Abstract
Purpose
Numerous studies have found that probiotics benefit the intestinal barrier. However, the prophylactic effects of probiotics on the intestinal barrier, i.e., if probiotics exert protective effects in healthy individuals to defend them against harmful elements, have seldomly been reported. The present study aimed to investigate the possible mechanisms of potential strains with the function of preventing intestinal barrier damage.
Methods
This study investigated nine potential probiotic strains using in vitro and in vivo models on their intestinal barrier-protecting properties. Transcriptomic was then employed to decipher the underlying mechanisms of action of the strains.
Results
The results showed that the strains, to varying degrees, regulated the ratio of interleukin (IL)-10 and IL-12 in peripheral blood mononuclear cells (PBMCs), increased the transepithelial electrical resistance (TEER) values, and decreased Caco-2 cell monolayers permeability. Correspondingly, the strains showed different prophylactic efficacies in protecting mice from dextran sulfate sodium (DSS)-induced intestinal barrier damage. Remarkably, Bifidobacterium bifidum FL-228.1 (FL-228.1) showed the best prophylactic efficacies in protecting mice from DSS-induced intestinal barrier damage. Further research suggested that FL-228.1 exerted its prophylactic effects by enhancing mucin 2 (Muc2) production and Claudin (Cldn)-4 in the colon. Furthermore, the transcriptomic and protein-protein interactions (PPI) analyses indicated that the inhibition of NLRP3 and the activation of PPARγ and TLR2 could be involved in protecting the intestinal barrier by FL-228.1.
Conclusion
Bifidobacterium bifidum FL-228.1 may be developed as a promising probiotic for the prevention of intestinal barrier damage via PPARγ/NLRP3/ TLR2 pathways by enhancing Muc2 and Cldn-4.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The intestinal barrier is a dynamic entity mainly consisting of epithelial cells, the mucus layer, immune cells in the lamina propria, and commensal bacteria in the lumen [1]. The intestinal barrier function is essential in protecting the host from the invasion of the commensal bacteria or other harmful pathogens in the lumen. Impairment of the intestinal barrier increases the risk of intestinal diseases and metabolic diseases such as inflammatory bowel disease (IBD) [2], necrotizing enterocolitis [3], obesity [4], and diabetic kidney disease [5]. Therefore, the intestinal barrier has been considered a promising prophylactic and therapeutic target in treating intestinal conditions such as IBD [6]. Particularly, tight junction (TJ) proteins, which connect the epithelial cells, and the mucus, are two crucial factors of intestinal barrier function. Altered TJs morphological complexity and a decrease in its content could lead to increased intestinal permeability, which could permit infiltration of pro-inflammatory molecules, contributing to inflammation and tissue damage [7]. The mucus, mainly composed of Mucin 2 (Muc2), plays a vital role in preventing commensal microbes and other pathogens from reaching the epithelium. Animal studies have shown that Muc2 deficiency was prone to colonic inflammation and could maintain the damaging effects of inflammation during colitis [8]. Mice lacking Muc2 have altered intestinal flora and increased susceptibility to dextran sulfate sodium (DSS)-induced colitis [9]. Furthermore, human studies have found that the decrease of Muc2 contributes to the pathogenesis of ulcerative colitis (UC) [10].
Probiotics have been proven to protect or restore the intestinal barrier by regulating TJs, mucus production, and immune systems [11]. Probiotics can repair intestinal barrier damage by inhibiting the DSS-induced decrease in TJs content and the trinitrobenzene sulfonic (TNBS)-induced decrease in zonula occludens (ZO)-1, Occludin and claudin (Cldn)-5 [12, 13]. Studies have also shown that probiotics could directly alter epithelial barrier function by affecting the structure of TJs, such as preventing the reduction of TJs phosphorylation without significant changes in total levels [14]. Meanwhile, probiotics can enhance epithelial barrier function by increasing the secretion of Muc2 [15, 16]. As for the involved pathways, studies showed that probiotics enhanced the activation of AMP-activated protein kinase (AMPK) and inhibited nuclear factor-κB (NF-κB) activation in regulating TJs by stimulating toll-like receptor 2 (TLR2) [17, 18]. The well-known Muc2 regulation pathways are the TLR pathways and the mitogen-activated protein kinase (MAPK) signaling pathways, including c-Jun N-terminal kinase (JNK) and NF-κB [16, 19]. In addition, upregulation of the expression of atonal homolog 1 (Atoh1) [20] and activation of the EGF receptor (EGFR) [21] in intestinal epithelial cells were also found to be associated with increased Muc2 expression. However, as probiotics have strain-specific traits, the mechanisms of their actions are not entirely understood [22]. For example, Archana Chandran et al. [15] showed that the Muc2 was differentially expressed in the host colon under the same conditions stimulated by two Lactobacillus plantarum strains, but the mechanism was not elucidated. Although numerous studies have demonstrated that probiotics could enhance intestinal barrier function, most studies have focused on the therapeutic effects or the efficacies of probiotics in restoring the injured intestine. Few studies have investigated the prophylactic roles that probiotics play in enhancing the barrier function to defend against possible damages, let alone the involved mechanisms.
This study screened probiotics for preventing intestinal barrier damage by combining in vitro and in vivo approaches. Furthermore, the involved genes and pathways in the preventive effect of the potential probiotics were further analyzed by transcriptomics and verified by quantitative PCR. This study may provide scientific guidance for further use of probiotics to prevent intestinal barrier damage and inflammation.
Materials and methods
Bacteria and pretreatment
Strains used in this study including Bifidobacterium bifidum FL-228.1 (FL-228.1), Lacticaseibacillus rhamnosus MN45 (MN45), Lacticaseibacillus paracei K14 (K14), Lacticaseibacillus paracei M5 (M5), Lacticaseibacillus paracei YZX38 (YZX38), Lacticaseibacillus casei YRL577 (YRL577), Lacticaseibacillus casei K11 (K11), and Lactiplantibacillus plantarum YZX21 (YZX21) were stored in the Functional Dairy and Probiotics Engineering Laboratory of Ocean University of China and reported in our previous study [23,24,25,26]. Bifidobacterium animalis subsp. lactis BB12 (BB12, isolated from a commercial probiotic powder purchased from Puractive) was used as a reference strain as it is a widely used probiotic and shows protective effects on the intestine [27]. The strains were subcultured in De Man, Rogosa, Sharpe (MRS) for 48 h at 37 ℃ anaerobically. Then, the bacteria were collected by centrifugation (6000 r/min, 15 min, 4 ℃) and washed twice with precooling phosphate-buffered solution (PBS) and then re-suspended in the Dulbecco’s modified Eagle’s medium (DMEM; Solarbio) for in vitro experiments (2 × 107 CFU/mL) or PBS for in vivo experiments (1 × 108 CFU/mL).
Isolation and treatment of peripheral blood mononuclear cells (PBMCs)
Human venous blood was obtained from four healthy informed donors upon approved agreement by authorized medical staff, and PBMCs were isolated according to previous reports [28, 29]. Then, cells were cultured in a humidified incubator in the Roswell Park Memorial Institute 1640 (RPMI1640) medium containing 10% heat-inactivated fetal bovine serum (FBS, purchased from Gibco) under an atmosphere of 5% CO2 in air at 37 °C. PBMCs were incubated in 24-well plates with a final concentration of 1 × 106 cells/mL in the presence or absence of live-bacterial cell suspensions (1 × 107 CFU/mL) at 37 °C in 5% CO2 for 48 h. The levels of interleukin (IL)-10 and interleukin (IL)-12 in the culture supernatant were measured using enzyme-linked immunosorbent assays (ELISA, Absin, Shanghai, China).
Caco-2 cells culturing
The Caco-2 cells were purchased from the Type culture collection cell bank of the Chinese Academy of Science (Shanghai, China). The determination of intestinal permeability was performed according to previous studies [30]. Caco-2 cells (2 × 105 cells/cm2) were seeded in 12-well cell culture chambers (LABSELECT, China) on Polyester membrane filters (pore size 0.4 μm, surface area 1.12 cm2), and the medium (DMEM; Solarbio; containing 10% fetal bovine serum, 100 U/mL penicillin and 100 U/mL streptomycin) was changed every other day. Every two days, the transepithelial electric resistance (TEER) was measured using a transmembrane cell resistance meter (Kingtech, Beijing, China). After the TEER value of the Caco-2 monolayers reached 400 Ω cm2, the Caco-2 monolayers were washed twice with PBS and incubated with or without the strain suspensions prepared above in the apical part of the insert for 24 h. TEER values were measured before and after the intervention of strains. Afterward, Fluorescein Isothiocyanate-dextran (FITC-dextran) (1 mg/mL FD4; Sigma) was added to the apical medium of the Caco-2 monolayers for 6 h of incubation, and then the fluorescence intensity of the bottom medium was measured at 480 nm excitation wavelength and 520 nm emission wavelength.
Animal experiment
A total of 60 specific pathogen-free (SPF) female BALB/C mice (6 weeks old, purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd.) were acclimated to the environment for 7 days (24 ± 1 ℃ temperature, 50% ± 10% relative humidity). All animal protocols used were pre-approved by the Laboratory Animal Ethics Committee of the College of Food Science and Engineering of Ocean University of China (permission number: SPXY2021112601). The mice were randomly divided into five groups (n = 12) after the following acclimation: the control group (Control), the post-treatment group (Post), and the strains-treatment groups (MN45, K11, and FL-228.1). MN45, K11, and FL-228.1 received 0.2 mL bacterial suspension (1 × 108 CFU/mL) via oral gavage, respectively, while Control and Post groups received PBS solution (0.2 mL). After 3 weeks [31], the oral gavage was stopped, and six mice from each group were killed after ether anesthesia [32] for the following measurements and transcriptomic analysis. Subsequently, except Control group, the remaining mice in the other groups were administered 2% (w/v) DSS (molecular weight, 30–50 kDa; YEASEN, Shanghai, China) dissolved in drinking water for 7 days [33].
Determination of indicators of inflammation and intestinal permeability
The serum of the mice was separated by centrifugation (4 ℃, 8000 r/min, 10 min). The levels of tumor necrosis factor-α (TNF-α), IL-10, IL-12, lipopolysaccharide (LPS), and D-lactic acid (D-LA) in the serum were determined using ELISA kits (Calvin, Suzhou, China) according to the manufacturer's instructions.
Histological evaluation
The colon length was measured, and a portion of the colon tissue was taken and fixed with 4% formalin. Hematoxylin and eosin (H&E) staining were performed according to standard methods. Goblet cells were stained with Alcian Blue and Periodic Acid-Schiff (AB-PAS) as directed by the manufacturer, and goblet cells were counted using Image J according to previous reports [34, 35].
Muc2 determination by immunofluorescence in mice colon
Colon tissue slides were deparaffinized and rehydrated. Tissue sections were antigenically repaired with ethylene diamine tetraacetic acid (EDTA) antigen retrieval buffer (pH 8.0) and then washed three times with PBS (pH 7.4). The blocking solution was added to cover the marked tissue. The Muc2 primary antibody (1:100 dilution, Servicebio) was added to the tissue sections and incubated overnight at 4 °C. Afterwards, the corresponding secondary antibody (1: 3000 dilution, Servicebio) was added to the tissue sections and incubated at room temperature for 50 min in dark condition. 4,6-Diamidino-2-phenylindole (DAPI) was added dropwise to the section for counterstaining, and after that, a spontaneous fluorescence quenching reagent was added to the section. The quantitative immunofluorescence staining analysis was performed using Image J software [36].
Transcriptome analysis
The total RNA of the colon of mice from the FL-228.1 (mice not administered DSS) and Control groups was extracted by TRIzol reagent. The purity and concentrations of the obtained RNA were determined using NanoDrop2000. The integrity of the RNA was detected by agarose gel electrophoresis. The Illumina Truseq™ RNA Sample Prep Kit was used to construct libraries, and the sequencing of these libraries was completed on the Illumina HiSeq platform. Gene Ontology (GO) functional enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis, and network of protein–protein interactions (PPI) were performed with the online Majorbio Cloud Platform [37].
Quantitative real-time PCR
A total RNA extraction kit (Solarbio, Beijing, China) was used to extract total RNA according to the instructions. RNA extraction from colon tissue of the mice was first performed by adding 1 mL of lysate and two grinding beads to make the tissue fully ground on the Tissue Grinder (Servicebio, Wuhan, China). After the concentration determination and normalization, RNA was reversely transcribed into cDNA using a reverse transcription kit (Toyobo, Shanghai, China). Specific primer sequences are shown in Table 1. Using Gapdh as a reference gene [3], the mRNA expressions were determined by qPCR with a CFX96 Realtime PCR system (Bio-rad Co., Hercules, CA, USA).
Statistical analysis
IBM SPSS Statistics 23 was used for statistical analysis, and one-way analysis of variance (ANOVA) followed by a Tukey–Kramer posttest was used to analyze the differences between the groups. A two-tailed unpaired Student’s t test was employed to compare gene expressions between Control and FL-228.1 group in the validation of transcriptome analysis. When P < 0.05, significant differences were found. Data were combined from at least three independent experiments unless otherwise stated. Data were expressed as mean ± SEM.
Results
The strains showed varying abilities to regulate cytokines
The PBMCs were co-incubated with the strains for 24 h, and the levels of IL-10 and IL-12 were determined (Fig. 1). Compared with Control group, different strains altered the secretion of IL-10 and IL-12 to varying degrees. The FL-228.1, K11, and BB12 groups significantly increased (P < 0.05) the IL-10/IL-12 value compared with Control group (Fig. 1C), indicating that different strains differed in their abilities to regulate cytokine secretion and immune balance. Notably, the reference strain BB12 showed the highest IL-10/IL-12 value, followed by the strains FL-228.1 and K11, but strain FL-228.1 was better than K11.
Cytokines regulation effects of strains in vitro. Levels of anti-inflammatory cytokines IL-10 (A), inflammatory cytokines IL-12 (B), and cytokines IL-10/IL-12 (C) in the supernatant of PBMC cells induced by strains (1 × 107 CFU/mL). Data were presented as mean ± SD of three independent experiments. a–f means with different superscript letters are significant differences (P < 0.05)
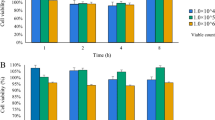
The strains could reduce the permeability of Caco-2 monolayers
As shown in Fig. 2, the YRL577, FL-228.1, BB12, and MN45 groups could significantly increase (P < 0.05) TEER value of the Caco-2 monolayers (Fig. 2A), and all strains significantly decreased (P < 0.05) FITC-dextran transmittance (Fig. 2B) compared with Control group. By combining these two indicators, strains YRL577, FL-228.1, and MN45 showed better or similar performances compared to the reference strain BB12 in reducing the permeability of the monolayers. Therefore, strains FL-228.1, YRL577, and MN45 were considered to have the potential to enhance intestinal barrier function.
Epithelial barrier protection effects of strains in vitro. TEER (A) and FITC-dextran (B) values of Caco-2 monolayers under the intervention of strains (1 × 107 CFU/mL), expressed as % change of the control. Data were presented as mean ± SD of three independent experiments. a–h means with different superscript letters are significant differences (P < 0.05)
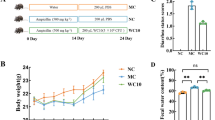
Specific strains showed prophylactic effects on DSS-induced damage in mice
The animal experiment procedure is shown in Fig. 3A. After 7 days of DSS induction, the weight loss rate in the Post group was significantly (P < 0.05) higher than that in Control group. The strains treatment could ameliorate the weight loss, especially the FL-228.1 and K11 (Fig. 3B). Overall, the disease activity index (DAI) scores [38] in FL-228.1, K11, and MN45 groups were significantly lower than that of the Post group (Fig. 3C). Remarkably, the colon length shortening was suppressed considerably by FL-228.1 (Fig. 3D, E). These results indicated that pre-administration with the strains, especially FL-228.1, alleviated the symptoms of DSS-induced damage in the mice intestine, showing significant prophylactic efficacies. Meanwhile, there were no significant changes in serum cytokines except the IL-12 in the MN45 group (Fig. 3F–H) after the intragastric administration for 21 days in the mice. However, intriguingly, the strains prevented, to varying degrees, the imbalance between the pro-inflammatory and anti-inflammatory cytokines caused by the DSS administration by inhibiting the decrease of IL-10 and the increase of TNF-α in mice serum. The mucosal layers and morphological changes of colon tissues were assessed by AB-PAS and H&E staining. The crypt status of the Post group showed extensive atrophy, and the mucous layer was destroyed seriously with goblet cell depletion and inflammatory cell infiltration. Nevertheless, the pre-administration of the strains inhibited DSS-induced damage to the mucous layer to varying degrees (Fig. 3I–K). Noticeably, the pre-administration of the strains, especially FL-228.1, showed promising anti-inflammatory and intestinal damage prevention effects in vivo.
Preventive effect of strains on DSS-induced damage. The exact method of administration (A); Change in body weight (B); DAI score (C); Macroscopic pictures of colons (D); Colon length (E); IL-10 (F), IL-12 (G) and TNF-α (H) in the supernatant of mice; AB-PAS staining of colonic tissue (I); H&E staining of colonic tissue (J); Number of goblet cells (K). Data were presented as mean ± SD (n = 6). *P < 0.05, **P < 0.01, ***P < 0.001
The strains protected intestinal barrier function from DSS damage
LPS and D-lactic acid (D-LA) levels in the blood were used to assess intestinal permeability. As shown in Fig. 4A, LPS concentration was significantly reduced in FL-228.1, K11, and MN45 groups compared with the Post group. The D-LA concentration showed similar results except the K11 group (Fig. 4B). The mRNA expressions of the intestinal barrier-associated proteins and cytokine including Muc2, ZO-1, Occludin, and IL-10 in the colon tissue were measured. The results showed that DSS significantly decreased their expressions, whereas the strains mitigated the decrease (Fig. 4D–F). Consistently, the strain FL-228.1 showed the best efficacy. Furthermore, in line with the levels of gene expressions, the secretion of Muc2 protein in the FL-228.1 group was less affected by DSS induction (Fig. 4G, H). Markedly, the strain FL-228.1 exhibited better protection of the intestinal barrier.
Strains reduced intestinal permeability and improved barrier function. LPS (A) and D-LA (B) in the DSS-induced mice serum; Relative mRNA expression ratio of IL-10 (C), Muc2 (D), ZO-1 (E), and Occludin (F); Representative picture of immunofluorescence staining (G); Red staining represents Muc2-positive staining and the nuclei were stained by DAPI, the average fluorescence intensity of mucin in each group (H). Data were presented as means ± SD. n = 4 to 6 mice per group. *P < 0.05, **P < 0.01, ***P < 0.001
The prophylactic effect may result from the enhanced intestinal barrier function
After 21 d of intervention of strains in mice, the mRNA expressions of the intestinal barrier-associated proteins, including Occludin, ZO-1, and Muc2 in the colon tissue, were measured, and the results showed that the strains increased the expressions in varying degrees (Fig. 5A–C). LPS concentrations in FL-228.1 and K11 were significantly lower than those in Control, while LPS concentrations in the MN45 group significantly increased (Fig. 5D). The concentration of D-LA in FL-228.1, MN45, and K11 was downregulated considerably (Fig. 5E). Compared with other groups, the secretion of Muc2 protein in the colon of FL-228.1 mice was significantly increased (Fig. 5F, G). These findings suggest that pretreatment with strain FL-228.1 could enhance intestinal barrier function somehow.
Effects of pretreatment with strains on colonic state and intestinal barrier function. Relative mRNA expression ratio of Occludin (A), ZO-1 (B), and Muc2 (C); LPS (D) and D-LA (E) in mice serum; Red staining represents Muc2-positive staining, and the nuclei were stained by DAPI (F); Average fluorescence intensity of mucin in each group (G). Data were presented as means ± SD. n = 4 to 6 mice per group. *P < 0.05, **P < 0.01, ***P < 0.001
RNA-sequencing revealed the signaling pathways regulated by FL-228.1 in enhancing intestinal barrier function.
To further elucidate the protective mechanism of the strain FL-228.1 on intestinal barrier function, genome-wide transcriptional profiling was performed on whole colon tissues of mice intervened with the strain FL-228.1 for 21 days. The transcriptome analysis showed significant differences between Control and FL-228.1 groups (Fig. 6A). The DESeq2 software with negative binomial distribution was used to analyze the raw data (\(\left| {\log 2{\text{FC}}} \right| \ge 2,P < 0.05\)), and the processed data were used as scatter plots of expression differences. Compared to Control, FL-228.1 had 1196 differentially expressed genes (DEGs), including 506 up-regulated genes and 690 down-regulated genes (Fig. 6B). Pathway and gene enrichment analyses were performed using GO and KEGG based on the 1196 DEGs. The results showed that the GO terms bile acid and bile salt transport, immunoglobulin production, cellular lipid catabolic process, acute inflammatory response, and metabolic processes of long-chain fatty acids were significantly enriched in the FL-228.1 group (Fig. 6C). Of note, bile acid bile salt transport, lipid, and fatty acid metabolic processes were significantly upregulated. As for the KEGG analysis, Amebiasis, NF-κB, calcium signaling, peroxisome proliferator-activated receptor (PPAR), cytokine–cytokine receptor interaction, IgA-producing intestinal immune network, lipid and protein digestion and absorption, and bile secretion pathways were significantly enriched in the FL-228.1 group (Fig. 6D). Interestingly, the PPAR signaling pathway down-regulated in IBD inflammatory tissues was significantly up-regulated here, while the generally up-regulated pathways NF-κB and NOD-like receptor thermal protein domain-associated protein 3 (NLRP3) in IBD were significantly down-regulated here. Specifically, 12 key up-regulated genes (Prlr, Tnfsf10, Pparg, Ppara, Muc2, Hmgcs2, Nr1h4, Abcg2, Abst, Lep, Lpl, Cldn4) and six down-regulated genes (Il12a, Il18, Nlrp3, Cldn2, Vcam1, Lta) were screened from these pathways. These genes involved in fatty acid metabolism, bile acid transport, inflammatory response, and regulation of the content of TJs and mucins have been reported to be closely related to intestinal barrier function. Then, seven genes were selected for qPCR validation, which were among the differentially expressed genes (DEGs) revealed by the transcriptomic analysis and are closely related to intestinal barrier function. As shown in Fig. 6E, the seven gene expression changes were consistent with the transcriptomic results. The selected DEGs were then subjected to PPI network analysis, and the genes Tlr2, Nrlp3, Il18, Il10, Pparg, Cldn-2, Cldn-4, and Muc2 showed more interactions and stronger associations revealed by the inter-gene connectivity (Fig. 6F). These results suggested that the strain FL-228.1 may protect intestinal barrier function by regulating the expression of Muc2 and TJs via the PPARγ/NLRP3/TLR2 pathways.
Transcriptomic analysis of sample between FL-228.1 and Control. Principal coordinates analysis (A); DEGs between Control and FL-228.1 (B); Bubble diagram of top 40 ranked GO enrichment analysis of DEGs (C); Bubble diagram of top 40 ranked KEGG pathway enrichment analysis of DEGs (D); qPCR verification results (E); PPI network analysis of DEGs (F). Note: The horizontal axis represents the Rich factor, the enrichment degree was stronger with a bigger Rich factor, and the size of dots indicates the number of genes in GO term or KEGG pathway. PPI shows the different colors of nodes are divided according to the number of interactions. Data were presented as mean ± SD (n = 4). *P < 0.05, **P < 0.01, ***P < 0.001
Discussion
The intestinal barrier is a dynamic and multilayer system. Therefore, the dysfunction of the intestinal barrier generally implicates multiple elements such as the epithelium, immune cells, cytokines, and gut microbiota. Probiotic treatment for intestinal barrier dysfunction has been widely studied, with different strains targeting different elements. For example, two recent studies have shown that Lacticaseibacillus strains could maintain intestinal homeostasis and ameliorate DSS-induced colitis by regulating the gut microbiota and increasing mucin content [39, 40]. However, it is worth noting that the intestinal barrier elements are not independent, but, on the contrary, they are implicative of each other. For instance, IL-10 is generally considered to be an intestinal repair cytokine that regulates the growth and differentiation of natural killer cells [41] and epithelial cells [42], which can promote the expression of intestinal TJs and inhibit the expression of various inflammatory cytokines to maintain immune homeostasis. Moreover, one study showed that IL-10 also had a preventive effect before the damage occurred [43]. In contrast, IL-12 and TNF-α are considered destructive cytokines because they increase intestinal permeability when overproduced. Anti-IL-12 and anti-TNF-α therapies are currently effective ways to treat IBD [44, 45]. This study evaluated nine strains for their efficacies in enhancing intestinal barrier function in vitro and vivo. Considering the crucial roles played by the two cytokines in intestinal barrier function, they were used for our study’s in vitro screening of probiotics, as described in previous studies [46, 47]. The commercial probiotic BB12, which was previously reported to have the ability to stimulate higher levels of IL-10 production in PBMCs and markedly ameliorates DSS-induced colitis [27, 29], was used as the reference strain. In this study, we found that strains FL-228.1, K11, and BB12 could increase IL-10/IL-12 ratio in PBMCs, which suggested that the strains possess anti-inflammatory abilities. It is worth mentioning that the strains did not increase the IL-10/IL-12 ratio in vivo. The discrepancy may be explained by that the in vivo environment is more complex than the in vitro environment, which allows the whole organism to have a better regulatory capacity. Meanwhile, strains YRL577, FL-228.1, BB12, and MN45 effectively increased TEER values and reduced FITC-dextran transmission in Caco-2 monolayers, demonstrating their capability to enhance barrier function. Taking these two aspects into consideration, strain YRL577 failed to increase IL-10/IL-12 ratio, thus the three strains, FL-228.1, K11, and MN45, were subjected to in vivo studies to verify their preventive effects on intestinal barrier damage in DSS-induced mice. Large amounts of luminal LPS and D-LA can enter the blood after impairment of intestinal barrier function. Therefore, blood levels of LPS and D-LA have been employed to reflect changes in intestinal permeability [48]. In this study, the 7 days administration of DSS caused tremendous intestinal barrier damage, as increased DAI scores, shortened colon, loss of goblet cells, and inflammatory infiltrates were observed. Interestingly, the pre-administration of strains for 21 days could ameliorate the intestinal damage caused by DSS administration, partly consistent with the in vitro results. Furthermore, our results are also consistent with a recent study in that Lactobacillus intervention in advance ameliorated DSS-induced colonic length reduction and alleviated subsequent colonic inflammation [49]. The results indicated that although the strains were stopped for administration before the DSS induction, they still exerted protective effects on the intestinal barrier function, especially FL-228.1. This may be explained by the enhanced intestinal barrier function by the strains, indicated by the decreased intestinal permeability and increased secretion of Muc2. On the other hand, the relatively stable cytokines may also be contributive factors.
To investigate the possible mechanisms of action by which strain FL-228.1 exerted its prophylactic effects, transcriptomic analysis was performed in the colon tissue of mice from Control and FL-228.1 groups. Using RNA sequencing, the changes in gene expressions and the relationship between these genes and the intestinal barrier function were analyzed. Surprisingly, Cldn-4 was the only TJ-encoding gene significantly up-regulated by FL-228.1. Notably, the gene expression of Muc2 was upregulated considerably in FL-228.1, in line with its enhanced secretion observed in this study. The GO, KEGG functional enrichment analysis revealed that the genes Tlr2, Cldn-4, Muc2, Nrlp3, Il18, and Pparg were significantly altered, and the PPI analysis showed that they possessed strong interactions with each other. TLR2 reduces susceptibility to intestinal damage and inflammation by controlling TJs-associated intestinal epithelial barrier integrity [50]. Probiotics can regulate TJs content and distribution to protect intestinal barrier function by acting on the TLR2 pathway [18]. In addition, some studies had reported that probiotics could work on the TLR2 pathway to increase the secretion of Muc2 and thus prevent leaky gut and inflammation [19], which may explain the effects of FL-228.1 in this study. It has been shown that Nlrp3 activation stimulated the expression of Il18, which significantly decreased ZO-1 and E-cadherin expression [51]. IL-18 signal transduction prevents the maturation of goblet cells before colitis, leading to decreased colonic mucus. It also exacerbates goblet cell depletion during colitis [52, 53], which is by the downregulation of the Nlrp3 and Il18 genes in our research. Meanwhile, studies have revealed that the activation of PPARγ inhibited NLRP3 inflammasome, resulting in anti-inflammatory effects [54]. Furthermore, it has been shown that the expression of Muc2 was significantly reduced after the knockdown of Pparg [55]. In this study, FL-228.1 could dramatically activate the PPAR signaling pathway in mice's colonic tissues. Considering all the above, the strain FL-228.1 may have increased the expression of Muc2 and Cldn-4 by upregulating Tlr2 and Pparg and downregulating Nlrp3 and Il18, and these changes, at least partly, prevented the DSS-induced intestinal barrier damage. However, further research and verifications are warranted to elucidate the underlying mechanisms.
In conclusion, in this study, in vitro, and in vivo investigations were performed to explore potential probiotics for protecting intestinal barrier function. Unlike previous studies, we focused on the prophylactic efficacy of the strains. Our study found that the pre-administration of the strain FL-228.1 exhibited a significant protective effect on intestinal barrier function. The mechanisms of action of strain FL-228.1 on preventing intestinal barrier function damage involved Muc2 and Cldn-4 regulation via PPARγ/NLRP3/ TLR2 pathways.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author on reasonable request.
References
Martel J, Chang S, Ko Y, Hwang T, Young JD, Ojcius DM (2022) Gut barrier disruption and chronic disease. Trends Endocrinol Metab 334:247–265. https://doi.org/10.1016/j.tem.2022.01.002
Takamura T, Harama D, Fukumoto S, Nakamura Y, Shimokawa N, Ishimaru K, Ikegami S, Makino S, Kitamura M, Nakao A (2011) Lactobacillus bulgaricus OLL1181 activates the aryl hydrocarbon receptor pathway and inhibits colitis. Immunol Cell Biol 897:817–822. https://doi.org/10.1038/icb.2010.165
Nanthakumar NN, Fusunyan RD, Sanderson I, Walker WA (2000) Inflammation in the developing human intestine: a possible pathophysiologic contribution to necrotizing enterocolitis. Proc Natl Acad Sci U S A 9711:6043–6048. https://doi.org/10.1073/pnas.97.11.6043
Wang Y, Yang J, Wang W, Sanidad KZ, Cinelli MA, Wan D, Hwang SH, Kim D, Lee KSS, Xiao H, Hammock BD, Zhang G (2020) Soluble epoxide hydrolase is an endogenous regulator of obesity-induced intestinal barrier dysfunction and bacterial translocation. P Natl Acad Sci Usa. https://doi.org/10.1073/pnas.1916189117/-/DCSupplemental
Zhang M, Yang L, Zhu M, Yang B, Yang Y, Jia X, Feng L (2022) Moutan Cortex polysaccharide ameliorates diabetic kidney disease via modulating gut microbiota dynamically in rats. Int J Biol Macromol 206:849–860. https://doi.org/10.1016/j.ijbiomac.2022.03.077
Kobayashi T, Siegmund B, Le Berre C, Wei SC, Ferrante M, Shen B, Bernstein CN, Danese S, Peyrin-Biroulet L, Hibi T (2020) Ulcerative colitis. Nat Rev Dis Primers. https://doi.org/10.1038/s41572-020-0205-x
Suzuki T (2013) Regulation of intestinal epithelial permeability by tight junctions. Cell Mol Life Sci 70:631–659
Van der Sluis M, De Koning BAE, De Bruijn ACJM, Velcich A, Meijerink JPP, Van Goudoever JB, Büller HA, Dekker J, Van Seuningen I, Renes IB, Einerhand AWC (2006) Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 1311:117–129. https://doi.org/10.1053/j.gastro.2006.04.020
Leon-Coria A, Kumar M, Workentine M, Moreau F, Surette M, Chadee K (2021) Muc2 mucin and nonmucin microbiota confer distinct innate host defense in disease susceptibility and colonic injury. Cell Mol Gastroenterol Hepatol 111:77–98. https://doi.org/10.1016/j.jcmgh.2020.07.003
van der Post S, Jabbar KS, Birchenough G, Arike L, Akhtar N, Sjovall H, Johansson MEV, Hansson GC (2019) Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut 6812:2142–2151. https://doi.org/10.1136/gutjnl-2018-317571
Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA (2019) Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastro Hepat 1610:605–616. https://doi.org/10.1038/s41575-019-0173-3
Zaylaa M, Al Kassaa I, Alard J, Peucelle V, Boutillier D, Desramaut J, Dabboussi F, Pot B, Grangette C (2018) Probiotics in IBD: combining in vitro and in vivo models for selecting strains with both anti-inflammatory potential as well as a capacity to restore the gut epithelial barrier. J Funct Foods 47:304–315. https://doi.org/10.1016/j.jff.2018.05.029
Cuffaro B, Assohoun ALW, Boutillier D, Súkeníková L, Desramaut J, Boudebbouze S, Salomé-Desnoulez S, Hrdý J, Waligora-Dupriet A, Maguin E, Grangette C (2020) In vitro characterization of gut microbiota-derived commensal strains: Selection of parabacteroides distasonis strains alleviating TNBS-Induced colitis in mice. Cells 99:2104. https://doi.org/10.3390/cells9092104
Resta-Lenert S, Barrett KE (2003) Live probiotics protect intestinal epithelial cells from the effects of infection with enter invasive Escherichia coli (EIEC). Gut 527:988–997. https://doi.org/10.1136/gut.52.7.988
Chandran A, Duary RK, Grover S, Batish VK (2013) Relative expression of bacterial and host specific genes associated with probiotic survival and viability in the mice gut fed with Lactobacillus plantarum Lp91. Microbiol Res 1689:555–562. https://doi.org/10.1016/j.micres.2013.04.010
Sun M, Liu Y, Song Y, Gao Y, Zhao F, Luo Y, Qian F, Mu G, Tuo Y (2020) The ameliorative effect of Lactobacillus plantarum-12 on DSS-induced murine colitis. Food Funct 116:5205–5222. https://doi.org/10.1039/d0fo00007h
Shi M, Yue Y, Ma C, Dong L, Chen F (2022) Pasteurized Akkermansia muciniphila ameliorate the LPS-induced intestinal barrier dysfunction via modulating AMPK and NF-κB through TLR2 in Caco-2 Cells. Nutrients 144:764. https://doi.org/10.3390/nu14040764
Rose EC, Odle J, Blikslager AT, Ziegler AL (2021) Probiotics, prebiotics and epithelial tight junctions: a promising approach to modulate intestinal barrier function. Int J Mol Sci 2213:6729. https://doi.org/10.3390/ijms22136729
Wang S, Ahmadi S, Nagpal R, Jain S, Mishra SP, Kavanagh K, Zhu X, Wang Z, Mcclain DA, Kritchevsky SB, Kitzman DW, Yadav H (2020) Lipoteichoic acid from the cell wall of a heat killed Lactobacillus paracasei D3–5 ameliorates aging-related leaky gut, inflammation and improves physical and cognitive functions: from C Elegans to mice. Geroscience 421:333–352. https://doi.org/10.1007/s11357-019-00137-4
Li Y, Zhang T, Guo C, Geng M, Gai S, Qi W, Li Z, Song Y, Luo X, Zhang T, Wang N (2020) Bacillus subtilis RZ001 improves intestinal integrity and alleviates colitis by inhibiting the notch signaling pathway and activating ATOH-1. Pathog Dis. https://doi.org/10.1093/femspd/ftaa016
Wang L, Cao H, Liu L, Wang B, Walker WA, Acra SA, Yan F (2014) Activation of epidermal growth factor receptor mediates mucin production stimulated by p40, a Lactobacillus rhamnosus GG-derived protein. J Biol Chem 28929:20234–20244. https://doi.org/10.1074/jbc.M114.553800
Mujagic Z, de Vos P, Boekschoten MV, Govers C, Pieters HHM, de Wit NJW, Bron PA, Masclee AAM, Troost FJ (2017) The effects of Lactobacillus plantarum on small intestinal barrier function and mucosal gene transcription; a randomized double-blind placebo-controlled trial. Sci Rep-Uk. https://doi.org/10.1038/srep40128
Lu Y, Zhang J, Yi H, Zhang Z, Zhang L (2019) Screening of intestinal peristalsis-promoting probiotics based on a zebrafish model. Food Funct 104:2075–2082. https://doi.org/10.1039/c8fo02523a
Lu Y, Zhang Z, Liang X, Chen Y, Zhang J, Yi H, Liu T, Yang L, Shi H, Zhang L (2019) Study of gastrointestinal tract viability and motility via modulation of serotonin in a zebrafish model by probiotics. Food Funct 1011:7416–7425. https://doi.org/10.1039/c9fo02129a
Wang SM, Zhang LW, Fan RB, Han X, Yi HX, Zhang LL, Xue CH, Li HB, Zhang YH, Shigwedha N (2014) Induction of HT-29 cells apoptosis by lactobacilli isolated from fermented products. Res Microbiol 1653:202–214. https://doi.org/10.1016/j.resmic.2014.02.004
Cui Q, Tian X, Liang X, Zhang Z, Wang R, Zhou Y, Yi H, Gong P, Lin K, Liu T, Zhang L (2022) Bifidobacterium bifidum relieved DSS-induced colitis in mice potentially by activating aryl hydrocarbon receptor. Food Funct. https://doi.org/10.1039/D1FO04219J
Xie J, Yu Q, Nie S, Fan S, Xiong T, Xie M (2015) Effects of Lactobacillus plantarum NCU116 on intestine mucosal immunity in immunosuppressed mice. J Agr Food Chem 6351:10914–10920. https://doi.org/10.1021/acs.jafc.5b04757
Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, Slagter M, van der Velden DL, Kaing S, Kelderman S, van Rooij N, van Leerdam ME, Depla A, Smit EF, Hartemink KJ, de Groot R, Wolkers MC, Sachs N, Snaebjornsson P, Monkhorst K, Haanen J, Clevers H, Schumacher TN, Voest EE (2018) Generation of tumor-reactive t cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell 1746:1586–1598. https://doi.org/10.1016/j.cell.2018.07.009
Sheikhi A, Shakerian M, Giti H, Baghaeifar M, Jafarzadeh A, Ghaed V, Heibor MR, Baharifar N, Dadafarin Z, Bashirpour G (2016) Probiotic yogurt culture bifidobacterium animalis subsp. Lactis BB-12 and lactobacillus acidophilus LA-5 modulate the cytokine secretion by peripheral blood mononuclear cells from patients with ulcerative colitis. Drug Res (Stuttg) 666:300–305. https://doi.org/10.1055/s-0035-1569414
Wu X, Huang X, Chen R, Li T, Ye H, Xie W, Huang Z, Cao G (2019) Paeoniflorin prevents intestinal barrier disruption and inhibits lipopolysaccharide (LPS)-induced inflammation in Caco-2 cell monolayers. Inflammation 426:2215–2225. https://doi.org/10.1007/s10753-019-01085-z
Shi C, Chen H, Liang Y, Xia Y, Yang Y, Yang J, Zhang J, Wang S, Liu J, Qin H (2014) Combined probiotic bacteria promotes intestinal epithelial barrier function in interleukin-10-gene-deficient mice. World J Gastroenterol 2016:4636–4647. https://doi.org/10.3748/wjg.v20.i16.4636
Tsai P, Schlichtig A, Ziegler E, Ernst H, Haberstroh J, Stelzer HD, Hackbarth H (2015) Effects of different blood collection methods on indicators of welfare in mice. Lab Anim 448:301–310. https://doi.org/10.1038/laban.738
Sivakumar PV, Westrich GM, Kanaly S, Garka K, Born TL, Derry JM, Viney JL (2002) Interleukin 18 is a primary mediator of the inflammation associated with dextran sulphate sodium induced colitis: blocking interleukin 18 attenuates intestinal damage. Gut 506:812–820. https://doi.org/10.1136/gut.50.6.812
Arena ET, Rueden CT, Hiner MC, Wang S, Yuan M, Eliceiri KW (2017) Quantitating the cell: turning images into numbers with ImageJ Wiley interdisciplinary reviews. Dev Biol. https://doi.org/10.1002/wdev.260
Mao T, Su CW, Ji Q, Chen CY, Wang R, Vijaya KD, Lan J, Jiao L, Shi HN (2021) Hyaluronan-induced alterations of the gut microbiome protects mice against Citrobacter rodentium infection and intestinal inflammation. Gut Microbes 131:1972757. https://doi.org/10.1080/19490976.2021.1972757
Wu Y, Li J, Ding W, Ruan Z, Zhang L (2021) Enhanced intestinal barriers by puerarin in combination with tryptophan. J Agr Food Chem 6951:15575–15584. https://doi.org/10.1021/acs.jafc.1c05830
Hao S, Li S, Wang J, Zhao L, Yan Y, Cao Q, Wu T, Liu L, Wang C (2018) Transcriptome analysis of Phycocyanin-mediated inhibitory functions on non-Small cell lung cancer a549 cell growth. Mar Drugs 1612:511. https://doi.org/10.3390/md16120511
Wu D, Chen S, Ye X, Ahmadi S, Hu W, Yu C, Zhu K, Cheng H, Linhardt RJ, He Q (2022) Protective effects of six different pectic polysaccharides on DSS-induced IBD in mice. Food Hydrocoll 127:107209. https://doi.org/10.1016/j.foodhyd.2021.107209
Han M, Liao W, Si X, Bai C, Gai Z (2022) Protective effects of Lacticaseibacillus rhamnosus Hao9 on dextran sulphate sodium-induced ulcerative colitis in mice. J Appl Microbiol 1333:2039–2049. https://doi.org/10.1111/jam.15665
Wu Y, Li A, Liu H, Zhang Z, Zhang C, Ma C, Zhang L, Zhang J (2022) Lactobacillus plantarum HNU082 alleviates dextran sulfate sodium-induced ulcerative colitis in mice through regulating gut microbiome. Food Funct 1319:10171–10185. https://doi.org/10.1039/D2FO02303B
Hermans L, De Pelsmaeker S, Denaeghel S, Cox E, Favoreel HW, Devriendt B (2021) β-Glucan-Induced IL-10 secretion by monocytes triggers porcine NK cell cytotoxicity. Front Immunol 12:634402. https://doi.org/10.3389/fimmu.2021.634402
Powell DN, Swimm A, Sonowal R, Bretin A, Gewirtz AT, Jones RM, Kalman D (2020) Indoles from the commensal microbiota act via the AHR and IL-10 to tune the cellular composition of the colonic epithelium during aging. Proc Natl Acad Sci 11735:21519–21526. https://doi.org/10.1073/pnas.2003004117
Zheng L, Kelly CJ, Battista KD, Schaefer R, Lanis JM, Alexeev EE, Wang RX, Onyiah JC, Kominsky DJ (1950) Colgan SP (2017) Microbial-Derived butyrate promotes epithelial barrier function through IL-10 receptor-dependent repression of Claudin-2. J Immunol 1998:2976–2984. https://doi.org/10.4049/jimmunol.1700105
Moschen AR, Tilg H, Raine T (2019) IL-12, IL-23 and IL-17 in IBD: immunobiology and therapeutic targeting. Nat Rev Gastroenterol Hepatol 163:185–196. https://doi.org/10.1038/s41575-018-0084-8
Danese S (2012) New therapies for inflammatory bowel disease: from the bench to the bedside. Gut 616:918–932. https://doi.org/10.1136/gutjnl-2011-300904
de Roock S, van Elk M, van Dijk MEA, Timmerman HM, Rijkers GT, Prakken BJ, Hoekstra MO, de Kleer IM (2009) Lactic acid bacteria differ in their ability to induce functional regulatory T cells in humans. Clin Exp Allergy. https://doi.org/10.1111/j.1365-2222.2009.03344.x
Ashraf R, Vasiljevic T, Day SL, Smith SC, Donkor ON (2014) Lactic acid bacteria and probiotic organisms induce different cytokine profile and regulatory T cells mechanisms. J Funct Foods 6:395–409. https://doi.org/10.1016/j.jff.2013.11.006
Tao F, Xing X, Wu J, Jiang R (2021) Enteral nutrition modulation with n-3 PUFAs directs microbiome and lipid metabolism in mice. PLoS ONE 163:e248482. https://doi.org/10.1371/journal.pone.0248482
Zhang P, Li B, Mu J, Liu D, Zhang G, Mao X, Huang K, Waldron KJ, Chen X (2022) The therapeutic and preventive effects of a canine-origin VB12-producing Lactobacillus on DSS-induced colitis in mice. J Anim Physiol an N. https://doi.org/10.1111/jpn.13767
Cario E, Gerken G, Podolsky DK (2007) Toll-Like receptor 2 controls mucosal inflammation by regulating epithelial barrier function. Gastroenterology 1324:1359–1374. https://doi.org/10.1053/j.gastro.2007.02.056
Maroni L, Agostinelli L, Saccomanno S, Pinto C, Giordano DM, Rychlicki C, De Minicis S, Trozzi L, Banales JM, Melum E, Karlsen TH, Benedetti A, Baroni GS, Marzioni M (2017) Nlrp3 activation induces IL-18 synthesis and affects the epithelial barrier function in reactive cholangiocytes. Am J Pathol 1872:366–376. https://doi.org/10.1016/j.ajpath.2016.10.010
Nowarski R, Jackson R, Gagliani N, de Zoete MR, Palm NW, Bailis W, Low JS, Harman CCD, Graham M, Elinav E, Flavell RA (2015) Epithelial IL-18 equilibrium controls barrier function in colitis. Cell 1636:1444–1456. https://doi.org/10.1016/j.cell.2015.10.072
Hand TW (2015) Interleukin-18: the bouncer at the mucosal bar. Cell 1636:1310–1312. https://doi.org/10.1016/j.cell.2015.11.041
Yang C, Wu C, Lin T, Cheng Y, Chang C, Lee K, Tsai P, Tsai Y (2021) Inhibitory effect of PPARγ on NLRP3 inflammasome activation. Theranostics 115:2424–2441. https://doi.org/10.7150/thno.46873
Li D, Feng Y, Tian M, Ji J, Hu X, Chen F (2021) Gut microbiota-derived inosine from dietary barley leaf supplementation attenuates colitis through PPARγ signaling activation. Microbiome. https://doi.org/10.1186/s40168-021-01028-7
Funding
This work was supported by the Project of Taishan Industry Leading Talent of Shandong Province (LJNY202101), and the National Natural Science Foundation of China (Grant number: 31901664).
Author information
Authors and Affiliations
Contributions
LZ and TL were in charge of conceptualization, funding acquisition and writing-review and editing; Investigating, methodology, visualization, and writing-original draft preparation were done by RW; MK, QC and XT were in charge of methodology; Data curation was done by R. W. and YZ; HY, PG, KL and ZZ were in charge of writing-review and editing. All authors have edited and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest relevant to this work.
Ethical approval
The Laboratory Animal Ethics Committee of the College of Food Science and Engineering of Ocean University of China.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, R., Kuerman, M., Cui, Q. et al. Protective effects of Bifidobacterium bifidum FL-228.1 on dextran sulfate sodium-induced intestinal damage in mice. Eur J Nutr 62, 1267–1280 (2023). https://doi.org/10.1007/s00394-022-03064-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00394-022-03064-x