Abstract
Seminal studies from the early 20th century defined the structural changes associated with development and regeneration of the gills in goldfish at the gross morphological and cellular levels using standard techniques of light and electron microscopy. More recently, investigations using cell lineage tracing, molecular biology, immunohistochemistry and single-cell RNA-sequencing have pushed the field forward and have begun to reveal the cellular and molecular processes that orchestrate cell proliferation and regeneration in the gills. The gill is a multifunctional organ that mediates an array of important physiological functions, including respiration, ion regulation and excretion of waste products. It is comprised of unique cell types, such as pavement cells, ionocytes, chemoreceptors and undifferentiated stem or progenitor cells that regulate growth and replenish cell populations. The gills develop from the embryonic endoderm and are rich in cell types derived from the neural crest. The gills have the capacity to remodel themselves in response to environmental change, such as in the case of ionocytes, chemoreceptors and the interlamellar cell mass, and can completely regenerate gill filaments and lamellae. Both processes of remodeling and regeneration invariably involve cell proliferation. Although gill regeneration has been reported in only a limited number of fish species, the process appears to have many similarities to regeneration of other organs in fish and amphibians. The present article reviews the studies that have described gill development and growth, and that demonstrate a suite of genes, transcription factors and other proteins involved in cell proliferation and regeneration in the gills.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The gills of fish are multifunctional organs that mediate an impressive array of physiological processes. Among them are exchange of the respiratory gases, O2 and CO2, control of respiration by peripheral chemoreceptors, osmoregulation, excretion of waste products, and even undergoing structural changes to facilitate acclimatization to changing environmental conditions (Evans et al. 2005; Dymowska et al. 2012; Nilsson et al. 2012). The gills develop during embryonic and larval stages and continue to grow throughout life (Wilson and Laurent 2002; Morgan 1974a,b; Stolper et al. 2019). As an organ continuously exposed to the aquatic environment, the gills are recognized as an important pathway for uptake of toxic compounds and are vulnerable to water-borne diseases—both significant factors in affecting fish health (Evans et al. 2005; Marcos-López and Rodger 2020). The gills are also homologues of the mammalian lung (reviewed in Cadiz and Jonz 2020) and are often investigated as a convenient model system in respiratory physiology.
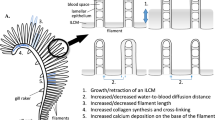
Although numerous studies over the last few decades have been devoted to investigating the development and growth of the gills (Morgan 1974a,b; Kimmel et al. 1995; Jonz and Nurse 2005), there are many questions that remain regarding the molecular mechanisms and signaling pathways that drive formation and growth of the gills. The gills possess a competent capacity for wound healing (Burkhardt-Holm et al. 1999; Dutta et al. 1996; Hemalatha and Banerjee 1997), as do many other tissues, but in some species they are also capable of regeneration following resection or amputation (Fig. 1; Schäfer 1936; Jonz et al. 2015; Stolper et al. 2019; Mierzwa et al. 2020; Nguyen and Jonz 2021; Cadiz et al. 2024; Ghanizadeh-Kazerouni et al. 2024). This stands in stark contrast to the limited, facultative regenerative potential of the mammalian lungs (Cadiz and Jonz 2020). A refined understanding of the cellular and molecular basis of growth and structural homeostasis of the fish gill is therefore needed to better appreciate the functional details of such a significant organ in fish biology. Moreover, of the more than 30,000 fish species described, gill regeneration has only been characterized in a limited number of species, suggesting that much more remains to be discovered. This brief review focuses specifically on studies that have helped to define mechanisms of post-embryonic growth, cell proliferation, structural plasticity, and regeneration in the gills. This includes recent studies that have exploited state-of-the-art techniques, as well as seminal studies that bear historical significance.

Modified from Stolper et al. (2019) with permission
Gill regeneration in three species of teleost fish. a Schäfer demonstrated regeneration of a gill filament stump and gradual replacement of the filament tip and lamellae in goldfish. Staging from left to right shows the day of resection to approximately 7 days post-resection. The horizontal line indicates the level of resection. From Schäfer (1936). Gill filament regeneration in zebrafish is shown at 7 (b) and 161 (c) days post-resection (dpr). The same gill filaments are shown at each stage with the dashed line indicating the level of resection. Scale bar = 50 μm and applies to both panels. d After 160 days of regeneration, most of the resected tissue was replaced. b-d are modified from Mierzwa et al. (2020) with permission. e In gill filaments from medaka, homeostatic and growth domains were defined in the gill filaments, both of which are replaced during regeneration.
Organization and cell types of the gill
The anatomy and organization of fish gills are well described and have been reviewed previously (Hughes 1984; Laurent 1984; Evans et al. 2005). In teleosts, four pairs of gill arches are present. Each of these extend two parallel rows of supportive filaments, called hemibranchs. The gill filaments have a unique structure that includes a rod-like column of chondrocytes, which provides support and prevents collapse of the gills during breathing. The filaments give rise to numerous respiratory lamellae, the units of gas exchange, and possess an extensive vascular system that receives deoxygenated blood via afferent arteries of the gill arches and filaments, and returns oxygenated blood back to the systemic circulation via efferent arteries of the filaments and gill arches (Laurent 1984; Olson 2002). During respiration, blood flows through a vascular sinus of each lamella, formed by pillar cells, in a manner referred to as counter-current exchange, which maximizes blood oxygenation.
The gills are composed of a wide range of cell types, many of which are unique to the gills and contribute to its diverse functionality (Wilson and Laurent 2002; Evans et al. 2005). An epithelium lines the gill filaments and lamellae. The epithelium is composed primarily of squamous pavement cells, which facilitate gas and ion transport. In the filament epithelium, ionocytes (or mitochondrion-rich cells) transport ions, such as Na+, Cl−, Ca2+, H+ and HCO3− to maintain homeostasis of internal fluids (Dymowska et al. 2012). These cells may be numerous and comprise up to 10% of cells in the gill epithelium, depending on species or environment. Mucous (or goblet) cells are found in the filament epithelium and may secrete mucous components, such as glycoproteins or lipids, that protect the epithelial surface. Also found within the gill filament epithelium are neuroepithelial or neuroendocrine cells (NECsFootnote 1), which fulfill the role of O2 and CO2/H+ chemoreceptors to regulate changes in ventilation rate during hypoxia (low O2) and hypercapnia (elevated CO2) (Jonz et al. 2004; Burleson et al. 2006; Qin et al. 2010; Porteus et al. 2014). The gills also possess an extensive nervous system that contains both sensory and motor branches (Nilsson 1984; Sundin and Nilsson 2002).
A growing number of studies are using single-cell RNA-sequencing to study cells of the fish gill, and this technique has so far been performed in species such as Atlantic salmon (Salmo salar, West et al. 2021), zebrafish (Danio rerio, Pan et al. 2022; Fabian et al. 2022) and tilapia (Oreochromis niloticus, Zheng and Wang 2023). These studies reveal the transcriptomic profiles of individual cell types within the gills by identifying expression of hundreds or thousands of genes across as many as 20 distinct cell types. These include the cell types mentioned above, as well as blood cells, immune cells, fibroblasts, endothelial cells and undifferentiated or stem cells.
Cell proliferation in the gill
Many cell types in the gills undergo proliferation, though the genes or factors responsible for their return to a proliferative state may be only partially understood. One of the best known examples of proliferation in the gill is that of the ionocytes that control the transport of ions across the epithelium (Evans et al. 2005; Dymowska et al. 2012; Zimmer et al. 2021). In rainbow trout (Oncorhynchus mykiss) acclimated to water of relatively low ionic concentration, ionocytes doubled their fractional surface area in the gill epithelium, owing to both proliferation and an increase in cell size, and effectively increased the blood-to-water diffusion distance (Greco et al. 1996). In killifish (Fundulus heteroclitus) transferred from seawater to freshwater, an increase in density of a specific subtype of ionocyte that facilitates the transition to freshwater was observed, without an increase in total ionocyte number (Katoh and Kaneko 2003). A similar phenomenon occurs in Atlantic salmon, where the numbers of ionocytes expressing specific isoforms of the Na+/K+-ATPase changes during migration from freshwater to seawater (McCormick et al. 2013). Moreover, studies have shown that ionocyte populations are also modified by thermal changes. In goldfish (Carassius auratus) acclimated from 25 °C to 7 °C, more ionocytes were found within the interlamellar cell mass (ILCM) as an apparent result of increased proliferation and migration of existing ionocytes from other regions of the gill (Mitrovic and Perry 2009).
Regulation of ionocyte proliferation in the gills has been described. In developing fish, which may lack fully-formed or functional gills, ionocytes may also be found in the skin. Studies investigating the origins of skin ionocytes in zebrafish (Danio rerio) have demonstrated that the transcription factor, Krüppel-like factor 4 (Klf4), regulates ionocyte progenitor cell populations in the epidermis (Chen et al. 2019); and that foxi3a and foxi1, genes encoding forkhead transcription factors, play a role in differentiation of vacuolar-type H+-ATPase ionocytes (Esaki et al. 2007, 2009). In zebrafish, gcm2, encoding the transcription factor, glial cells missing 2, is also involved in differentiation of multiple subtypes of ionocytes in the gills and skin (Chang et al. 2009; Shono et al. 2011; Kumai et al. 2015). The ionocyte fate of epidermal precursors appears to be mediated by Delta-Notch competitive lateral inhibition (Hsiao et al. 2007; Janicke et al. 2007).
Some species of fish are capable of dramatically altering their gill morphology as a response to temperature or hypoxia (Nilsson et al. 2012). The ILCM, a mass of cells that occupies the space between adjacent respiratory lamellae, is reversibly modified, thereby regulating the surface area of the gill epithelium that may be exposed to water. This phenomenon was first described in crucian carp (Carassius carassius), where exposure to hypoxia led to a reduction in the ILCM driven by apoptosis and reduced cell proliferation, allowing for a higher capacity of O2 uptake (Sollid et al. 2003). Subsequent studies further showed in goldfish and crucian carp that a reduction in temperature (e.g. from 25 °C to 7.5 °C in goldfish) produced an increase in the ILCM (Sollid et al. 2005); and in killifish acclimated to warm temperatures, regression of the ILCM was observed (McBryan et al. 2016). The ILCM is also increased upon air exposure in the amphibious mangrove killifish (Kryptolebias marmoratus, Ong et al. 2007). Although the regulation of cell populations within the ILCM is not yet clearly defined, in the grass carp (Ctenopharyngodon idella), an increase in expression of Bcl2, Bax and caspase 3, genes involved in regulating apoptosis, was associated with regression of the ILCM (Xu et al. 2002).
Neuroepithelial cells, the chemosensors of the gills that mediate ventilatory responses to hypoxia and hypercapnia, also proliferate. This process may underlie long-term changes in the ventilatory response to acute hypoxia observed in fish following acclimation to chronic hypoxia (Burleson et al. 2002; Jonz et al. 2004; Vulesevic et al. 2006). In response to acclimation to hypoxia for at least two weeks, the number and size of gill NECs in zebrafish was shown to increase significantly using techniques of immunohistochemistry and flow cytometry (Jonz et al. 2004; Pan et al. 2021), whereas hyperoxia reduced the number of NECs found in gill tissue (Vulesevic et al. 2006). Studies on NECs of the skin in zebrafish, which display chemoreceptor-like properties, have demonstrated that hypoxia acts as a mitogen to promote proliferation in as little as 24 h. Experiments using 5-bromo-2’-deoxyuridine (BrdU) revealed an increased uptake by hypoxic NECs undergoing DNA replication—an indicator of progression through the cell cycle (Dean et al. 2017). In addition, single-cell RNA-sequencing in the gills of adults acclimated to hypoxia identified an increased expression of numerous genes known to be involved in cell cycle regulation and proliferation (Table 1; Pan et al. 2022). In gill NECs, hypoxia increased expression of epas1, the gene encoding the endothelial PAS domain protein 1 (also called hypoxia-inducible factor [HIF]-2α). HIFs are a family of proteins comprised of multiple isoforms that mediate the cellular response to changes in O2 and are active under hypoxia (Semenza 2012). In teleosts, HIFs (particularly HIF-1α) have been implicated in O2 homeostasis, the cardioventilatory response to hypoxia, and hypoxia tolerance; and numerous studies have demonstrated increased levels of HIF isoforms as a response to hypoxia in many species, such as zebrafish, goldfish, killifish and rainbow trout, amongst others (reviewed by Mandic et al. 2021). Important for the present discussion, HIFs appear to play a role in regulation of cell proliferation in the gill. In the carotid body, the primary O2 sensors in mammals, chronic exposure to hypoxia induces cell proliferation and hyperplasia as a means of regulating the size and O2 sensitivity of the carotid body during acclimatization (Ortega-Sáenz and López-Barneo 2020). In mice exposed to chronic hypoxia, HIF-2α, but not HIF-1α, was necessary for cell proliferation in the carotid body (Hodson et al. 2016). HIF-2α also appears to be critical for acute O2 sensing in the mouse carotid body (Moreno-Domínguez et al. 2020). Within this context, transcriptomic analysis showing high differential expression of both paralogs of epas1 in gill NECs compared to other cell types, and the increased expression of epas1 in NECs following chronic hypoxia (Pan et al. 2022), highlights the potential importance of HIF-2α in regulating populations of O2 chemoreceptors in vertebrates as a means of promoting morphological changes responsible for O2 sensing during physiological adaptation to hypoxia.
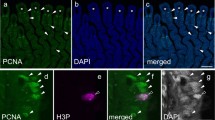
The progenitors or stem cells that eventually give rise to most cell types of the gills remain unknown. There is, however, plenty of evidence of stem cells in the gills, and it is generally understood that these may give rise to a variety of cell types. It has been appreciated for decades that undifferentiated or progenitor cells lie deep within the epithelium (Laurent and Dunel 1980; Wilson and Laurent 2002). It has been proposed, for example, that NECs with absent or variable immunoreactivity for the neurotransmitter, serotonin (5-hydroxytryptamine, 5-HT), may reflect the degree of differentiation of this specific cell type during cell renewal (Bailly et al. 1992; Jonz and Nurse 2003). More recent studies in zebrafish showed that the gene, sall4, was expressed in the gill filaments and branchial arches (Jackson et al. 2013). sall4 encodes a transcription factor involved in regulating stem cell pluripotency and self-renewal (Zhang et al. 2006; Yang et al. 2008) and has been implicated in reprogramming of differentiated cells (Neff et al. 2011). Regulation by sall4 may therefore provide a mechanism for supporting cell turnover in the gills by maintaining stem cell populations, or inducing dedifferentiation of cells to a state that would lead to proliferation. Other studies in zebrafish have identified a constitutive population of stem or mitotic cells that express the proliferating cell nuclear antigen (PCNA; Mierzwa et al. 2020). These cells were described within the interlamellar regions of the gill filament epithelium as well as the distal tip of the filaments. In addition, three separate populations of stem or progenitor cells were characterized by RNA-sequencing in the zebrafish gill based on their transcriptomic profiles (Pan et al. 2022). Stem cells positive for Sox-9 immunohistochemistry were also identified in the pseudobranch, a reduced gill-like non-respiratory organ, in molly fish (Poecilia sphenops, Mokhtar et al. 2023).
Perhaps the most informative description of cell proliferation and maintenance in the fish gill was provided by Stolper et al. (2019). In the gills of medaka (Oryzias latipes), they broadly defined two categories of stem cells that were limited to different regions of the gill. Growth stem cells were found in the branchial arches and at the distal tips of the filaments, where filaments are known to grow (Morgan 1974b); whereas homeostatic stem cells were embedded in tissue at the base of the lamellae along the longitudinal axis of the filament and generated cells that migrated away to other areas (Fig. 1e). Growth cells were further divided into four fate-restricted subtypes. Essentially, post-embryonic growth in the medaka gill appears to depend on the coordinated activity of these four stem cell subtypes, which maintain non-overlapping lineages, and each having independent embryonic origins (Stolper et al. 2019). Homeostatic stem cells, on the other hand, were shown to be important in generating new cells as they are lost, as in the case of pillar cells. Interestingly, homeostatic stem cells could be induced to generate a new growth domain in the gill filaments following tissue ablation.
Another cell type involved in development and cellular homeostasis of the gills are those that migrate from the neural crest. In vertebrates, neural crest cells (NCCs) are multipotent cells that are specified during embryonic stages at the neural plate and are present during formation of the neural tube. Subsequently, NCCs migrate away from this site and give rise to a diverse range of cell types, including those found in the central and peripheral nervous systems, muscle, the endocrine system, connective tissue, bone and skin (Gilbert 1988; Hall 2009). Earlier studies demonstrated that in the pharyngeal arches (which give rise to gill filaments in fish) the cartilaginous regions and blood vessels have neural crest origin (Landacre 1921; Le Lièvre and Le Douarin 1975). In zebrafish, it was further shown that smooth muscle of the gill filaments and the pillar cells of the lamellae are also derived from NCCs, based on their expression of sox10 (Mongera et al. 2013). Single-cell RNA-sequencing recently corroborated these findings and further demonstrated that the cartilaginous and filament tip regions of the gills are derived from gill-specific neural crest progenitors and require expression of gata3 and fgf10 (Fabian et al. 2022). Furthermore, expression of ascl1a was found in O2-chemoreceptive gill NECs, also located at the tips of gill filaments, as well as in intrinsic filament neurons (Pan et al. 2022). Ascl1 (achaete-scute homolog 1) is a member of the basic helix-loop-helix family of proteins involved in neurogenesis and is an autonomic neural crest transcription factor. Originally named Mash1 for the mammalian homolog of asc genes, it is required for formation of O2 chemoreceptors in the mammalian carotid body (Kameda 2005), a structure of neural crest origin (Pearse et al. 1973; Pardal et al. 2007). Expression of ascl1a is required for development of serotonergic cells within taste buds of the gill arches in zebrafish (Kapsimali et al. 2011), but whether ascl1a is critical for formation of serotonergic NECs in the gill filaments has yet to be resolved. Nevertheless, lineage tracing studies suggest that NECs are not derived from the neural crest, but are produced locally from the endoderm (Hockman et al. 2017).
Gill regeneration
The first report of regeneration of the gills in fish was provided by Schäfer (1936). He described formation of the Kiemenblattstumpf, a “gill leaf [or filament] stump”, in goldfish following removal of the filament tip. In his article, Schäfer described active cellular proliferation distal to the site of resection and eventual replacement of gill structure, including gill filaments and lamellae (Fig. 1a). Since that time, many studies have documented the ability of the gill to heal in response to injury or infection (e.g. Burkhardt-Holm et al. 1999; Dutta et al. 1996; Hemalatha and Banerjee 1997), but relatively few studies have observed the entire process of regeneration in the gill, or described the mechanism or processes that support regeneration. Regeneration in fish and amphibians generally involves three major phases: (1) wound healing, (2) formation of a blastema (or stump) may occur, and (3) redevelopment, which includes the proliferation of blastema cells, their differentiation and final tissue reconstruction (Nye et al. 2003; Poss et al. 2003; Münch et al. 2013; Stocum 2017; Saito et al. 2019; Cadiz and Jonz 2020).
More recent studies have used model vertebrates to investigate gill regeneration. Regeneration in the zebrafish gill was first identified by the presence of a blastema, similar to Schäfer’s gill stump, after resection of a gill filament (Jonz et al. 2015). Subsequent morphological and immunohistochemical studies in zebrafish revealed that 24 h after 0.2–0.3 mm of the filament tip was resected, undifferentiated cells were present in the blastema and expressed PCNA, indicating that they were mitotic (Fig. 1b,c; Mierzwa et al. 2020). Cell types, such as NECs, ionocytes and the vasculature were replaced during regeneration, and neurons intrinsic to the gill filaments began to extend axons to reinnervate NECs (Mierzwa et al. 2020; Nguyen and Jonz 2021). Significant replacement of these differentiated cell populations occurred about a week after resection, and it took approximately 160 days for 85% of the resected filament tip to be replaced (Fig. 1d; Mierzwa et al. 2020). Gill regeneration was also reported in medaka, where the entire growth domain of the filament tip was removed, and it was replaced about a month later (Fig. 1e; Stolper et al. 2019). As noted in the previous section, stem cells of the homeostatic domain in medaka can be induced to produce new tissue (Stolper et al. 2019). The same appears to be true in zebrafish. After complete amputation of gill filaments at their base, which included both homeostatic and growth domains, the filaments regenerated (F. Nguyen and M. Jonz, unpublished observations). A more recent study further demonstrated, in Atlantic salmon, that resection of 50% of the length of gill filaments resulted in a greater degree of tissue regeneration, compared to resection of 30% of filament length (Ghanizadeh-Kazerouni et al. 2024). The capacity of the gills to regenerate may therefore be correlated with the extent of injury or amputation.
Whereas the morphological changes observed during gill regeneration are well described, the molecular pathways orchestrating replacement of gill tissue are just beginning to be revealed. Fortunately, some aspects of the later stages of regeneration (i.e. tissue reconstruction) may be similar to events that occur during embryonic development. It is therefore possible to refer to the literature on gill development as a guide to understanding how gills regenerate. Some of the first extensive studies on gill development in fish were provided by Morgan in rainbow trout (1974a, 1974b). She showed complete development of the gill arches, filaments, lamellae, as well as internal structures, such as the vasculature, basement membrane and a differentiated epithelium, using light and electron microscopy (Fig. 2). Whereas gill development in trout began at approximately six days following incubation of fertilized eggs, corresponding to completion of gastrulation (at 10ºC, Morgan 1974a), in other species the gills may begin to develop sooner. In zebrafish, the gills begin to develop around the time of hatching at 48–72 h post-fertilization (at 28.5ºC, Kimmel et al. 1995) and resemble adult gills by approximately 14 days (Jonz and Nurse 2005). It was previously believed that the gills were derived from embryonic ectoderm (Goette 1901; Morgan 1974a), but more recent studies involving lineage tracing indicate that the gills are endoderm-derived (Warga and Nüsslein-Volhard 1999; Gillis and Tidswell 2017; Hockman et al. 2017).
Developmental stages of the gill in rainbow trout. a Consecutive stages (1–4) of the development of the gill arch, filaments and vasculature. Staging corresponds to approximately 30 days of development. ga, gill arch; pba, primary branchial artery; ff, filament folds; fa, filament artery; eba, efferent branchial artery; aba, afferent branchial artery. From Morgan 1974a with permission. b Consecutive stages (a-c) in transverse section showing development of a gill filament and lamella. Staging corresponds to approximately 30–100 days of development. afa, afferent filament artery; efa, efferent filament artery; c, collagen; mc, marginal channel; cl, column; d, desmsome; ed, endothelium; bm, basement membrane; e, epithelium; gr, granulocyte; pc, pillar cell; bs, blood spaces. From Morgan 1974b with permission
But what are the genes or factors responsible for directing formation and regeneration of the gills? In zebrafish, expression of gcm genes produce transcription factors required for formation of the pharyngeal arches and gill filament buds (gcm2, Hogan et al. 2004; gcmb, Hanaoka et al. 2004). Single-cell RNA-sequencing in adult zebrafish gills demonstrated continued expression of gcm genes in endothelial cells, ionocytes and progenitor cells (Pan et al. 2022), suggesting the potential of these genes in maintaining or potentially restructuring the gills during regeneration. Moreover, parathyroid hormone was shown to play an essential role in ionocyte differentiation by maintaining expression of gcm2 (Kwong and Perry 2015). In another study, microarray analysis was used to generate transcriptomic profiles of gills from zebrafish exposed to zinc (Zheng et al. 2010). The authors demonstrated upregulation of transcription factors, jun and gata, thought to stimulate stem cell differentiation and reactivate developmental pathways in the gill, although morphological changes in the gills were not confirmed in that study. However, a subsequent investigation identified increased expression of jun and gata in cell types, such as fibroblasts, neurons, endothelial cells, NECs and neurons following chronic exposure to hypoxia (Table 1; Pan et al. 2022), suggesting that these genes may mediate proliferation of these cell types in the adult gill. In addition, gata3 expression was previously shown to be required for gill bud formation in zebrafish (Sheehan-Rooney et al. 2013).
Wnt/β-catenin signaling, which mediates cell proliferation and fate decisions during embryogenesis and contributes to tissue regeneration (Poss et al. 2000; Logan and Nusse 2004; Tal et al. 2010), is another candidate for mediating gill regeneration. Wnt/β-catenin signaling via Lef1 was required for gill filament growth in developing zebrafish (Shimizu et al. 2012). Retinoic acid (RA) is involved in anterior-posterior patterning of the pharyngeal arches in vertebrates (DeLaurier 2019), and has also been implicated in formation of the gill arches in the little skate (Leucoraja erinacea) by acting through regulation of Fgf8 and Shh (Gillis et al. 2009). fgf8a is implicated in development of the branchial arches and head in zebrafish (Crump et al. 2004; Gebuijs et al. 2019). Collectively, Wnt, Fgf and RA signaling are well known to regulate blastema formation, proliferation and outgrowth during zebrafish fin regeneration (Gemberling et al. 2013). Interestingly, many of the above factors may play a role in gill regeneration. Increased expression of fgf8a and shha was localized to regenerating tips of gill filaments in zebrafish, and regrowth of the regenerating filament tip was reduced following chemical inhibition of fibroblast growth receptor 1 (FGFR1) activity (Cadiz et al. 2024). Furthermore, transcriptomic analysis in the gills of adult zebrafish localized shha specifically to NECs, and expression of genes encoding FGF receptors and the γ subunit of the RA receptor (rarga) were found in other cell types (Pan et al. 2022), though these genes have not yet been linked with regeneration.
Other important signaling pathways implicated in pharyngeal arch formation in zebrafish include the hox genes, which direct anterior-posterior identity of the pharyngeal arches, as well as end1, dlx and notch pathways, which control dorsal-ventral identity of the pharyngeal arches (reviewed by DeLaurier 2019). Many of these genes continue to be expressed in the adult zebrafish gill filaments. For example, expression of hoxb3a was found in gill NECs and fibroblasts, dlx genes were found in multiple cell types, and notch was expressed in pavement cells (Pan et al. 2022). Moreover, intense expression of her6 was localized to the regenerating filament tip in zebrafish gills, and chemical inhibition of the Notch signaling cascade almost entirely inhibited gill filament regeneration (Cadiz et al. 2024). her6 encodes a Notch effector protein that is involved in blastema cell proliferation during fin regeneration (Grotek et al. 2013). RNA-sequencing indicated her6 expression in endothelial cells and fibroblasts in the zebrafish gill, and her6 expression increased in the gills following in vivo exposure to hypoxia (Table 1; Pan et al. 2022). These data argue strongly for a role for Notch signaling in gill regeneration by mediating blastema cell proliferation.
In addition to the literature on gill development and regeneration of other organs in fish, studies on the regeneration of the external gills of the axolotl (Ambystoma mexicanum) are of interest to the present discussion. As a neotenic amphibian that does not undergo metamorphosis, the axolotl retains its external gills, which play a role in respiration (Gahlenbeck and Bartels 1970; Rosenkilde and Ussing 1996). While few studies have investigated gill regeneration in axolotls, Saito et al. (2019) demonstrated the importance of bone morphogenetic protein (BMP) and FGF in orchestrating gill regeneration. The authors demonstrated expression of Bmp2 and Fgf2 in the gills and showed that pharmacological blockade of BMP and FGF receptor activity inhibited gill regeneration (Saito et al. 2019). In zebrafish, bmp2b was also expressed in regenerating filament tips, and as mentioned above for FGF receptors, blocking activation of BMP receptors reduced gill regeneration (Cadiz et al. 2024). Additionally, other studies in axolotls have demonstrated that RA and thyroid hormones also influence gill regeneration (Crawford and Vincenti 1998; Lazcano et al. 2023).
Conclusion and future directions
The gill represents an organ of critical importance to fish biology. There is clear evidence that it is capable of profound cell proliferation, which underlies replacement of unique cell populations, significant structural remodeling in response to environmental change, and even regeneration following resection or amputation. This review has demonstrated that many of the genes and signaling pathways involved in gill development and organogenesis in zebrafish and other models may also be involved in gill regeneration. Specifically, the Notch, Wnt and Shh pathways may be particularly important in reconstruction of gill tissue during regeneration, as well as other factors, such as BMP, FGF and RA. Continued use of model vertebrates and other developmental or regenerative systems will provide important information for understanding gill regeneration in fish.
Challenges that remain for future exploration to better understand gill regeneration may include investigation of how stem cells are recruited to replace lost tissue, and how stem cell activity is controlled to ensure differentiation of specific cell types. And, more generally, studies may address whether gill regeneration occurs in all fish species, or if it is confined to specific taxa; and if there are limitations on gill regeneration. One idea proposes that because aquatic vertebrates, such as fish and amphibians, have complex life cycles that involve metamorphosis during early development, they retain the unique genetic potential that allows them to activate organ regeneration during later adult stages (Alibardi 2019). We may therefore expect that gill regeneration is widespread in fishes, making this an important field for future study.
Notes
It is important to note that gill neuroepithelial cells (NECs), which are respiratory chemoreceptors, are distinct from neuroepithelial cells of the same name that are important in development of the vertebrate nervous system.
References
Alibardi L (2019) Organ regeneration evolved in fish and amphibians in relation to metamorphosis: speculations on a post-embryonic developmental process lost in amniotes after the water to land transition. Ann Anat 222:114–119
Bailly Y, Dunel-Erb S, Laurent P (1992) The neuroepithelial cells of the fish gill filament: indolamine-immunocytochemistry and innervation. Anat Rec 233:143–161
Burkhardt-Holm P, Oulmi Y, Schroeder A, Storch V, Braunbeck T (1999) Toxicity of 4-chloroaniline in early life stages of zebrafish (Danio rerio): II. Cytopathology and regeneration of liver and gills after prolonged exposure to waterborne 4-chloroani- line. Arch Environ Contam Toxicol 37:85–102
Burleson ML, Carlton AL, Silva PE (2002) Cardioventilatory effects of acclimatization to aquatic hypoxia in channel catfish. Resp Physiol Neurohiol 131:223–232
Burleson ML, Mercer SE, Wilk-Blaszczak MA (2006) Isolation and characterization of putative O2 chemoreceptor cells from the gills of channel catfish (Ictalurus punctatus). Brain Res 1092:100–107
Cadiz L, Jonz MG (2020) A comparative perspective on lung and gill regeneration. J Exp Biol 223:jeb226076
Cadiz L, Reed M, Monis S, Akimenko MA, Jonz MG (2024) Identification of signalling pathways involved in gill regeneration in zebrafish. J Exp Biol 227(2):jeb246290
Chang WJ, Horng JL, Yan JJ, Hsiao CD, Hwang PP (2009) The transcription factor, glial cell missing 2, is involved in differentiation and functional regulation of H+-ATPase-rich cells in zebrafish (Danio rerio). Am J Physiol Regul Integr Comp Physiol 296:R1192–1201
Chen YC, Liao BK, Lu YF, Liu YH, Hsieh FC, Hwang PP, Hwang SL (2019) Zebrafish Klf4 maintains the ionocyte progenitor population by regulating epidermal stem cell proliferation and lateral inhibition. PLoS Genet 15(4):e1008058
Crawford K, Vincenti DM (1998) Retinoic acid and thyroid hormone may function through similar and competitive pathways in regenerating axolotls. J Exp Zool 282:724–738
Crump JG, Maves L, Lawson ND, Weinstein BM, Kimmel CB (2004) An essential role for Fgfs in endodermal pouch formation influences later craniofacial skeletal patterning. Development 131:5703–5716
Dean BW, Rashid TJ, Jonz MG (2017) Mitogenic action of hypoxia upon cutaneous neuroepithelial cells in developing zebrafish. Dev Neurobiol 77:789–801
DeLaurier A (2019) Evolution and development of the fish jaw skeleton. Wiley Interdiscip Rev Dev Biol 8(2):e337
Dutta HM, Munshi JS, Roy PK, Singh NK, Adhikari S, Killius J (1996) Ultrastructural changes in the respiratory lamellae of the catfish, Heteropneustes fossilis after sublethal exposure to malathion. Environ Pollut 92:329–341
Dymowska AK, Hwang PP, Goss GG (2012) Structure and function of ionocytes in the freshwater fish gill. Respir Physiol Neurobiol 184:282–292
Esaki M, Hoshijima K, Kobayashi S, Fukuda H, Kawakami K, Hirose S (2007) Visualization in zebrafish larvae of Na+ uptake in mitochondria-rich cells whose differentiation is dependent on foxi3a. Am J Physiol Regul Integr Comp Physiol 292:R470–R480
Esaki M, Hoshijima K, Nakamura N, Munakata K, Tanaka M, Ookata K, Asakawa K, Kawakami K, Wang W, Weinberg ES, Hirose S (2009) Mechanism of development of ionocytes rich in vacuolar-type H+-ATPase in the skin of zebrafish larvae. Dev Biol 329:116–129
Evans DH, Piermarini PM, Choe KP (2005) The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev 85:97–177
Fabian P, Tseng KC, Thiruppathy M, Arata C, Chen HJ, Smeeton J, Nelson N, Crump JG (2022) Lifelong single-cell profiling of cranial neural crest diversification in zebrafish. Nat Commun 13(1):13
Gahlenbeck H, Bartels H (1970) Blood gas transport properties in gill and lung forms of the axolotl (Ambystoma mexicanum). Respir Physiol 9:175–182
Gebuijs IGE, Raterman ST, Metz JR, Swanenberg L, Zethof J, Van den Bos R, Carels CEL, Wagener FADTG, Von den Hoff JW (2019) Fgf8a mutation affects craniofacial development and skeletal gene expression in zebrafish larvae. Biol Open 8(9):bio039834
Gemberling M, Bailey TJ, Hyde DR, Poss KD (2013) The zebrafish as a model for complex tissue regeneration. Trends Genet 29:611–620
Ghanizadeh-Kazerouni E, Wilson JM, Jones SRM, Brauner CJ (2024) Characteristics of a gill resection – regeneration model in freshwater laboratory-reared Atlantic salmon (Salmo salar). Aquaculture 579:740210
Gilbert SF (1988) Developmental Biology, 2nd edn. Sinauer, Sunderland
Gillis JA, Tidswell ORA (2017) The origin of vertebrate gills. Curr Biol 27:729–732
Gillis JA, Dahn RD, Shubin NH (2009) Shared developmental mechanisms pattern the vertebrate gill arch and paired fin skeletons. Proc Natl Acad Sci USA 106:5720–5724
Goette A (1901) Über die Kiemen Der Fische. Z Wiss Zool 69:533–577
Greco AM, Fenwick JC, Perry SF (1996) The effects of soft-water acclimation on gill structure in the rainbow trout Oncorhynchus mykiss. Cell Tissue Res 285:75–82
Grotek B, Wehner D, Weidinger G (2013) Notch signaling coordinates cellular proliferation with differentiation during zebrafish fin regeneration. Development 140:1412–1423
Hall BK (2009) The neural crest and neural crest cells in Vertebrate Development and Evolution. Springer, New York
Hanaoka R, Ohmori Y, Uyemura K, Hosoya T, Hotta Y, Shirao T, Okamoto H (2004) Zebrafish gcmb is required for pharyngeal cartilage formation. Mech Dev 121:1235–1247
Hemalatha S, Banerjee TK (1997) Histopathological analysis of sublethal toxicity of zinc chloride to the respiratory organs of the airbreathing catfish Heteropneustes fossilis (Bloch). Biol Res 30:11–21
Hockman D, Burns AJ, Schlosser G, Gates KP, Jevans B, Mongera A, Fisher S, Unlu G, Knapik EW, Kaufman CK, Mosimann C, Zon LI, Lancman JJ, Dong PDS, Lickert H, Tucker AS, Baker CV (2017) Evolution of the hypoxia-sensitive cells involved in amniote respiratory reflexes. Elife 6:e21231
Hodson EJ, Nicholls LG, Turner PJ, Llyr R, Fielding JW, Douglas G, Ratnayaka I, Robbins PA, Pugh CW, Buckler KJ, Ratcliffe PJ, Bishop T (2016) Regulation of ventilatory sensitivity and carotid body proliferation in hypoxia by the PHD2/HIF-2 pathway. J Physiol 594:1179–1195
Hogan BM, Hunter MP, Oates AC, Crowhurst MO, Hall NE, Heath JK, Prince VE, Lieschke GJ (2004) Zebrafish gcm2 is required for gill filament budding from pharyngeal ectoderm. Dev Biol 276:508–522
Hsiao C, You M, Guh Y, Ma M, Jiang Y, Hwang P (2007) A positive regulatory loop between foxi3a and foxi3b is essential for specification and differentiation of zebrafish epidermal ionocytes. PLoS ONE 2:e302
Hughes GM (1984) General anatomy of the gills. In: Hoar WS, Randall DJ (eds) Fish physiology, vol XA. Academic, San Diego, pp 1–72
Jackson R, Braubach OR, Bilkey J, Zhang J, Akimenko MA, Fine A, Croll RP, Jonz MG (2013) Expression of sall4 in taste buds of zebrafish. Dev Neurobiol 73:543–558
Janicke M, Carney TJ, Hammerschmidt M (2007) Foxi3 transcription factors and notch signaling control the formation of skin ionocytes from epidermal precursors of the zebrafish embryo. Dev Biol 307:258–271
Jonz MG, Nurse CA (2003) Neuroepithelial cells and associated innervation of the zebrafish gill: a confocal immunofluorescence study. J Comp Neurol 461:1–17
Jonz MG, Nurse CA (2005) Development of oxygen sensing in the gills of zebrafish. J Exp Biol 208:1537–1549
Jonz MG, Fearon IM, Nurse CA (2004) Neuroepithelial oxygen chemoreceptors of the zebrafish gill. J Physiol 560:737–752
Jonz MG, Zachar PC, Da Fonte DF, Mierzwa AS (2015) Peripheral chemoreceptors in fish: a brief history and a look ahead. Comp Biochem Physiol Mol Integr Physiol 186:27–38
Kameda Y (2005) Mash1 is required for glomus cell formation in the mouse carotid body. Dev Biol 283:128–139
Kapsimali M, Kaushik A-L, Gibon G, Dirian L, Ernest S, Rosa FM (2011) Fgf signaling controls pharyngeal taste bud formation through miR-200 and Delta-Notch activity. Development 138:3473–3484
Katoh F, Kaneko T (2003) Short-term transformation and long-term replacement of branchial chloride cells in killifish transferred from seawater to freshwater, revealed by morphofunctional observations and a newly established ‘time-differential double fluorescent staining’ technique. J Exp Biol 206:4113–4123
Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310
Kumai Y, Kwong RW, Perry SF (2015) A role for transcription factor glial cell missing 2 in Ca2+ homeostasis in zebrafish, Danio rerio. Pflügers Arch 467:753–765
Kwong RW, Perry SF (2015) An essential role for parathyroid hormone in gill formation and differentiation of ion-transporting cells in developing zebrafish. Endocrinology 156:2384–2394
Landacre FL (1921) The fate of the NC in the head of the Urodeles. J Comp Neurol 33:1–43
Laurent P (1984) Gill internal morphology. In: Hoar WS, Randall DJ (eds) Fish physiology, vol XA. Academic, San Diego, pp 73–183
Laurent P, Dunel S (1980) Morphology of gill epithelia in fish. Am J Physiol 238:R147–159
Lazcano I, Olvera A, Pech-Pool SM, Sachs L, Buisine N, Orozco A (2023) Differential effects of 3,5-T2 and T3 on the gill regeneration and metamorphosis of the Ambystoma mexicanum (axolotl). Front Endocrinol 14:1208182
Le Lièvre CS, Le Douarin NM (1975) Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. J Embryol Exp Morphol 34:125–154
Logan CY, Nusse R (2004) The wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20:781–810
Mandic M, Joyce W, Perry SF (2021) The evolutionary and physiological significance of the Hif pathway in teleost fishes. J Exp Biol 224:jeb231936
Marcos-López M, Rodger HD (2020) Amoebic gill disease and host response in Atlantic salmon (Salmo salar L.): a review. Parasite Immunol 42:e12766
McBryan TL, Healy TM, Haakons KL, Schulte PM (2016) Warm acclimation improves hypoxia tolerance in Fundulus heteroclitus. J Exp Biol 219:474–484
McCormick SD, Regish AM, Christensen AK, Björnsson BT (2013) Differential regulation of sodium-potassium pump isoforms during smolt development and seawater exposure of Atlantic salmon. J Exp Biol 216:1142–1151
Mierzwa AS, Nguyen F, Xue M, Jonz MG (2020) Regeneration of the gill filaments and replacement of serotonergic neuroepithelial cells in adult zebrafish (Danio rerio). Respir Physiol Neurobiol 274:103366
Mitrovic D, Perry SF (2009) The effects of thermally induced gill remodeling on ionocyte distribution and branchial chloride fluxes in goldfish (Carassius auratus). J Exp Biol 212:843–852
Mokhtar DM, Sayed RKA, Zaccone G, Alesci A, Hussein MM (2023) The potential role of the pseudobranch of molly fish (Poecilia sphenops) in immunity and cell regeneration. Sci Rep 13(1):8665
Mongera A, Singh AP, Levesque MP, Chen YY, Konstantinidis P, Nüsslein-Volhard C (2013) Genetic lineage labeling in zebrafish uncovers novel neural crest contributions to the head, including gill pillar cells. Development 140:916–925
Moreno-Domínguez A, Ortega-Sáenz P, Gao L, Colinas O, García-Flores P, Bonilla-Henao V, Aragonés J, Hüttemann M, Grossman LI, Weissmann N, Sommer N, López-Barneo J (2020) Acute O2 sensing through HIF2α-dependent expression of atypical cytochrome oxidase subunits in arterial chemoreceptors. Sci Signal 13(615):eaay9452
Morgan M (1974a) The development of gill arches and gill blood vessels of the rainbow trout, Salmo Gairdneri. J Morphol 142:351–363
Morgan M (1974b) Development of secondary lamellae of the gills of the trout, Salmo Gairdneri (Richardson). Cell Tissue Res 151:509–523
Münch J, González-Rajal A, de la Pompa JL (2013) Notch regulates blastema proliferation and prevents differentiation during adult zebrafish fin regeneration. Development 140:1402–1411
Neff AW, King MW, Mescher AL (2011) Dedifferentiation and the role of sall4 in reprogramming and patterning during amphibian limb regeneration. Dev Dyn 240:979–989
Nguyen F, Jonz MG (2021) Replacement of mitochondrion-rich cells during regeneration of the gills and opercular epithelium in zebrafish (Danio rerio). Acta Histochem 123:151738
Nilsson S (1984) Innervation and pharmacology of the gills. In: Hoar WS, Randall DJ (eds) Fish physiology, vol XA. Academic, San Diego, pp 185–227
Nilsson GE, Dymowska A, Stecyk JA (2012) New insights into the plasticity of gill structure. Respir Physiol Neurobiol 184:214–222
Nye HLD, Cameron JA, Chernoff EAG, Stocum DL (2003) Regeneration of the urodele limb: a review. Dev Dyn 226:280–294
Olson KR (2002) Vascular anatomy of the fish gill. J Exp Zool 293:214–231
Ong KJ, Stevens ED, Wright PA (2007) Gill morphology of the mangrove killifish (Kryptolebias Marmoratus) is plastic and changes in response to terrestrial air exposure. J Exp Biol 210:1109–1115
Ortega-Sáenz P, López-Barneo J (2020) Physiology of the carotid body: from molecules to disease. Annu Rev Physiol 82:127–149
Pan W, Scott AL, Nurse CA, Jonz MG (2021) Identification of oxygen-sensitive neuroepithelial cells through an endogenous reporter gene in larval and adult transgenic zebrafish. Cell Tissue Res 384:35–47
Pan W, Godoy RS, Cook DP, Scott AL, Nurse CA, Jonz MG (2022) Single-cell transcriptomic analysis of neuroepithelial cells and other cell types of the gills of zebrafish (Danio rerio) exposed to hypoxia. Sci Rep 12(1):10144
Pardal R, Ortega-Sáenz P, Durán R, López-Barneo J (2007) Glia-like stem cells sustain physiologic neurogenesis in the adult mammalian carotid body. Cell 131:364–377
Pearse AG, Polak JM, Rost FW, Fontaine J, Le Lièvre C, Le Douarin N (1973) Demonstration of the neural crest origin of type I (APUD) cells in the avian carotid body, using a cytochemical marker system. Histochemie 34:191–203
Porteus CS, Abdallah SJ, Pollack J, Kumai Y, Kwong RW, Yew HM, Milsom WK, Perry SF (2014) The role of hydrogen sulphide in the control of breathing in hypoxic zebrafish (Danio rerio). J Physiol 592:3075–3088
Poss KD, Shen J, Keating MT (2000) Induction of lef1 during zebrafish fin regeneration. Dev Dyn 219:282–286
Poss KD, Keating MT, Nechiporuk A (2003) Tales of regeneration in zebrafish. Dev Dyn 226:202–210
Qin Z, Lewis JE, Perry SF (2010) Zebrafish (Danio rerio) gill neuroepithelial cells are sensitive chemoreceptors for environmental CO2. J Physiol 588:861–872
Rosenkilde P, Ussing AP (1996) What mechanisms control neoteny and regulate induced metamorphosis in urodeles? Int J Dev Biol 40:665–673
Saito N, Nishimura K, Makanae A, Satoh A (2019) Fgf- and bmp- signaling regulate gill regeneration in Ambystoma mexicanum. Dev Biol 452:104–113
Schäfer W (1936) Die Regeneration der Kiemen und Flossenstrahlen beim Goldfisch (Carassius auratus). Jena Z Naturw 70:303–358
Semenza GL (2012) Hypoxia-inducible factors in physiology and medicine. Cell 148:399–408
Sheehan-Rooney K, Swartz ME, Zhao F, Liu D, Eberhart JK (2013) Ahsa1 and Hsp90 activity confers more severe craniofacial phenotypes in a zebrafish model of hypoparathyroidism, sensorineural deafness and renal dysplasia (HDR). Dis Model Mech 6:1285–1291
Shimizu N, Kawakami K, Ishitani T (2012) Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/β-catenin signaling-reporter transgenic zebrafish. Dev Biol 370:71–85
Shono T, Kurokawa D, Miyake T, Okabe M (2011) Acquisition of glial cells missing 2 enhancers contributes to a diversity of ionocytes in zebrafish. PLoS ONE 6(8):e23746
Sollid J, De Angelis P, Gundersen K, Nilsson GE (2003) Hypoxia induces adaptive and reversible gross morphological changes in crucian carp gills. J Exp Biol 206:3667–3673
Sollid J, Weber RE, Nilsson GE (2005) Temperature alters the respiratory surface area of crucian carp Carassius carassius and goldfish Carassius auratus. J Exp Biol 208:1109–1116
Stocum DL (2017) Mechanisms of urodele limb regeneration. Regeneration 4:159–200
Stolper J, Ambrosio EM, Danciu DP, Buono L, Elliott DA, Naruse K, Martínez-Morales JR, Marciniak-Czochra A, Centanin L (2019) Stem cell topography splits growth and homeostatic functions in the fish gill. Elife 8:e43747
Sundin L, Nilsson S (2002) Branchial innervation. J Exp Zool 293:232–248
Tal TL, Franzosa JA, Tanguay RL (2010) Molecular signaling networks that choreograph epimorphic fin regeneration in zebrafish - a mini-review. Gerontology 56:231–240
Vulesevic B, McNeill B, Perry SF (2006) Chemoreceptor plasticity and respiratory acclimation in the zebrafish Danio rerio. J Exp Biol 209:1261–1273
Warga RM, Nüsslein-Volhard C (1999) Origin and development of the zebrafish endoderm. Development 126:827–838
West AC, Mizoro Y, Wood SH, Ince LM, Iversen M, Jørgensen EH, Nome T, Sandve SR, Martin SAM, Loudon ASI, Hazlerigg DG (2021) Immunologic profiling of the Atlantic salmon gill by single nuclei transcriptomics. Front Immunol 12:669889
Wilson JM, Laurent P (2002) Fish Gill morphology: inside out. J Exp Zool 293:192–213
Xu XN, Chen SL, Jiang ZX, Nissa MU, Zou SM (2002) Gill remodeling increases the respiratory surface area of grass carp (Ctenopharyngodon idella) under hypoxic stress. Comp Biochem Physiol Mol Integr Physiol 272:111278
Yang J, Chai L, Fowles TC, Alipio Z, Xu D, Fink LM, Ward DC, Ma Y (2008) Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murine-embryonic stem cells. Proc Natl Acad Sci USA 105:19756–19761
Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y, Yang J, Ma Y, Chai L, Ng HH, Lufkin T, Robson P, Lim B (2006) Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol 8:1114–1123
Zheng S, Wang WX (2023) Physiological and immune profiling of tilapia Oreochromis Niloticus gills by high-throughput single-cell transcriptome sequencing. Fish Shellfish Immunol 141:109070
Zheng D, Kille P, Feeney GP, Cunningham P, Handy RD, Hogstrand C (2010) Dynamic transcriptomic profiles of zebrafish gills in response to zinc supplementation. BMC Genomics 11:553
Zimmer AM, Goss GG, Glover CN (2021) Reductionist approaches to the study of ionoregulation in fishes. Comp Biochem Physiol B Biochem Mol Biol 255:110597
Funding
Research support was provided by a grant from the Natural Sciences and Engineering Research Council of Canada (grant no. 05571).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author has no competing interests to declare.
Additional information
Communicated by Bernd Pelster.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jonz, M.G. Cell proliferation and regeneration in the gill. J Comp Physiol B (2024). https://doi.org/10.1007/s00360-024-01548-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00360-024-01548-2