, after lentiviral expression of SMN in the mutant GEMIN5H913R neurons, we found a higher number of significant AS events with an increased inclusion event per each splicing category compared to controls (Fig. 7b). Additionally, we found a lower number of significant AS events exhibiting a decreased inclusion per splicing category after lentiviral SMN expression in mutant GEMIN5H913R neurons compared to controls (Fig. 7b). Exon skipping resulted in a 7.3% increase in inclusions followed by an increase in A5’SS (5.3%), A3’SS (10.2%), and RI (17.45%) AS events in GEMIN5H913R neurons with SMN vs. control compared to GEMIN5H913R neurons with EGFP vs. control (Fig. 7c). Additionally, Exon skipping resulted in a lower percentage of decreased inclusions (7.4%) followed by a lower percentage of significant A5’SS (5.4%), A3’SS (7.1%), and RI (17.42%) events in GEMIN5H913R neurons with SMN vs. control compared to GEMIN5H913R neurons with EGFP vs. control (Sup Fig. 8). Out of the five AS categories, we identified 919 total genes that exhibited a significant increase in inclusion (or rescued splicing event) with 575 genes of those genes exhibiting rescued exon skipping events after lentiviral SMN (Fig. 7d). Gene ontology analysis was applied to assess the biologically relevant pathways impacted by the genes that exhibited an increase in inclusion after SMN expression. The most affected biological pathways involved DNA synthesis, DNA replication, serine/threonine-protein kinases, WD-repeat domain, nucleotide binding, and transcriptional regulation (Fig. 7e). Collectively, since SMN upregulation exhibited the largest effect on exon skipping, we further analyzed increased inclusion events in this category. We identified three transcripts in this data set that were also differentially spliced in response to biallelic mutations in GEMIN510. Using rmatssashimplot software, we visualized the exon skipping events of these three genes, FNBP1 (Fig. 7f), EHBP1 (Fig. 7g), and SPTSSB (Fig. 7h), and found them all to result in increased exon inclusion after SMN upregulation. Of note, FNBP1 and SPTSSB have recently been implicated in the motor neuron disease amyotrophic lateral sclerosis (ALS) [40, 64] and have not been linked with SMA or other neurodevelopment diseases. Overall, SMN expression significantly restored the inclusion of alternatively spliced isoforms in mutant GEMIN5-derived iPSC neurons.
Discussion
In our current study, we sought to identify novel regulators of GEMIN5-mediated disease by further investigating the association between GEMIN5 and its interacting partners of the SMN complex. GEMIN5 is a part of the SMN complex that is essential for RNA metabolism. The SMN complex members are thought to be highly conserved throughout all metazoans [29]. Utilizing a loss-of-function model of GEMIN5-mediated disease, we conducted a candidate gene screen of Drosophila SMN complex members and identified Survival Motor Neuron protein as a regulator of GEMIN5-mediated neurodegeneration in vivo (Fig. 1). Genetic modulation of Drosophila Smn via overexpression significantly ameliorated the external eye degeneration caused by loss of Gemin5 protein. In addition, we also observed a significant reduction in Smn expression in Gemin5 KD animals. In turn, upregulation of Smn significantly suppressed motor impairments and developmental defects observed in loss of Gemin5 animals, further supporting our findings that SMN acts a positive regulator of GEMIN5-mediated disease.
It is hypothesized that neuronal loss attributed to SMA arises through defects in SMN complex function resulting in incomplete snRNP assembly leading to downstream consequences including a significant decrease in the levels of spliceosomal snRNPs [13, 22, 51, 67]. Recent advancements toward the development of SMN protein-upregulating therapies to suppress snRNP assembly defects have shown remarkable success in ameliorating SMA patient phenotypes [37, 56, 58, 68, 70]. Currently, little is known about how SMN-upregulating therapies directly influence the GEMIN components of the SMN complex. Recent studies have shown that ASO approaches to increase functional SMN protein result in increased expression of GEMIN2, GEMIN6, and GEMIN8 [57, 66]. However, the molecular effect of SMN overexpression on the impact of GEMIN5 has yet to be explored.
To help fill this gap, we examined the expression pattern of endogenous GEMIN5 in HEK293T cells using two methods of SMN upregulation (lentiviral expression of SMN and inclusion of exon 7 SMN2 via ASO N1). We observed that the expression levels of GEMIN5 were significantly increased in response to ectopic expression of SMN at both the mRNA and protein levels (Fig. 2). More importantly, ectopic expression of other SMN complex members, GEMIN2 and GEMIN4, had no effect on the expression levels of GEMIN5. Interestingly, upregulation of SMN, via the FDA-approved Nusinersen ASO, significantly increased the expression of GEMIN5 at the mRNA and protein levels (Fig. 2). These findings agree with the previous conclusions that highlight the interdependence of constituent levels within the members of the SMN complex [30, 54]. Specifically, GEMIN5 has been shown to bind to the mRNA of SMN and regulate its expression, suggesting that these two proteins may autoregulate each other’s levels [71]. In addition, other DNA/RNA-binding proteins other than GEMIN5 have also been shown to influence the levels of SMN in an independent manner. Changes in the expression of ZPR1, PLS3, and CORO1C have all been shown to influence SMN expression and act as disease modifiers of Spinal Muscular Atrophy [15, 24,25,26]. Our findings of increased GEMIN5 expression upon upregulation of SMN prompted us to conduct reciprocal experiments and investigate the levels of GEMIN5 in SMA iPSC-derived motor neurons. In motor neurons derived from SMA patients with loss-of-function mutations in SMN, GEMIN5 was significantly reduced in response to over a 75% decrease in SMN protein. In addition, GEMIN5 exhibited a strong positive correlation with the expression levels of SMN in type 1 and type 2 SMA motor neurons (Fig. 3). These findings suggest a strong associate between both proteins and provide further evidence of SMN being a regulator of GEMIN5 expression.
GEMIN5 is a multifunctional RNA-binding protein that harbors 13 N-terminal tryptophan aspartic (WD) repeat domains, a middle linker dimerization domain, and two C-terminal RNA-binding sites [35]. The WD repeats of GEMIN5 recognize precursors of small nuclear RNA (snRNA) via a specific AU5-6 sequence, so they can be assembled into snRNPs via delivery of the SMN complex [4, 74]. We previously reported individuals with biallelic mutations in GEMIN5 that appear to cause loss-of-function by decreased expression of both GEMIN5 and SMN [28, 48]. We hypothesize that the broad clinical spectrum and variable disease course across patients could be caused by the differences in decreased levels of endogenous GEMIN5 and its interacting partner SMN. Further, we found that mutations in GEMIN5 disrupt the interaction between GEMIN5 and SMN (Sup Fig. 9). This suggests a destabilization of the two proteins leading to decreased expression which may contribute to the phenotypes observed in GEMIN5 patients. Since we observed increased SMN expression regulates GEMIN5 in HEK293T cells, we asked if upregulation of SMN exerted similar effects on GEMIN5 expression in iPSC neurons derived from GEMIN5 patients exhibiting over a 70% decrease in GEMIN5 protein expression. Surprisingly, we observed that lentiviral expression of SMN and upregulation of SMN via ASO N1 both increased the endogenous expression levels of GEMIN5 in mutant GEMIN5 iPSC neuronal cells (Fig. 4). We hypothesize that the increase in GEMIN5 expression resulting from an increase in SMN is due to the presence of the Tudor domain of SMN, as the Tudor domain is required for efficient GEMIN5 binding and upregulation (Fig. 5). Furthermore, the significant increase in GEMIN5 protein levels in the mutant GEMIN5-derived iPSC neurons could be a result of increased GEMIN5 mRNA in response to both methods of SMN upregulation (Sup Fig. 5). It is possible that upregulation of SMN in mutant GEMIN5 iPSC-derived neurons results in a positive impact on the interaction between GEMIN5 and its ability to recruit SMN and other complex members for snRNP biogenesis. Additionally, SMN may exert protective effects against GEMIN5 protein degradation and enhances protein stability, as lentiviral SMN expression increased the half-life of GEMIN5 and SMN protein in mutant GEMIN5-derived iPSC neurons.
Although SMN-mediated suppression of GEMIN5 neurodegeneration is novel and interesting, the molecular mechanism(s) behind these observations remain perplexing. We identified Smn as a genetic modifier of GEMIN5 neuropathologies (Rig KD) in vivo, where Rig RNA expression was genetically disrupted resulting in external eye degeneration and developmental defects. These Rig KD phenotypes were rescued by Smn upregulation and raise multiple possibilities about the potential protective effects of Smn. It is possible that loss-of-function (LOF) GEMIN5 defects are ameliorated in response to restoration of functional Smn levels, as loss of GEMIN5 results in a significant decrease in SMN expression in mutant GEMIN5 iPSC-derived neurons and in flies. Alternatively, SMN expression may be sufficient to bypass GEMIN5 LOF effects and offer beneficial effects through an alternative biological pathway. A more mechanistic understanding of the impact of SMN expression on suppressing GEMIN5-mediated neurodegeneration in vivo needs further exploration for testing of therapeutic interventions.snRNP biogenesis and CB formation strongly contribute to and influence the organization of the human genome, as defects in snRNP core assembly and CB depletion have been shown to perturb RNA splicing kinetics [52, 65]. In the nucleus, harbors one of the key structures termed, CBs, which are the primary site of modification and final biogenesis of newly constructed U snRNPs that are imported into the nucleus from the cytoplasm via the SMN complex. More importantly, individuals with SMA exhibit a significant decrease or complete absence of the main CB marker coilin, which is a key pathological hallmark of SMA [62]. Coilin and SMN interact directly to organize and localize snRNP assembly factors into CBs for final modification [63]. We previously reported that homozygous mutant GEMIN5 variants exhibit a significant reduction in snRNP assembly, resulting in global splicing defects in mutant GEMIN5 iPSC-derived neurons compared to controls [28]. We hypothesized that these deficiencies could be attributed to defects in mature importation of U snRNPs into CBs. To test this hypothesis, we assessed for the presence of U snRNP formation in mutant GEMIN5 iPSC neurons and Gemin5 KD animals in vivo. We observed a significant loss in nuclear Sm expression (U snRNP marker) in both mutant GEMIN5 iPSC neurons and Gemin5 KD larvae that was partially rescued upon expression of SMN. Furthermore, we observed a significant loss in the number of CBs and percentage of neurons harboring CBs in mutant GEMIN5 iPSC neurons and in vivo. Interestingly, ectopic expression of SMN has been previously shown to stimulate the formation of CBs [19, 34]. Thus, we transduced mutant GEMIN5 iPSC neurons with lentiviral SMN and observed a recovery of CBs back to the level of control neurons (Fig. 6). In addition, the loss of CB numbers in neurons or disruption of CB organization may reduce the efficiency of snRNP assembly reactions that occur in these nuclear structures [36, 60], thus leading to a significant disruption of pre-mRNA splicing processes in neurons. Using an in vitro reconstitution approach, we reconstituted the snRNP-core and found that upregulation of SMN significantly rescued snRNP-core assembly formation in mutant GEMIN5 iPSC neurons (Fig. 6). Finally, we investigated the downstream consequences of defective snRNP biogenesis in mutant GEMIN5 neurons. We show that lentiviral expression of SMN significantly influences the alternative splicing defects exhibited in GEMIN5 iPSC neurons, while resulting in a significantly higher proportion of retained splice events (Fig. 7). We document that upregulating low levels of SMN protein in GEMIN5 iPSC neurons and a loss-of-function model of GEMIN5-mediated disease in vivo, restores defective snRNP components and results in a significant reduction of alternative splicing events.
In conclusion, we have demonstrated that SMN may be a positive genetic modifier of GEMIN5 loss-of-function-mediated neurodegeneration. We highlight the novel involvement of investigating SMN-upregulating therapies in the context of other diseases outside of SMA. Our results are the first to show that SMN modifies GEMIN5-mediated neuropathologies and demonstrate that SMN-upregulating therapies in GEMIN5-mediated disease could be an effective therapeutic strategy for restoring the SMN complex defects exhibited by mutant GEMIN5.
Data availability
The data that support the findings of this study are all included throughout the manuscript and available on request from the corresponding author.
Abbreviations
- ALS:
-
Amyotrophic lateral sclerosis
- ASO:
-
Antisense oligonucleotide
- CB:
-
Cajal body
- iPSC:
-
Induced pluripotent stem cell
- SMA:
-
Spinal muscular atrophy
- SMN:
-
Survival motor neuron
References
Anderson EN, Gochenaur L, Singh A, Grant R, Patel K, Watkins S et al (2018) Traumatic injury induces stress granule formation and enhances motor dysfunctions in ALS/FTD models. Hum Mol Genet 27:1366–1381. https://doi.org/10.1093/hmg/ddy047
Anhuf D, Eggermann T, Rudnik-Schoneborn S, Zerres K (2003) Determination of SMN1 and SMN2 copy number using TaqMan technology. Hum Mutat 22:74–78. https://doi.org/10.1002/humu.10221
Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J et al (2006) The SMN complex: an assembly machine for RNPs. Cold Spring Harb Symp Quant Biol 71:313–320. https://doi.org/10.1101/sqb.2006.71.001
Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G (2006) The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell 23:273–279. https://doi.org/10.1016/j.molcel.2006.05.036
Boulisfane N, Choleza M, Rage F, Neel H, Soret J, Bordonne R (2011) Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum Mol Genet 20:641–648. https://doi.org/10.1093/hmg/ddq508
Burghes AH, Beattie CE (2009) Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 10:597–609. https://doi.org/10.1038/nrn2670
Calucho M, Bernal S, Alias L, March F, Vencesla A, Rodriguez-Alvarez FJ et al (2018) Correlation between SMA type and SMN2 copy number revisited: An analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscul Disord 28:208–215. https://doi.org/10.1016/j.nmd.2018.01.003
Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M (1999) The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol 147:715–728. https://doi.org/10.1083/jcb.147.4.715
Castello A, Fischer B, Hentze MW, Preiss T (2013) RNA-binding proteins in Mendelian disease. Trends Genet 29:318–327. https://doi.org/10.1016/j.tig.2013.01.004
Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M et al (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448:151–156. https://doi.org/10.1038/nature05954
Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B (2002) Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet 70:358–368. https://doi.org/10.1086/338627
Francisco-Velilla R, Azman EB, Martinez-Salas E (2019) Impact of RNA-Protein Interaction Modes on Translation Control: The Versatile Multidomain Protein Gemin5. Bioessays 41:e1800241. doi: https://doi.org/10.1002/bies.201800241
Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L (2007) Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One 2:e921. doi: https://doi.org/10.1371/journal.pone.0000921
Garbes L, Heesen L, Holker I, Bauer T, Schreml J, Zimmermann K et al (2013) VPA response in SMA is suppressed by the fatty acid translocase CD36. Hum Mol Genet 22:398–407. https://doi.org/10.1093/hmg/dds437
Genabai NK, Kannan A, Ahmad S, Jiang X, Bhatia K, Gangwani L (2017) Deregulation of ZPR1 causes respiratory failure in spinal muscular atrophy. Sci Rep 7:8295. https://doi.org/10.1038/s41598-017-07603-z
Gleixner AM, Verdone BM, Otte CG, Anderson EN, Ramesh N, Shapiro OR et al (2022) NUP62 localizes to ALS/FTLD pathological assemblies and contributes to TDP-43 insolubility. Nat Commun 13:3380. https://doi.org/10.1038/s41467-022-31098-6
Gubitz AK, Mourelatos Z, Abel L, Rappsilber J, Mann M, Dreyfuss G (2002) Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J Biol Chem 277:5631–5636. https://doi.org/10.1074/jbc.M109448200
Guo W, Naujock M, Fumagalli L, Vandoorne T, Baatsen P, Boon R et al (2017) HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat Commun 8:861. https://doi.org/10.1038/s41467-017-00911-y
le Hao T, Fuller HR, le Lam T, Le TT, Burghes AH, Morris GE (2007) Absence of gemin5 from SMN complexes in nuclear Cajal bodies. BMC Cell Biol 8:28. https://doi.org/10.1186/1471-2121-8-28
Heesen L, Peitz M, Torres-Benito L, Holker I, Hupperich K, Dobrindt K et al (2016) Plastin 3 is upregulated in iPSC-derived motoneurons from asymptomatic SMN1-deleted individuals. Cell Mol Life Sci 73:2089–2104. https://doi.org/10.1007/s00018-015-2084-y
Irwin M, Tare M, Singh A, Puli OR, Gogia N, Riccetti M et al (2020) A Positive Feedback Loop of Hippo- and c-Jun-Amino-Terminal Kinase Signaling Pathways Regulates Amyloid-Beta-Mediated Neurodegeneration. Front Cell Dev Biol 8:117. https://doi.org/10.3389/fcell.2020.00117
Jablonka S, Holtmann B, Meister G, Bandilla M, Rossoll W, Fischer U et al (2002) Gene targeting of Gemin2 in mice reveals a correlation between defects in the biogenesis of U snRNPs and motoneuron cell death. Proc Natl Acad Sci U S A 99:10126–10131. https://doi.org/10.1073/pnas.152318699
Jin W, Wang Y, Liu CP, Yang N, Jin M, Cong Y et al (2016) Structural basis for snRNA recognition by the double-WD40 repeat domain of Gemin5. Genes Dev 30:2391–2403. https://doi.org/10.1101/gad.291377.116
Kannan A, Bhatia K, Branzei D, Gangwani L (2018) Combined deficiency of Senataxin and DNA-PKcs causes DNA damage accumulation and neurodegeneration in spinal muscular atrophy. Nucleic Acids Res 46:8326–8346. https://doi.org/10.1093/nar/gky641
Kannan A, Cuartas J, Gangwani P, Branzei D, Gangwani L (2022) Mutation in senataxin alters the mechanism of R-loop resolution in amyotrophic lateral sclerosis 4. Brain 145:3072–3094. https://doi.org/10.1093/brain/awab464
Kannan A, Jiang X, He L, Ahmad S, Gangwani L (2020) ZPR1 prevents R-loop accumulation, upregulates SMN2 expression and rescues spinal muscular atrophy. Brain 143:69–93. https://doi.org/10.1093/brain/awz373
Kapeli K, Martinez FJ, Yeo GW (2017) Genetic mutations in RNA-binding proteins and their roles in ALS. Hum Genet 136:1193–1214. https://doi.org/10.1007/s00439-017-1830-7
Kour S, Rajan DS, Fortuna TR, Anderson EN, Ward C, Lee Y et al (2021) Loss of function mutations in GEMIN5 cause a neurodevelopmental disorder. Nat Commun 12:2558. https://doi.org/10.1038/s41467-021-22627-w
Lanfranco M, Cacciottolo R, Borg RM, Vassallo N, Juge F, Bordonne R et al (2017) Novel interactors of the Drosophila Survival Motor Neuron (SMN) Complex suggest its full conservation. FEBS Lett 591:3600–3614. https://doi.org/10.1002/1873-3468.12853
Lanfranco M, Vassallo N, Cauchi RJ (2017) Spinal Muscular Atrophy: From Defective Chaperoning of snRNP Assembly to Neuromuscular Dysfunction. Front Mol Biosci 4:41. https://doi.org/10.3389/fmolb.2017.00041
Lanson NA Jr, Maltare A, King H, Smith R, Kim JH, Taylor JP et al (2011) A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum Mol Genet 20:2510–2523. https://doi.org/10.1093/hmg/ddr150
Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L et al (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155–165. https://doi.org/10.1016/0092-8674(95)90460-3
Liu Q, Fischer U, Wang F, Dreyfuss G (1997) The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 90:1013–1021. https://doi.org/10.1016/s0092-8674(00)80367-0
Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Soderberg O, Stromblad S, Wiman KG, Farnebo M (2010) WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol 8:e1000521. doi: https://doi.org/10.1371/journal.pbio.1000521
Martinez-Salas E, Embarc-Buh A, Francisco-Velilla R (2020) Emerging Roles of Gemin5: From snRNPs Assembly to Translation Control. Int J Mol Sci 21. doi: https://doi.org/10.3390/ijms21113868
Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK (2009) Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell 17:639–647. https://doi.org/10.1016/j.devcel.2009.10.017
Mercuri E, Pera MC, Scoto M, Finkel R, Muntoni F (2020) Spinal muscular atrophy - insights and challenges in the treatment era. Nat Rev Neurol 16:706–715. https://doi.org/10.1038/s41582-020-00413-4
Morris GE (2008) The Cajal body. Biochim Biophys Acta 1783:2108–2115. https://doi.org/10.1016/j.bbamcr.2008.07.016
Moses K, Rubin GM (1991) Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev 5:583–593. https://doi.org/10.1101/gad.5.4.583
Nakamura R, Misawa K, Tohnai G, Nakatochi M, Furuhashi S, Atsuta N et al (2020) A multi-ethnic meta-analysis identifies novel genes, including ACSL5, associated with amyotrophic lateral sclerosis. Commun Biol 3:526. https://doi.org/10.1038/s42003-020-01251-2
Nussbacher JK, Tabet R, Yeo GW, Lagier-Tourenne C (2019) Disruption of RNA Metabolism in Neurological Diseases and Emerging Therapeutic Interventions. Neuron 102:294–320. https://doi.org/10.1016/j.neuron.2019.03.014
Otter S, Grimmler M, Neuenkirchen N, Chari A, Sickmann A, Fischer U (2007) A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J Biol Chem 282:5825–5833. https://doi.org/10.1074/jbc.M608528200
Panopoulos AD, Ruiz S, Yi F, Herrerias A, Batchelder EM, Izpisua Belmonte JC (2011) Rapid and highly efficient generation of induced pluripotent stem cells from human umbilical vein endothelial cells. PLoS One 6:e19743. doi: https://doi.org/10.1371/journal.pone.0019743
Passon N, Pozzo F, Molinis C, Bregant E, Gellera C, Damante G et al (2009) A simple multiplex real-time PCR methodology for the SMN1 gene copy number quantification. Genet Test Mol Biomarkers 13:37–42. https://doi.org/10.1089/gtmb.2008.0084
Pineiro D, Fernandez-Chamorro J, Francisco-Velilla R, Martinez-Salas E (2015) Gemin5: A Multitasking RNA-Binding Protein Involved in Translation Control. Biomolecules 5:528–544. https://doi.org/10.3390/biom5020528
Prior TW, Krainer AR, Hua Y, Swoboda KJ, Snyder PC, Bridgeman SJ et al (2009) A positive modifier of spinal muscular atrophy in the SMN2 gene. Am J Hum Genet 85:408–413. https://doi.org/10.1016/j.ajhg.2009.08.002
Raj A, Chimata AV, Singh A (2020) Motif 1 Binding Protein suppresses wingless to promote eye fate in Drosophila. Sci Rep 10:17221. https://doi.org/10.1038/s41598-020-73891-7
Rajan DS, Kour S, Fortuna TR, Cousin MA, Barnett SS, Niu Z, Babovic-Vuksanovic D, Klee EW, Kirmse B, Innes M, Rydning SL, Selmer KK, Vigeland MD, Erichsen AK, Nemeth AH, Millan F, DeVile C, Fawcett K, Legendre A, Sims D, Schnekenberg RP, Burglen L, Mercier S, Bakhtiari S, Martinez-Salas E, Wigby K, Lenberg J, Friedman JR, Kruer MC, Pandey UB (2022) Autosomal Recessive Cerebellar Atrophy and Spastic Ataxia in Patients With Pathogenic Biallelic Variants in GEMIN5. Front Cell Dev Biol 10:783762. doi: https://doi.org/10.3389/fcell.2022.783762
Ramesh N, Daley EL, Gleixner AM, Mann JR, Kour S, Mawrie D et al (2020) RNA dependent suppression of C9orf72 ALS/FTD associated neurodegeneration by Matrin-3. Acta Neuropathol Commun 8:177. https://doi.org/10.1186/s40478-020-01060-y
Saida K, Tamaoki J, Sasaki M, Haniffa M, Koshimizu E, Sengoku T et al (2021) Pathogenic variants in the survival of motor neurons complex gene GEMIN5 cause cerebellar atrophy. Clin Genet 100:722–730. https://doi.org/10.1111/cge.14066
Sapaly D, Delers P, Coridon J, Salman B, Letourneur F, Dumont F et al (2020) The Small-Molecule Flunarizine in Spinal Muscular Atrophy Patient Fibroblasts Impacts on the Gemin Components of the SMN Complex and TDP43, an RNA-Binding Protein Relevant to Motor Neuron Diseases. Front Mol Biosci 7:55. https://doi.org/10.3389/fmolb.2020.00055
Sawyer IA, Dundr M (2016) Nuclear bodies: Built to boost. J Cell Biol 213:509–511. https://doi.org/10.1083/jcb.201605049
Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN et al (2014) rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci U S A 111:E5593–E5601. https://doi.org/10.1073/pnas.1419161111
Shpargel KB, Matera AG (2005) Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci U S A 102:17372–17377. https://doi.org/10.1073/pnas.0508947102
Singh A, Chan J, Chern JJ, Choi KW (2005) Genetic interaction of Lobe with its modifiers in dorsoventral patterning and growth of the Drosophila eye. Genetics 171:169–183. https://doi.org/10.1534/genetics.105.044180
Singh NN, Howell MD, Androphy EJ, Singh RN (2017) How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther 24:520–526. https://doi.org/10.1038/gt.2017.34
Singh NN, Lawler MN, Ottesen EW, Upreti D, Kaczynski JR, Singh RN (2013) An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy. Nucleic Acids Res 41:8144–8165. https://doi.org/10.1093/nar/gkt609
Singh RN, Seo J, Singh NN (2020) RNA in spinal muscular atrophy: therapeutic implications of targeting. Expert Opin Ther Targets 24:731–743. https://doi.org/10.1080/14728222.2020.1783241
Sleeman JE, Ajuh P, Lamond AI (2001) snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci 114:4407–4419
Smolinski DJ, Wrobel B, Noble A, Zienkiewicz A, Gorska-Brylass A (2011) Periodic expression of Sm proteins parallels formation of nuclear Cajal bodies and cytoplasmic snRNP-rich bodies. Histochem Cell Biol 136:527–541. https://doi.org/10.1007/s00418-011-0861-8
Stabley DL, Harris AW, Holbrook J, Chubbs NJ, Lozo KW, Crawford TO et al (2015) SMN1 and SMN2 copy numbers in cell lines derived from patients with spinal muscular atrophy as measured by array digital PCR. Mol Genet Genomic Med 3:248–257. https://doi.org/10.1002/mgg3.141
Tapia O, Bengoechea R, Palanca A, Arteaga R, Val-Bernal JF, Tizzano EF et al (2012) Reorganization of Cajal bodies and nucleolar targeting of coilin in motor neurons of type I spinal muscular atrophy. Histochem Cell Biol 137:657–667. https://doi.org/10.1007/s00418-012-0921-8
Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ et al (2001) Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol 154:293–307. https://doi.org/10.1083/jcb.200104083
Waegaert R, Dirrig-Grosch S, Parisot F, Keime C, Henriques A, Loeffler JP, Rene F (2020) Longitudinal transcriptomic analysis of altered pathways in a CHMP2B(intron5)-based model of ALS-FTD. Neurobiol Dis 136:104710. doi: https://doi.org/10.1016/j.nbd.2019.104710
Wang Q, Sawyer IA, Sung MH, Sturgill D, Shevtsov SP, Pegoraro G et al (2016) Cajal bodies are linked to genome conformation. Nat Commun 7:10966. https://doi.org/10.1038/ncomms10966
Winkelsas AM, Grunseich C, Harmison GG, Chwalenia K, Rinaldi C, Hammond SM et al (2021) Targeting the 5’ untranslated region of SMN2 as a therapeutic strategy for spinal muscular atrophy. Mol Ther Nucleic Acids 23:731–742. https://doi.org/10.1016/j.omtn.2020.12.027
Winkler C, Eggert C, Gradl D, Meister G, Giegerich M, Wedlich D et al (2005) Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev 19:2320–2330. https://doi.org/10.1101/gad.342005
Wirth B (2021) Spinal Muscular Atrophy: In the Challenge Lies a Solution. Trends Neurosci 44:306–322. https://doi.org/10.1016/j.tins.2020.11.009
Wirth B, Brichta L, Schrank B, Lochmuller H, Blick S, Baasner A et al (2006) Mildly affected patients with spinal muscular atrophy are partially protected by an increased SMN2 copy number. Hum Genet 119:422–428. https://doi.org/10.1007/s00439-006-0156-7
Wirth B, Karakaya M, Kye MJ, Mendoza-Ferreira N (2020) Twenty-Five Years of Spinal Muscular Atrophy Research: From Phenotype to Genotype to Therapy, and What Comes Next. Annu Rev Genomics Hum Genet 21:231–261. https://doi.org/10.1146/annurev-genom-102319-103602
Workman E, Kalda C, Patel A, Battle DJ (2015) Gemin5 Binds to the Survival Motor Neuron mRNA to Regulate SMN Expression. J Biol Chem 290:15662–15669. https://doi.org/10.1074/jbc.M115.646257
Xu C, Ishikawa H, Izumikawa K, Li L, He H, Nobe Y et al (2016) Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly. Genes Dev 30:2376–2390. https://doi.org/10.1101/gad.288340.116
Yong J, Golembe TJ, Battle DJ, Pellizzoni L, Dreyfuss G (2004) snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol Cell Biol 24:2747–2756. https://doi.org/10.1128/MCB.24.7.2747-2756.2004
Yong J, Kasim M, Bachorik JL, Wan L, Dreyfuss G (2010) Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol Cell 38:551–562. https://doi.org/10.1016/j.molcel.2010.03.014
Acknowledgements
The authors would like to thank Dr. Joseph Gall for gifting the Drosophila anti-coilin antibody. A.S. is supported by 1RO1EY032959-01 from NIH, Schuellein Chair Endowment Fund, and STEM Catalyst Grant from the University of Dayton.
Funding
This work was supported by the Children’s Neuroscience Institute Research grant (D.S.R. and U.B.P.), Children’s Hospital of Pittsburgh of the UPMC Health system (T.R.F.), and by the German Research Foundation [Wi 945/17–1 (ID 398410809), Wi 945/19–1 (ID 417989143), SFB1451 (ID 431549029), and GRK1960 (ID 233886668)], and the European Union’s Horizon 2020 Marie Skłodowska-Curie [ID 956185 (SMABEYOND] (B.W.).
Author information
Authors and Affiliations
Contributions
Design of this study: T.R.F, S.K., A.V.C., A.M.B., E.N.A., A.S., and U.B.P.; bioinformatics and statistical analysis: T.R.F, S.K., O.C., C.O’B., and D.R.; wrote the manuscript: T.R.F., S.K., and U.B.P.; critically read and edited the manuscript: T.R.F., S.K., A.M.B., C.W., B.W., A.S., and U.B.P. All authors reviewed and approved the manuscript prior to submission.
Corresponding author
Ethics declarations
Conflict of interest
The authors report no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fortuna, T.R., Kour, S., Chimata, A.V. et al. SMN regulates GEMIN5 expression and acts as a modifier of GEMIN5-mediated neurodegeneration. Acta Neuropathol 146, 477–498 (2023). https://doi.org/10.1007/s00401-023-02607-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-023-02607-8