Abstract
Purpose
This study investigated the expression of interleukin 32 (IL-32) in hepatoblastoma, the most common primary pediatric liver tumor, and its possible roles in tumorigenesis.
Methods
IL-32 expression was investigated in two hepatoblastoma cell lines (Hep G2 and HuH 6) in the steady state and after co-culture with macrophages by RNA-seq analysis and RT-qPCR, and after stimulation with chemotherapy. Cultured macrophages were stimulated by IL-32 isoforms followed by RT-qPCR and western blot analysis. IL-32 immunohistochemical staining (IHC) was performed using specimens from 21 hepatoblastoma patients. Clustering analysis was also performed using scRNA-seq data downloaded from Gene Expression Omnibus.
Results
The IL-32 gene is expressed by hepatoblastoma cell lines; expression is upregulated by paracrine cell–cell communication with macrophages, also by carboplatin and etoposide. IL-32 causes protumor activation of macrophages with upregulation of PD-L1, IDO-1, IL-6, and IL-10. In the patient pool, IHC was positive only in 48% of cases. However, in the downloaded dataset, IL-32 gene expression was negative.
Conclusion
IL-32 was detected in hepatoblastoma cell lines, but not in all hepatoblastoma patients. We hypothesized that stimulation such as chemotherapy might induce expression of IL-32, which might be a critical mediator of chemoresistance in hepatoblastoma through inducing protumor activation in macrophages.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatoblastoma (HB) is known to be the most common primary liver malignant tumor in the pediatric age group. Presentation typically occurs before the age of 3 years with a median age of 1.5 years old [1]. The histological subtypes of HB represent a wide spectrum: embryonal, fetal, macrotrabecular, small cell undifferentiated, cholangioblastic, mixed stromal and teratoid [2], reflecting different stages in the differentiation pathway from human fetal liver multipotent progenitor cells [3]. Many cases remain unresectable even after repeated cycles of chemotherapy.
The tumor microenvironment (TME), which plays an important role in regulating tumor progression, comprises tumor-infiltrating immune cells, such as T lymphocytes, dendritic cells and tumor-associated macrophages (TAMs) [4], which are the most abundant. TAMs have a pro-tumor function and secrete numerous cytokines (e.g., TGF-β, PDGF and EGF) that stimulate the proliferation and invasion of tumor cells [5], as well as enzymes that promote tumor angiogenesis, such as Cyclooxygenase-2 and MMP-2,7,9 [6]. Recent studies have focused on the immunosuppressive functions of TAMs, as they express a variety of immunosuppressive factors, such as PD-L1, PD-L2, IDO-1 and IL-10. TAMs-targeted therapy is now a reality, and its efficacy has been improved with the application of nanotechnology in medicine [7]. At present, many clinical trials are being conducted in which TAMs-targeted therapy is being combined with conventional chemo, radio, or other immunotherapies (e.g., checkpoint inhibitors), with promising outcomes [7].
Although many studies have demonstrated the clinical significance of TAMs in solid tumors, only a few have described the role of TAMs in HB. Increased TAMs density has been reported in the embryonal component of HB, and IL-34 produced from the embryonal component was suggested to be involved in TAMs infiltration [8]. Further, Macrophage colony stimulating factor receptor (M-CSFR), which is an IL-34 receptor in tumor cells, was increased in the embryonal component and recurrent lesions. In vitro studies have shown that TAMs-derived factors enhance M-CSFR and IL-34 expression in tumor cells, and that these signals were potentially involved in tumor cell proliferation and chemoresistance. Thus, cell-to-cell interactions between TAMs and HB cells have been suggested to be associated with the progression of HB [9].
In the present study, we newly found that interleukin-32 (IL-32) expression in HB cell lines was increased by cell–cell communication with TAMs. IL-32 is a newly discovered proinflammatory cytokine, which is encoded in humans by the IL-32 gene. Although its definitive receptor remains unknown [10], IL-32 is expressed mainly by immune cells; natural killer cells (NKs) have the highest expression rates [11]. Macrophages express IL-32 only during inflammation or when stimulated with Lipopolysaccharide (LPS) or Mycobacterium tuberculosis (MTB) [12]. IL-32 is involved in the pathogenesis of a variety of inflammatory, autoimmune, and infectious diseases, and its role in tumors is controversial [13]. To the best of our knowledge, no studies have investigated its role in HB. Therefore, we examined IL-32 gene expression in HB cell lines and human samples in the present study. Since TAMs have been suggested to be affected by IL-32, an in vitro culture study using human monocyte-derived macrophages was also performed.
Methods
Ribonucleic acid sequencing (RNA-Seq)
Hep G2 cells were cultured alone then cocultured directly with macrophages for 48 h, followed by total RNA isolation using an RNAiso Plus kit (Takara Bio INC. Shiga, Japan) and Trizol extraction (Thermo Fisher Scientific, Waltham, MA, USA). A BioAnalyzer 2100 (Agilent Technologies, Inc. CA, USA) was used to confirm the RNA quality (RNA integrity number > 9). For RNA-seq data analysis, the resulting reads were aligned to the human genome (UCSC hg19) using STAR ver.2.6.0a (Dr. Alexander Dobin, Cold Spring Harbor Laboratory, NY, USA) after trimming to remove adapter sequences and low-quality ends using Trim Galore! v0.5.0 (cutadapt v1.16) (Babraham Bioinformatics, Cambridge, UK). The resulting data was deposited in the Gene Expression Omnibus (GEO) at the NCBI [Accession No. GSE212734], and gene expression levels were measured as transcripts per million using RSEM v1.3.1 (Dr. Bo Li and Dr. Colin Dewey, University of Wisconsin-Madison, Madison, WI, USA). Gene Ontology enrichment analysis and pathway enrichment analysis were performed using multiple databases: Metascape (http://metascape.org/gp/index.html#/main/step1).
Cell culture of macrophages
The THP-1 cell line was obtained from the American Type Culture Collection (Manassas, VA, USA). THP-1-derived macrophages were induced by adding phorbol-12-myristate-13-acetate to the culture media. In addition, blood samples were obtained from healthy donors to extract human monocyte-derived macrophages (HMDMs) according to a protocol approved by the Kumamoto University Hospital Review Board (#1169), as described previously [14]. Monocytes were isolated using a RosetteSep Human Monocyte Enrichment Cocktail (STEMCELL Technologies, Vancouver, Canada) and then plated in 12-well plates (2 × 105 cells/well; CellSeed, Tokyo, Japan). AIM-V medium (Thermo Fisher Scientific) supplemented with 2% human serum was used as a culture medium. Monocyte differentiation into macrophages was induced by adding macrophage-colony stimulating factor (100 ng/ml; Wako Pure Chemical Industries, Ltd., Tokyo, Japan) and granulocyte macrophage-colony stimulating factor (1 ng/ml; WAKO Pure Chemical Industries, Ltd.) for 7 days.
Cell cultures of HB cell lines
Hep G2 and HuH-6 cells were obtained from the Japanese Collection of Research Bioresources (JCRB) Cell Bank (Osaka, Japan). As the culture medium, D-MEM/Ham’s F-12 (WAKO Pure Chemical Industries, Ltd.) supplemented with 10% fetal bovine serum, 1% non-essential amino acids, and 1% penicillin/streptomycin, was used. Hep G2 and HuH-6 cells (3 × 106 cells/well) were cultured on 100-mm Petri dishes (Thermo Fisher Scientific) for 2 days.
Co-culture of Hep G2 cells with macrophages
For the direct co-culture of Hep G2 cells with macrophages, Hep G2 cells (3 × 106 cells/well) and macrophages (1 × 106 cells/well) were seeded on 100-mm dishes (Thermo Fisher Scientific), and co-cultured for 2 days. Conventional paraffin-embedded cell blocks were prepared as described previously [15].
For the indirect co-culture of Hep G2 cells with macrophages, Hep G2 cells (3 × 105 cells/well) were seeded on 6-well plates (Thermo Fisher Scientific), and macrophages (2 × 105 cells/well) were seeded on the upper side of a 6-well cell culture inserts with a 0.4-μm pore size (Falcon, Durham, NC, USA). The insert was placed into the well, and the cells were co-cultured for 2 days.
Stimulation of macrophages by IL-32 proteins
Cultured macrophages, either THP-1 derived or HMDMs, were stimulated by the addition of 100 ng/ml of recombinant human IL-32 proteins (IL-32 alpha: #3040-IL-050, IL-32 beta: #6769-IL-025, IL-32 gamma: 4690-IL-025/CF; R&D Systems, Inc. a Bio-Techne Corp. brand, Japan).
Real time quantitative polymerase chain reaction (RT-qPCR)
RNA extraction from cultured cells and RT-qPCR were performed as described previously [16] using the following primers: β-Actin, forward 5'-ATTCCTATGTGGGCGACGAG-3' and reverse 5'-AAGGTGTGGTGCCAGATTTTC-3'; IL-32 α, forward 5'-GCTGGAGGACGACTTCAAAGA-3' and reverse 5'-GGGCTCCGTAGGACTTGTCA-3'; IL-32 β, forward 5'-CAGTGGAGCTGGGTCATCTCA-3' and reverse 5'-GGGCCTTCAGCTTCTTCATGTCATCA-3'; IL-32 γ, forward 5'-ACGCCCGAATGGTAATGCT-3' and reverse 5'-CCACAGTGTCCTCAGTGTCACA-3'; PD-L1, forward 5'-GACCAGCACACTGAGAATCAACAC-3' and reverse 5'-TTGGAGGATGTGCCAGAGGTAG-3'; IDO-1, forward 5'-TGTTCTCATTTCGTGATGGAGACTG-3' and reverse 5'-AAAGTGTCCCGTTCTTGCATTTG-3'; IL-6, forward 5'-ATGTGTGAAAGCAGCAAAGAGG-3' and reverse 5'-GTGATGATTTTCACCAGGCAAG-3'; IL-10, forward 5'-GGTTGCCAAGCCTTGTCTGA-3' and reverse 5'-AGGGAGTTCACATGCGCCT-3'; Cluster of differentiation 163 (CD163), forward 5'-CGAGTTAACGCCAGTAAGG-3' and reverse 5'-GAACATGTCACGCCAGC-3'; VEGF, forward 5'-TCTTCAAGCCATCCTGTGTG-3' and reverse 5'-TCTGCATGGTGATGTTGGAC-3'; and TGF-β, forward 5'-AGCAACAATTCCTGGCGATAC-3' and reverse 5'-GCGAAAGCCCTCAATTTCC-3'.
Western blot analysis
The cell lysates were harvested using NP-40 lysis buffer and the protein concentration of samples was calculated using Bicinchoninic acid assay (BCA) (Thermo Fisher Scientific) protein assay reagent. Protein (50 μg) was electrophoresed on a 15% sodium dodecyl sulfate–polyacrylamide gel, and transferred to a polyvinylidene fluoride membrane (Millipore, Bedford, MA, USA). The following antibodies were used to examine PD-L1 and IDO-1 protein expression by macrophages before and after stimulation with IL-32 proteins: anti-PD-L1 antibody (0.1 μg/ml, clone E1L3N; Cell Signaling Technology, Inc., Massachusetts, USA), anti-IDO-1 antibody (0.1 μg/ml, clone ab55305; Abcam PLC, Cambridge, UK), and anti-actin antibody (0.1 μg/ml, clone sc-47778; Santa Cruz Biotechnology, Inc., TX, USA). Other details of the methods used for the western blot analysis were described previously [17].
Single-cell clustering analysis
Single cell RNA-seq (scRNA-seq) data of HB (GSE186975) were downloaded from (GEO). GSM5664587 to GSM5664593 were used for the analysis [18]. Cells captured in single-cell RNA sequencing analysis were clustered and analyzed using the Seurat (Version 4.3.0) package in R (Version 4.2.2)43. Cells with <200 genes, 500 transcripts, or a mitochondrial level of ≥ 20%, were filtered out as part of the quality control process. Samples were divided into two groups, “Tumor source” and “Normal source”, and a standard workflow using the sctransform (SCT) method was devised to normalize the gene expression for each cell and then merge the two groups directly. We calculated and returned the top 2000 most variably expressed genes among the cells, and principal components analysis (PCA) was performed for linear dimensional reduction; the first 100 principal components (PCs) were stored for subsequent analysis. To determine how many PCs to use for the cluster analysis, a JackStraw resampling method was implemented by performing permutations on a subset of data (1% by default) and rerunning the PCA for a total of 100 replicates to select statistically significant PCs for inclusion in K-nearest neighbors cluster analysis. For graph-based clustering, the first 50 PCs were selected followed by resolution-level selection using the “clustree” package (Version 0.5.0). As a result, a resolution of 1.2 was selected, yielding 20 cell clusters. The differentially expressed genes (DEGs) in each cluster were identified using the FindAllMarker function implemented in the Seurat package, and a corresponding p-value was estimated by Wilcoxon’s test followed by Bonferroni correction.
Cell viability assay
To estimate the Half maximal inhibitory concentration (IC50) of carboplatin (Wako Pure Chemical Industries, Ltd., Tokyo, Japan) and etoposide (Wako Pure Chemical Industries, Ltd., Tokyo, Japan), a Cell Counting Kit-8 (WST-8/CCK-8, Dojindo, Kumamoto, Japan) was used. HuH-6 and Hep G2 cells (5 × 103 cells/well) were preincubated on 96-well plates at 37 °C for 24 h, and then treated with 10–6, 10–5,10–4, 10–3, 10–2, 10–1, and 100 μM carboplatin, etoposide, or medium alone and incubated at 37 °C for a further 48 h. Subsequently, 10 μl/well of the Cell Counting Kit-8 solution was added, and the cells were incubated at 37 °C for 2 h. Absorbance was measured at 450 nm using a SpectraMax i3x multimode microplate reader (Molecular Devices, San Jose, CA, USA).
Human tissue specimens
A total of 21 primary HB tissue specimens, already stored as paraffin-embedded blocks, were used in the study. Patient characteristics are summarized in Table 1. The specimens were obtained previously from pediatric patients who underwent hepatic resections for HB at Kumamoto University Hospital in Kumamoto, Japan, over two decades between 2000 and 2021. The diagnosis of HB was confirmed by histopathological examination. Informed consent was obtained from the patients’ caregivers to be included in the study. The study design and proposal were approved by the Institutional Review Board (IRB) at Kumamoto University (IRB No. 2224).
Immunohistochemistry (IHC)
Cell blocks of HuH-6 cells cultured alone and Hep G2 cells that were co-cultured directly with macrophages were fixed with 10% neutral buffered formalin and embedded in paraffin. Tissue samples were already stored as paraffin-embedded blocks. Using a microtome, 3 μm thick tissue slices were prepared and deparaffinized with xylene and ethanol. The following primary antibodies were used: mouse monoclonal NCNP24 (WAKO Pure Chemical Industries, Ltd.) to detect Ionized calcium-binding adapter molecule-1 (1 μg/ml, Iba-1, a specific marker for pan-macrophages) and rabbit polyclonal ab37158 (1 μg/ml, Abcam) to detect IL-32. Horseradish peroxidase-labeled anti-mouse or anti-rabbit immunoglobulin antibody (Nichirei, Tokyo, Japan) was used as the secondary antibody, and positive signals were detected using 3,3’-diaminobenzidine (Nichirei).
Statistical analysis
ImageJ 1.53t software (National Institutes of Health, USA) was used to measure the pixel density for relative quantification of the western blot bands. GraphPad Prism 9.4.0 software (GraphPad, San-Diego, CA, USA) was used for statistical analysis. Continuous data were presented as (mean ± SD). One-way ANOVA with post-hoc Tukey’s HSD test were used to precisely compare differences between different groups’ means, Fischer’s exact test was used for categorical univariate analysis; only differences with P < 0.05 were considered to be statistically significant.
Results
IL-32 is expressed by HB cells and its expression is upregulated by cell–cell communication with TAMs
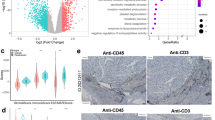
As described in the introduction, we previously demonstrated the significance of cell-to-cell communication between TAMs and tumor cells in HB progression [8]. Therefore, we attempted to clarify changes in gene expression in Hep G2 cells with or without direct co-culture with HMDMs by means of RNA-seq. Although numerous genes related to metabolic processes and signal activation were found to be up-regulated by co-culture (Fig. 1a–c), we focused on IL-32 genes in the present study.
RNA-Seq analysis for Hep G2 cells cultured alone and cocultured with macrophages a Replicate comparison scatter plot comparing genes of the two groups, with differences expressed in fold change (log2). IL-32 is indicated by red dot; b Gene set enrichment analysis with P-values expressed in (–log10); c Heat map of upregulated genes of interest, including IL-32
Then, RT-qPCR analysis was performed to investigate IL-32 gene expression in the steady state for HB cell lines as well as HMDMs. The findings showed that macrophages do not express IL-32 in the steady state. However, HB cell lines express IL-32 in the steady state, and fold change comparisons showed that expression in Hep G2 was markedly higher than in HuH 6, with results being the same for different isoforms of IL-32 [α, β and γ] (Fig. 2a). The findings of the RNA-seq analysis were confirmed by the RT-qPCR results obtained for Hep G2 cells that were either cultured alone or cocultured indirectly with macrophages. Specifically, fold change comparisons showed that IL-32 is upregulated by cell–cell communication with macrophages using a paracrine mechanism. Interestingly, the findings were similar to those obtained using THP-1-derived macrophages (Fig. 2b) or HMDMs (Fig. 2c).
RT-qPCR analysis of IL-32 (a) in the steady state for Hep G2, HuH-6 and HMDM (b and c) Indirect co-culture system for Hep G2 cells with macrophages; b THP-1-derived macrophages; c HMDM [2 (− ⊿⊿ Ct) method used to analyze relative change in gene expression. A one-way ANOVA followed by a Tukey’s post-hoc HSD test were used for multiple comparisons. P < 0.05 was considered statistically significant]
IL-32 potentially induces protumor activation of macrophages
The influence of IL-32 on TAMs was examined by RT-qPCR analysis of genes of interest. Among the isoforms used, the β isoform appears to exert the strongest influence on macrophages, and the results were similar to those obtained for the THP-1-derived macrophages (Fig. 3a) and HMDMs (Fig. 3b). The results showed that macrophages respond to IL-32 by upregulation of PD-L1, IDO-1, IL-6, and IL-10. However, changes in CD163, VEGF, and TGF-β were generally not significant/down regulated. Confirmatory western blot analysis was performed for PD-L1 and IDO-1 due to their well-known immunosuppressive and protumor effects, and the results were consistent with those of the RT-qPCR analysis (Fig. 3c).
RT-qPCR analysis of [PD-L1, IDO-1, VEGF, TGF-β, IL-6, IL-10 and CD163] for macrophages ± stimulation with IL-32 isoforms [α, β & γ] a THP-1-derived macrophages; b HMDM [2 (− ⊿⊿ Ct) method was used to analyze relative changes in gene expression. A one-way ANOVA followed by a Tukey’s post-hoc HSD test were used for multiple comparisons. P < 0.05 was considered statistically significant]; c Confirmatory western blot for PD-L1 and IDO-1
IL-32 gene expression was not detected in the tumor cells of resected HB specimens
IL-32 gene expression in HB samples was investigated using previously published scRNA-seq data open database, as described in the materials and methods section (Fig. 4a–c). Although IL-32 gene expression was not detected in hepatoblastoma tissue, it was observed in T cells and proliferating immune cells (Fig. 4d). The proliferating immune cells consisted of a mixed population of myeloid cells, T cells, and NK cells.
Single cell clustering analysis a Uniform manifold approximation and projection (UMAP) of 4,527 cells from one HB patient, annotated by cell type; b UMAP, annotated by cell source; c Dot-plot of all identified populations, each characterized by at least three known cell-type markers [average expression is indicated by the color gradient, and the percentage of marker expressed is represented by dot size]; d violin plot (left) and feature plot (right) of IL-32 expression
IL-32 was potentially induced by chemotherapy in HB
IL-32 gene expression was detected in HB cell lines, but not in resected HB cells. We hypothesized that the IL-32 gene was not expressed in tumor cells in the steady state, but that it was induced by chemotherapy. Therefore, cell lines were stimulated with carboplatin or etoposide and IL-32 gene expression was tested by RT-qPCR. Since the IC50 values obtained for carboplatin and etoposide in Hep G2 and HuH-6 cells were 0.46, 0.1, 0.44, and 0.04 μM, respectively, the cells were stimulated with carboplatin and etoposide at lower IC50 doses. Both agents up-regulated IL-32 gene expression, although IL-32β was more strongly induced by etoposide (Fig. 5).
RT-qPCR analysis of IL-32 in hepatoblastoma cell lines stimulated with IC50 values of chemotherapeutic agents a Hep G2 cells; b HuH-6 cells [2 (- ⊿⊿ Ct) method was used to analyze relative change in gene expression. A one-way ANOVA followed by a Tukey’s post-hoc HSD test were used for multiple comparisons. P < 0.05 was considered statistically significant]
IL-32 expression was observed in HB, but it was not associated with TAM infiltration
As a final step, IL-32 expression was examined by IHC. Il-32 expression was high in the cell block containing the Hep G2 and macrophage coculture, and low in the cell block containing HuH-6 cells, which was consistent with RT-qPCR analysis (Fig. 6a). Using 21 HB cases, IHC analysis showed that 52% of cases were negative for IL-32 and 48% of cases were positive. Low and high levels of IL-32 expression were seen in 70% and 30% of positive cases (Fig. 6b). There was no significant correlation between IL-32 expression and clinicopathological factors (Table 1). IHC analysis of Iba1 was performed in serial sections to examine the correlation between IL-32 expression and TAMs, however, no significant correlation was observed (Fig. 6b).
Immunohistochemical staining a IHC analysis of IL-32 for cell blocks of Hep G2 directly co-cultured with HMDM (positive control) and HuH-6 (negative control); b IHC for IL-32 and Iba1 for human tissue specimens [Magnification, 40 × /Scale bar, 20 µm]; c Scheme for proposed protumor effects of IL-32 in hepatoblastoma
Discussion
The findings of the present study showed that IL-32 was expressed in HB cell lines and up-regulated by cell–cell communication with macrophages. It is known that IL-32 expression is increased by stimulation with inflammatory cytokines such as TNF-α and IL-1β, infection, hypoxia, and oxidative stress [19]. Several stimuli can induce the expression of high levels of TNF-α and IL-1β in macrophages, and these cytokines might affect the expression of IL-32 in HB cell lines.
IL-32 expression has been observed in several kinds of cancer cells including lung cancer, and hepatocellular carcinoma. In hepatocellular carcinoma, IL-32α accelerated cancer cell progression via NF-κB and Bcl-2 signals [20]. In lung adenocarcinoma, an increase in IL-32 expression levels was associated with lymph node metastasis [21], and the activation of NF-κB signal, which induced production of matrix metalloproteinases, was suggested to be involved in cancer cell progression [22]. Thus, IL-32 was potentially associated with HB progression via NF-κB signal. Further studies are being planned to elucidate the autocrine mechanisms of IL-32 in HB by means of in vitro studies.
In the present study, IL-32 expression was enhanced by anticancer drugs. To the best of our knowledge, only one report has described the anticancer drug-induced IL-32 overexpression. Specifically, Zhang et al. demonstrated that cisplatin induced expression of Phospholipase A2-activating protein (PLAA) which up-regulated the IL-32 expression [23]. Goda et al. demonstrated the proapoptotic function of IL-32 in an in vitro cell culture study using the Tet-On/Off system in Hela cells [24]. However, the mechanisms by which IL-32 affects the chemosensitivity of HB cells remain unclear, and further studies are necessary to clarify this point.
IHC may be considered a limitation of our study due to the variability of the results. However, similar variability has been reported in gastric carcinoma [25], lung cancer [21], and multiple myeloma [26]. There are many reasons for such variability, including the probability that the IL-32 gene and the associated protein may not be expressed in the steady state, but only after stimulation (e.g., by chemotherapy), as shown in this study. Another possibility is the effect of long-term storage of paraffin sections on epitopes, which can produce false negative results [27]. More than 50% of our samples are more than a decade old, but this was due to the rarity of the examined tumors. The variability was also correlated with the histological subtype of the lung cancer [21]; HB is a heterogenous tumor with a spectrum of seven histological subtypes, however, such correlation would be meaningless in our study given the small number of patients. Despite these limitations, we included the IHC analysis in our study as it is necessary to investigate the target at the protein level as well as at the gene level [28].
In conclusion, IL-32 is potentially involved in chemoresistance through protumor activation of TAMs. IL-32 was detected in HB cell lines, but not in all HB patients, and we hypothesized that stimulation, such as chemotherapy, induced IL-32 expression in HB cells. As a result, IL-32 was considered to accelerate cell growth and suppress immune responses (Fig. 6c). In addition, our in vitro experiments showed that etoposide induced overexpression of IL-32 in cell lines. Further studies are necessary to confirm our hypothesis; however, IL-32 might be a critical mediator of chemoresistance in HB.
Data availability
The datasets generated and/or analyzed in the current study are available from the corresponding author on reasonable request.
Abbreviations
- Bcl-2:
-
B-cell lymphoma 2
- CD163:
-
Cluster of differentiation 163
- EGF:
-
Endothelial growth factor
- HB:
-
Hepatoblastoma
- HMDMs:
-
Human monocyte derived macrophages
- Iba1:
-
Ionized calcium-binding adapter molecule 1
- IC50:
-
Half maximal inhibitory concentration
- IDO-1/2:
-
Indoleamine dioxygenase-1/2
- IHC:
-
Immunohistochemical staining
- IL-1β/6/10/32/34:
-
Interleukin-1 beta/6/10/32/34
- Kyn pathway:
-
Kynurenine pathway
- LPS:
-
Lipopolysaccharide
- M-CSFR:
-
Macrophage colony stimulating factor receptor
- MMP-2,7,9:
-
Matrix metalloproteinases-2,7,9
- MTB:
-
Mycobacterium tuberculosis
- NF-κB:
-
Nuclear factor kappa-light-chain-enhancer of activated B cells
- NKs:
-
Natural killer cells
- PCNSL:
-
Primary central nervous system lymphoma
- PDGF:
-
Platelet-derived growth factor
- PD-L1/2:
-
Programmed death ligand 1/2
- PLAA:
-
Phospholipase A2-activating protein
- RNA-seq:
-
Ribonucleic acid sequencing
- RT-qPCR:
-
Real time quantitative polymerase chain reaction
- SCC:
-
Squamous cell carcinoma
- STAT3:
-
Signal transducer and activator of transcription 3
- TAMs:
-
Tumor-associated macrophages
- Tet-On/Off:
-
Tetracycline-On/Off
- TGF-β:
-
Tumor growth factor beta
- TME:
-
Tumor microenvironment
- TNF-α:
-
Tumor necrosis factor alpha
- VEGF:
-
Vascular endothelial growth factor
References
Spector LG, Birch J (2012) The epidemiology of hepatoblastoma. Pediatr Blood Cancer 59:776–779
López-Terrada D, Alaggio R, De Dávila MT, et al (2014) Towards an international pediatric liver tumor consensus classification: Proceedings of the Los Angeles COG liver tumors symposium. Modern Pathology 27:472–491
Cairo S, Armengol C, De Reyniès A et al (2008) Hepatic Stem-like Phenotype and Interplay of Wnt/β-Catenin and Myc Signaling in Aggressive Childhood Liver Cancer. Cancer Cell. https://doi.org/10.1016/j.ccr.2008.11.002
Dehne N, Mora J, Namgaladze D et al (2017) Cancer cell and macrophage cross-talk in the tumor microenvironment. Curr Opin Pharmacol 35:12–19
Yin M, Li X, Tan S et al (2016) Tumor-associated macrophages drive spheroid formation during early transcoelomic metastasis of ovarian cancer. J Clin Investig. https://doi.org/10.1172/JCI87252
Egawa M, Mukai K, Yoshikawa S et al (2013) Inflammatory Monocytes Recruited to Allergic Skin Acquire an Anti-inflammatory M2 Phenotype via Basophil-Derived Interleukin-4. Immunity. https://doi.org/10.1016/j.immuni.2012.11.014
Yang Q, Guo N, Zhou Y et al (2020) The role of tumor-associated macrophages (TAMs) in tumor progression and relevant advance in targeted therapy. Acta Pharm Sin B 10:2156–2170
Irie T, Yoshii D, Komohara Y et al (2022) IL-34 in hepatoblastoma cells potentially promote tumor progression via autocrine and paracrine mechanisms. Cancer Med. https://doi.org/10.1002/cam4.4537
Li L, Irie T, Yoshii D et al (2022) M-CSFR expression in the embryonal component of hepatoblastoma and cell-to-cell interaction between macrophages and hepatoblastoma. Med Mol Morphol. https://doi.org/10.1007/s00795-022-00323-y
Sloot YJE, Smit JW, Joosten LAB, Netea-Maier RT (2018) Insights into the role of IL-32 in cancer. Semin Immunol 38:24–32
Zaidan SM, Leyre L, Bunet R et al (1988) (2019) Upregulation of IL-32 Isoforms in Virologically Suppressed HIV-Infected Individuals: Potential Role in Persistent Inflammation and Transcription from Stable HIV-1 Reservoirs. J Acquir Immune Defic Syndr. https://doi.org/10.1097/QAI.0000000000002185
Netea MG, Azam T, Lewis EC et al (2006) Mycobacterium tuberculosis induces interleukin-32 production through a caspase-1/IL-18/interferon-γ-dependent mechanism. PLoS Med. https://doi.org/10.1371/journal.pmed.0030277
Shim S, Lee S, Hisham Y et al (2022) A Paradoxical Effect of Interleukin-32 Isoforms on Cancer. Front Immunol 13:422
Komohara Y, Kawauchi R, Makiyama E et al (2017) Selective depletion of cultured macrophages by magnetite nanoparticles modified with gelatin. Exp Ther Med. https://doi.org/10.3892/etm.2017.4640
Komohara Y, Hasita H, Ohnishi K et al (2011) Macrophage infiltration and its prognostic relevance in clear cell renal cell carcinoma. Cancer Sci. https://doi.org/10.1111/j.1349-7006.2011.01945.x
Komohara Y, Niino D, Saito Y et al (2013) Clinical significance of CD163+ tumor-associated macrophages in patients with adult T-cell leukemia/lymphoma. Cancer Sci. https://doi.org/10.1111/cas.12167
Fujiwara Y, Komohara Y, Ikeda T, Takeya M (2011) Corosolic acid inhibits glioblastoma cell proliferation by suppressing the activation of signal transducer and activator of transcription-3 and nuclear factor-kappa B in tumor cells and tumor-associated macrophages. Cancer Sci. https://doi.org/10.1111/j.1349-7006.2010.01772.x
Song H, Bucher S, Rosenberg K et al (2022) Single-cell analysis of hepatoblastoma identifies tumor signatures that predict chemotherapy susceptibility using patient-specific tumor spheroids. Nat Commun 13:4878. https://doi.org/10.1038/s41467-022-32473-z
Aass KR, Kastnes MH, Standal T (2021) Molecular interactions and functions of IL-32. J Leukoc Biol. https://doi.org/10.1002/JLB.3MR0620-550R
Kang YH, Park M-Y, Yoon D-Y et al (2012) Dysregulation of overexpressed IL-32α in hepatocellular carcinoma suppresses cell growth and induces apoptosis through inactivation of NF-κB and Bcl-2. Cancer Lett. https://doi.org/10.1016/j.canlet.2011.12.023
Sorrentino C, Di Carlo E (2009) Expression of IL-32 in human lung cancer is related to the histotype and metastatic phenotype. Am J Respir Crit Care Med. https://doi.org/10.1164/rccm.200903-0400OC
Zeng Q, Li S, Zhou Y et al (2014) Interleukin-32 contributes to invasion and metastasis of primary lung adenocarcinoma via NF-kappaB induced matrix metalloproteinases 2 and 9 expression. Cytokine. https://doi.org/10.1016/j.cyto.2013.09.017
Zhang F, Suarez G, Sha J et al (2009) Phospholipase A2-activating protein (PLAA) enhances cisplatin-induced apoptosis in HeLa cells. Cell Signal. https://doi.org/10.1016/j.cellsig.2009.02.018
Goda C, Kanaji T, Kanaji S et al (2006) Involvement of IL-32 in activation-induced cell death in T cells. Int Immunol. https://doi.org/10.1093/intimm/dxh339
Tsai CY, Wang CS, Tsai MM et al (2014) Interleukin-32 increases human gastric cancer cell invasion associated with tumor progression and metastasis. Clin Cancer Res. https://doi.org/10.1158/1078-0432.CCR-13-1221
Yan H, Dong M, Liu X et al (2019) Multiple myeloma cell-derived IL-32γ increases the immunosuppressive function of macrophages by promoting indoleamine 2,3-dioxygenase (IDO) expression. Cancer Lett. https://doi.org/10.1016/j.canlet.2019.01.012
Kim SW, Roh J, Park CS (2016) Immunohistochemistry for pathologists: protocols, pitfalls, and tips. J Pathol Transl Med. https://doi.org/10.4132/jptm.2016.08.08
Maggi LB, Weber JD (2013) Forget Transcription: Translation Is Where the Action Is. Mol Cell Biol. https://doi.org/10.1128/mcb.00231-13
Acknowledgements
We thank Mr. Takenobu Nakagawa and Mrs. Yuka Watanabe for their technical assistance.
Funding
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 20H03459 to Y.K., and No. 21K08622 to D.Y). This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Conceptualization and study design: AA, YK and TH; data collection: AA, LL, HH, TI, DY, HY, YF, SE, MH and SS; data analysis and interpretation: AA and YK; writing the manuscript (original draft preparation, review & editing): AA and YK; critical revision: TH.
Corresponding author
Ethics declarations
Conflict of interest
All of the authors declare that they have no conflicts of interest.
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. Informed consent to be included in the study was obtained from the patients’ caregivers. The study design and proposal were approved by the Institutional Review Board (IRB) at Kumamoto University (IRB No. 2224).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
383_2023_5557_MOESM1_ESM.tif
Supplementary file1 Image showing results of western blot analysis. Chemiluminescence (left) and chemiluminescence with molecular weight markers (right) for PD-L1 (a) and IDO-1 (b) are shown (TIF 310 KB)
383_2023_5557_MOESM2_ESM.tif
Supplementary file2 IC50 for carboplatin and etoposide. HepG2 (a) and HuH-6 (b) cell lines were cultured with various concentrations of carboplatin and etoposide for 48 hours and then the cell viability assay was performed (TIF 106 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Adawy, A., Li, L., Hirao, H. et al. Potential involvement of IL-32 in cell-to-cell communication between macrophages and hepatoblastoma. Pediatr Surg Int 39, 275 (2023). https://doi.org/10.1007/s00383-023-05557-0
Accepted:
Published:
DOI: https://doi.org/10.1007/s00383-023-05557-0