Abstract
Plants, being immobile, are vulnerable to a variety of environmental challenges, including abiotic stresses such as high temperatures, low temperatures, flooding, drought, heavy metal toxicity, and high salt levels, all of which can negatively impact plant growth and productivity. These stresses can cause a variety of plant responses, including the production of reactive oxygen species, damage to cell membranes, and decreased photosynthetic efficiency which can disrupt growth and development, by impacting biochemical, physiological, and molecular processes. Plants have evolved complex mechanisms to deal with these abiotic stresses. The way that plants perceive and respond to stress signals plays a crucial role in initiating the resistance mechanisms. Recent research has highlighted the complexity of the molecular processes involved in plant responses to abiotic stress, including signal perception, signaling cascades, gene expression, protein synthesis and post-translational modifications. This review provides an overview of how plants respond to major abiotic stresses, including cold, heat, drought, and salinity, on both at physiological and molecular level. We have also discussed the ways in which plants sense various stresses and use molecular signaling to enhance tolerance to environmental stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Abiotic stress is a major constraint for sedentary plants to achieve their maximum plant productivity. These stressors, including drought, salt, and extreme temperatures, can limit the growth, development, and finally reduce the agricultural output (Yasin et al. 2020). With the increasing global population, the impact of abiotic stress on plants has become the prime focus to achieve food security (Pareek et al. 2020; Chaudhry and Sidhu 2022). The full extent of the impact of abiotic stress on agricultural lands is challenging and, according to FAO, it is estimated that around 96.5% of rural land worldwide is affected by these stressors, and the proportion that each stress contributing to this overall figure is now known (FAO 2021).
Stress is a complex and multigenic phenomenon which impacts crop plants in theirdevelopment, growth, and yield. However, plants have developed mechanisms to defend themselves against stress, by adjusting their tolerance potential through a combination of molecular and cellular plasticity (Narendra and Sarvajeet 2016; Imran et al. 2021). Recent progress in understanding the molecular mechanisms of plant responses to abiotic stress has revealed a complexity of these processes including multiple levels sensing, signaling, gene expression, protein production, and post-translational modifications (Zhang et al. 2022). Comprehending the physiological and biochemical changes that occur during abiotic stress, as well as the interactions between different stressors, is essential to understand the cellular signaling, gene expression, communication between cells, and the overall coordination of stress responses in plants (Raza et al. 2023a, b, c, d). The identification of key molecular networks and the mechanisms of signal perception and transmission for activating stress adaptive responses is crucial for developing strategies for improving plant stress tolerance (Narendra and Sarvajeet 2016). Engineering stress tolerance in crops requires a deep understanding of how plants perceive and transmit signals in response to stress (Raza et al. 2021). The intent of this review is to comprehensively examine the mechanisms of signal perception, along with the physiological and molecular processes that plants employ in response to abiotic stress. Further, this review also sheds light on the intricate ways in which plants sense and respond to various abiotic stressors. Understanding these mechanisms is crucial for developing strategies to enhance crop resilience in the face of adverse environmental conditions, ultimately contributing to global food security and sustainable agriculture.
Response to Sensors of Abiotic Stress
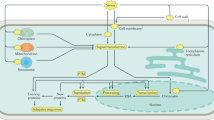
Abiotic stress responses in plants involve the activation of specific sensors, which are responsible for detecting and responding to various environmental stressors. These sensors are typically located in the cell membrane, and upon activation, trigger signaling cascades that lead to the transcription of stress-responsive genes, the production of stress-responsive proteins, and the activation of various metabolic pathways (Hotamisligil and Davis 2016). This eventually leads to the plant’s ability to tolerate or adapt to the stress factor. To cope with harsh environmental conditions, plants activate an acclimation response to increase their tolerance which is triggered by changes in various cellular processes such as the cytoskeleton, cellular organization, ion balance, and calcium influx in the membrane (Yasin 2015). Key signaling molecules involved in this process include ABA, calcium flux, phospholipids, and reactive oxygen species.
Abiotic Stress-Induced Mechanisms in Signaling Pathways
Calcium
The role of cytoskeleton organization and calcium influxes during early stages of the stress response pathway is well established (Yasin 2015; Kumar et al. 2023). The significance of calcium as a secondary messenger in the stress response has been proven through the use of chemical reagents that modify its concentration, such as N-(6-aminohexyl)-5-chloro-1-naphthalene-sulfonamide hydrochloride, an antagonist of calmodulin and Ca2+-dependent protein kinases, or the Ca2+ channel blocker La3+ (Ren et al. 2014; Ghosh et al. 2022). In addition, it has been found that calcium-selective cation channel that is mechano-sensitive and activated in response to low temperatures (Mitra 2015; Yasin 2015). In tobacco protoplasts, it has been proposed that membrane fluidity may control ion channel activity by causing cytoskeleton alteration and enhancing the cold-induced calcium influx after microtubules and actin microfilaments are disrupted (Kumar et al. 2023). A rapid increase in cytosolic Ca2+ and pH flux occurs in plants in response to abiotic stress as a result of both Ca2+influxes from extracellular storage and Ca2+ release from internal stores (Yasin 2015). Further transitory increases in Ca2+ are caused by signaling molecules like ABA.
Phospholipids
The perception and transmission of environmental cues rely heavily on the plasma membrane, and under stress, plants species have been shown to modify their phospholipid composition (Li et al. 2023; Liang et al. 2023). Because phosphatidylinositol 4-phosphate 5-kinase (PI5K) is activated during osmotic stress, plants produce more PIP2 (phosphatidylinositol 4,5-biphosphate), which is a precursor to second-message molecules (DeWald et al. 2001). Phosphatidic acid has been shown as a signaling lipid molecule that swiftly accumulates in response to different abiotic stresses and regulates a number of proteins involved in abscisic acid signaling (Li et al. 2023). PIP2 is a signal molecule that may play a role in several processes, including ion homeostasis and the recruitment of signal complexes to specific membrane sites (Kobrinsky et al. 2000). IP3 levels increasingly accumulate when plants are exposed to various stimuli, including hormones and stressors, (Gjindali and Johnson 2023). Elevated expression of genes that respond to ABA, such as RD29a, KIN1, COR15 A, HSP70, and ADH, implies that the expression of these genes is mediated by a different pathway independent of IP3 levels. Phosphatidic acid also serves as a second messenger (Singh et al. 2012; Yao and Xue 2018).
Reactive Oxygen Species
Reactive oxygen species (ROS) build up in response to all types of abiotic stress, act both as a damaging agent that contributes to stress injury and as secondary signals that trigger ROS scavengers and other therapeutic mechanisms (Yasin et al. 2014, 2020). Oxidative stress causes an upregulation of certain genes that are implicated in the response to osmotic stress. Many ABA-mediated effects, including temperature tolerance, Ca2+ channel activation, and stomatal closure, have been linked to ROS (Yasin et al. 2020). According to Xiong and Zhu (2002), ROS could be promptly recognized by the oxidation of conserved cysteine residues in signaling proteins like tyrosine phosphatases and triggered downstream from the signal cascade via Ca2+.
Reactive Oxygen Species (ROS), produced as natural byproducts of cellular metabolism are integral to plant physiology (Huang et al. 2019). However, under environmental stress like drought, extreme temperatures, salinity, or pathogen attacks, ROS levels can surge, leading to oxidative stress and cellular damage (Foyer and Noctor 2016). Remarkably, ROS also serve as crucial signaling molecules in stress responses (Mittler 2017). Moderate ROS levels activate signaling pathways, culminating in the expression of stress-responsive genes, enabling plants to cope with adversity. Key regulatory factors in ROS-mediated gene expression include Nuclear Factor-Y (NF-Y) (Li et al. 2005; Kavi Kishor et al. 2022) and APETALA2/Ethylene Response Factor (AP2/ERF) transcription factors (Liu and He 2017), Mitogen-Activated Protein Kinases (MAPKs) (Nuruzzaman et al. 2013), NAC (NAM, ATAF, and CUC) transcription factors (Davletova et al. 2005), and Jasmonic Acid (JA) and Abscisic Acid (ABA)-signaling components (Mittler and Blumwald 2010). ROS regulation also involves antioxidant enzymes such as superoxide dismutase (SOD), catalase (CAT), and peroxidases, which detoxify ROS, maintaining cellular redox balance (Yasin et al. 2014; Mittler 2017).
ROS, while essential for cellular processes, can be detrimental when overproduced due to environmental stress. Their dual role as harmful agents and signaling molecules underscores the significance of precise ROS regulation and the involvement of specific transcription factors and pathways. This suggests that ROS regulation and signaling holds promise for developing stress-tolerant crops and ensuring sustainable agriculture under changing environmental conditions.
Abscisic Acid
Under stressful conditions, plants produce a variety of hormones such as abscisic acid (ABA), ethylene, jasmonate, and salicylic acid, to cope with the stress. Among these hormones, ABA plays a key role in the signaling mechanism during abiotic stress (Raza et al. 2023c). Low-temperature, high-salt concentrations, and to a lesser extent, drought, can cause an increase in the production and accumulation of ABA. When stress is alleviated, the levels of ABA decrease but not necessarily ABA is deconstructed (Akhiyarova et al. 2023). Previous studies have suggested that ABA plays a crucial role in the response to osmotic stress in plants, while the response to cold stress is relatively ABA-independent (Ma et al. 2015). However, recent research has shown crosstalk between various stresses signaling pathways and the activation of ABA-independent genes, including LEA-like genes, in response to cold stress (Tuteja 2007; Joshi et al. 2010; Ding et al. 2013). These findings suggest that the role of ABA in stress responses is more complex than previously thought and that ABA-dependent and ABA-independent pathways may work together in a coordinated manner to regulate plant responses to stress.
ABA biosynthetic enzymes LOS5 and LOS6 have an impact on drought-induced ABA synthesis, as they both encode Mo cofactor sulfurase and are allelic to ABA3 and ABA1, respectively (Xiong and Zhu 2002). The activity of stress-responsive genes such as COR15, COR47, P5CS, RD22, and RD2A is greatly diminished or completely halted by certain genetic mutations (Chinnusamy et al. 2007). However, the expression of these genes remains unchanged when activated by DREB2A, a drought stress-specific transcription factor (Fujii et al. 2009). DREB2A interacts with cis-acting Dehydration-Responsive Element-Binding protein sequence, which triggers the activation of genes involved in coping with drought and salt stress in Arabidopsis thaliana. This suggests that DREB2A activity or the activity of other related factors may be controlled by ABA signaling (Cutler et al. 2010). ABA signaling also inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins, which may play a role in regulating DREB2A activity (Park et al. 2009).
Stress-Mediated Transcription Regulation
The stress response involves both transcriptional and post-transcriptional processes; moreover, many adaptive changes, such as lipid composition and sugar accumulation, may be derived at least in part from post-transcriptional activation processes (Aslam et al. 2022). The promoters of the COR15a and COR78 genes in Arabidopsis contain cis-regulatory regions that are responsible for regulating transcription in response to cold and drought conditions. These elements, known as either drought-responsive elements (DREs) or low-temperature responsive elements (LTREs), are found in all COR promoter regions and comprised a 9-bp consensus sequence with a 5-bp core motif (CCGAC) known as the C-repeat (CRT), for ABAbiosynthesis ABREs (ABA-responsive elements) (Fig. 1). In response to cold, high salt concentrations, and drought, DRE/LTREs (Dehydration-Responsive Element/ Low-Temperature-Responsive Element) promote gene expression, but not in response to exogenous application of ABA. Five different genes have been found to encode DRE-binding proteins, all of which can be binded to the DRE/CRT (C-repeat) and transcriptionally activate the expression of the COR gene. They have been divided into two groups, DREB1 and DREB2, based on their expression patterns (Chiraq et al. 2022).
The expression of the COR gene is affected by both cold and drought stress in various ways. Stress stimulus responses start when the binding proteins (DREB/CBF), which are all referred to as drought-responsive elements (DREs) or low-temperature responsive elements (LTREs) found in the promotor region of COR genes, receive the stress signals from transcription factors (like Myb-containing TF). As a reaction to drought stress, the ABA biosynthesis pathway will transmit the signals to the ABA-biosynthesis responsive elements (ABREs) to initiate the development of a tolerance mechanism
The expression of the three DREB1 genes, known as CBF (CRT binding factors), located in the tandem region of Arabidopsis chromosome 4 in the order DREB1B(CBF1), DREB1A(CBF3), and DREB1C(CBF2), and are particularly stimulated by cold, is only possible when these genes are expressed. Drought selectively increases the expression of the genes that code for the two DREB2 proteins. As a result of increased tolerance to dehydration brought on by salt or drought in Arabidopsis plants, cold-inducible promoters are not sensitive to autoregulation (Sakuma et al. 2006). The CBF/DREB1 gene transcripts and the CBF/DREB1 promoter are not present in the DRE/CRT sequence. However, after only 15 min of low-temperature exposure, the transcript level for three CBF genes increases and reaches a peak after 2 h. Recent studies suggest that in addition to transcriptional and post-transcriptional mechanisms, epigenetic modifications such as DNA methylation and histone modifications also play a crucial role in the regulation of the CBF/DREB1 regulon in Arabidopsis under low-temperature stress (Ghosh et al. 2022). It has been proposed that a putative transcription factor (TF) that is thought to be responsible for the transcriptional activation of CBF genes is constitutively expressed but remains inactive at warm temperatures. This TF, known as ICE, may be trapped in the cytoplasm or unable to bind DNA or activate transcription. The induction of the COR genes appears to involve a two-step cascade of transcriptional activators: Upon exposure to low temperatures, changes in either ICE or a related protein would activate ICE and trigger the induction of CBF/DREB1 expression, which in turn transcriptionally activates the COR genes. Additionally, ICE may also transcriptionally regulate other cold-adaptation genes unrelated to the CBF/DREB1 regulon.
According to the proposed function of HOS1, early components upstream of CBF/DREB1 appears to be susceptible to specific ubiquitination-mediated degradation. Cold-induced expression of the COR gene is enhanced in hos1 mutant plants, but induction by salt and ABA remains unchanged (Liu et al. 2019). Given that hos1 has higher CBF levels than the wild type, it has been suggested that hos1 may counteract the decline of CBF (Lee et al. 2001). The transcriptional adaptor ADA and the histone acetyl transferase GCN5 may also be required as cofactors for CBF-mediated transcription (Stockinger et al. 2001). It is hypothesized that mutations in these cofactors may influence the regulation of the COR genes without impacting the expression of the CBF. This gives us the impression of the Arabidopsis sfr6 mutant, which has reduced COR expression but unmodified CBF expression. The impact of SFR6 on the activity of CBF (post-transcriptionally or post-translationally) or a CBF cofactor is not yet known. COR15a, Kin1, and the stress-inducible expression of genes missing the DRE/CRT motif are examples of genes that cannot be induced to express in stressed sfr6 plants, though not all of them can (such as CBF1, CBF2, CBF3 & ATP5CS1). Sfr6 may play a role halfway between the transcription of CBF/DRE and the stimulation of the COR regulon, according to certain theories. Because the sfr6 mutation also affects the expression of the COR6 gene in response to osmotic stress and exogenous ABA, it is possible that sfr6 is involved in the post-transcriptional and post-translational activation of several TFs or that it is a co-activator for the transcription of the COR gene (Zhang et al. 2015).
The CBF/DREB1 regulatory system and its role in response to abiotic stress are believed to be conserved among the plant kingdom (Hu et al. 2022). The presence of a DRE/CRT region in the promoter of the ortholog of Arabidopsis COR15a, bn115 gene, in Brassica napus allows its cold-induced expression. Cold-induced expression of the wheat cold-inducible gene wcs120 has been described. In monocots (rice, barley, and rye) and dicots (alfalfa, rapeseed, and cucumber), but not in tomato and pepper, cold induces transcription from the wcs120 promoter. The genes for tomato COR homologs have not been discovered, possibly because tomatoes are subtropical plants that cannot acclimate to the cold. Although expression of A. thaliana CBF1 in tomatoes increases resistance to cold, drought, and oxidative stress, it does not enhance freezing tolerance (Hsieh et al. 2002). In rice, DREB genes have been identified, and their role in stress tolerance has been established (Mizoi et al. 2018).
Drought Stress
Drought is one of the major abiotic stresses that lead to significant yield loss in plants, such as rice (53–92%), wheat (57%), and maize (63–87%) (Daryanto et al. 2016) During drought conditions, plants regulate water homeostasis at the whole plant level through various physiological traits, which influence crop responses to drought (Fig. 2). The consequences of drought include poor germination, reduced seedling growth, reduced availability of nutrients, decreased photosynthesis activity, decreased plant fresh and dry weight, and dehydration (Raza et al. 2023b), which can reduce crop production by as much as 80% (Yasin et al. 2014, 2020).
Drought stress responses regulate and modulate physiological features: Under conditions of water scarcity, increased cell membrane permeability leads to cell membrane breakdown and electrolyte efflux (Yasin 2015). Increased stomatal conductance results in high transpiration, which helps maintain a cooler canopy temperature, a common characteristic of drought stress. Canopy temperature is often used as a surrogate indicator of stomatal conductance. To assess a plant's overall water status and create drought tolerance, leaf water potential and leaf relative water content are used as indicators of water supply and transpiration. The scavenging mechanism of antioxidants, such as SOD, APX, and CAT, is activated during drought stress leading to a decrease in ROS (Reactive Oxygen Species) (Yasin et al. 2014 and Yasin et al. 2020). Osmotic adjustment, or the active accumulation of organic solutes in response to increasing water deficit, is also observed. The fluorescence of chlorophyll A is also affected by water shortage. Water use efficiency, which measures the amount of water used per unit of dry matter production, also provides insight into a plant's ability to survive in a water-scarce environment, and some plants have adapted their anatomy to survive in such conditions
Distinctive molecular and cellular regulation mechanisms need to be in place to control both short-term (transpirational losses through guard cells) and long-term responses (stress resistance development in the whole plant) (Takahashi et al. 2020). Abscisic acid, a plant hormone, plays a key role in mediating drought stress responses and resistance through stomatal regulation and stress-responsive gene expression. Plants recognize water-deficit conditions and transmit the signal from the roots to the leaves to adapt to drought stress through the accumulation of abscisic acid and the mediation of drought stress responses through reactive oxygen species (ROS), hydraulic signals, electric currents, calcium waves, mRNA, and other phytohormones (Fig. 3). Yasin et al. (2014) have demonstrated different patterns of ROS scavenging by enzymes during abiotic stress among susceptible and tolerant horsegram accessions. Recent studies suggest that ncRNAs (Yasin et al. 2020) and peptides may also act as signaling molecules for mediating drought stress responses (Kim et al. 2021). Both ABA-dependent and ABA-independent regulatory pathways are involved in stomatal regulation to balance drought stress conditions (Takahashi et al. 2020). Accumulation of ABA is comparatively more in leaves than in the roots to regulate stomatal closure. The loss of turgor pressure caused by hydraulic stress activates ABA biosynthesis, which is regulated by the gene NINE CIS EPOXYCAROTENOID DIOXYGENASE 3 (NCED3) (Iuchi et al. 2001).
Schematic representation of signals transduction among the root, shoots, and leaf in response to drought stress: The acquisition of drought stress tolerance at the whole plant level is controlled by mobile signals such as hydraulic pressure, ROS/Ca2+ waves, peptides, and phytohormones. These signals travel between the root and shoot to regulate stomatal closure and mediate stress responses. The schematic may also depict how these signals, such as ABA, peptides, or ROS/Ca2+ waves, work together to coordinate drought stress tolerance at the cellular and molecular level
Under drought conditions, plants utilize a complex system of signals and receptors to coordinate stress responses across the root, shoot, and leaf. The long-distance peptide signal molecule (CLE25) CLAVATA3/EMBRYO SURROUNDING REGION-RELATED25, which is produced in the root and perceived by the BARELY ANY MERISTEM (BAM1 and BAM3) receptor-like protein kinases in the leaves, leading to increased expression of the NCED3 gene and ABA accumulation (Takahashi et al. 2018). Other signaling pathways include the involvement of transcription factors such as NGA1 and osmosensors like the A.thaliana histidine kinase 1 (AHK1) and mechano-sensitive channels like MCA1 and MCA2, which sense changes in cell wall tension and hydraulic pressure (Nakagawa et al. 2007; Yamanaka et al. 2010; Sato et al. 2014). These signaling systems allow plants to integrate water-deficit signals and optimize adaptations in different tissues.
ABA plays a vital role in drought stress resistance by triggering an increase in calcium through the activation of hydrogen peroxide (H2O2), which is a major ROS in guard cells. Drought stress response in guard cells is mediated by CLE9 gene (Zhang et al. 2019), which activates the accumulation of ROS and increases the activity of SLAC1 (SLOW ANION CHANNEL ASSOCIATED1) to control stomatal closure. Small molecular proteins known as late embryogenesis abundant (LEA) proteins help seeds during the development stage of drought stress by protecting other proteins from aggregation due to desiccation or osmotic stress during seed maturation. The activation of LEA-type genes, including the COR genes, seems to be regulated by a different signaling pathway. Phosphoinositol modulates the expression of LEA-like genes under drought stress and, because G-proteins may regulate the activity of phospholipase C in plants, G-protein-associated receptors may be responsible for the perception of secondary signals derived from these forms of stress. G-proteins have also been suggested as membrane-bound receptors for ABA (Takahashi et al. 2020).
Temperature Stress
The maintenance of cellular metabolism depends in part on temperature, and plants experience heat stress when temperatures increase over optimal ranges (Saini et al. 2022). Heat stress (Wahid et al. 2007) is an increase in temperature above a specific point for a length of time long enough to permanently harm plant health. Heat shock is the general term for a brief increase in temperature (10–15 °C). According to reports, rising temperatures reduce most cereals’ life cycles by causing senescence and shortening the growing season. A small temperature rise that staple crops (such as corn, wheat, and rice) can be detrimental during the floral transition, lowering crop productivity (Imran et al. 2021). Due to the heat shock, plants face several physiological and biochemical perturbations (Fig. 4).
Plant responses to heat stress. Upward pointing arrows indicate activated/upregulated, downward pointing arrows indicate deactivated/downregulated; Chloroplasts are particularly sensitive to heat stress and can experience a range of negative consequences, including inactivation of photosystem II (PSII), breakdown of chlorophyll, inactivation of Rubisco, and impaired protein translation. In response to heat stress, chloroplasts produce a large number of protein chaperones to protect PSII. In addition, chloroplasts are involved in retrograde signaling pathways that help protect cellular integrity and promote normal plant growth
Transcriptional Regulation of Heat Stress
Heat shock (HS) triggers the expression of several heat shock transcription factors (HSF) and heat shock protein (HSP) genes in plants (Saini et al. 2022). Both HSFs and HSPs play crucial roles in the plant’s response to heat stress and the establishment of thermotolerance (Ohama et al. 2017; Ren et al. 2019). The HSFs quickly activate the production of HSPs. The structural characteristics of the oligomerization domains of plant HSFs are used to classify them into three conserved evolutionary classes (A, B, and C). Class A HSFs are necessary for the activation of transcription, but Class B and C HSFs lack the necessary motifs made up of acidic amino acid residues and are unable to serve as activators (Scharf et al. 2012). When it comes to class A HSFs, HSFA1 is the primary transcriptional activator, triggering the production of additional heat-responsive transcription factors (TFs) such as HSFA2, HSFA7, HSFBs, DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 2A (DREB2A) and MULTIPROTEIN-BRIDGING FACTOR 1C (MBF1C) immediately. Interaction with HEAT SHOCK PROTEIN 70 (HSP70) and HSP90 during HS induces HSFA1 transactivation activity (Hahn et al. 2011). It is interesting to note that both HSFA1a and HSFA1b have a role in the early stages of HS-responsive gene expression (Li et al. 2010). As a heat-inducible transactivator, HSFA2 prolongs acquired thermotolerance in Arabidopsis by maintaining the expression of HSP genes (Li et al. 2017). To maintain thermotolerance, HSFA3 is regulated by DREB2A and DREB2C (Ohama et al. 2017; Sato et al. 2018). The key transcription factor DREB2A controls the transcription of HSFA3 directly through a co-activator complex that includes DNA POLYMERASE II SUBUNIT B3-1 (DPB3-1)/NF-YC10, NUCLEAR FACTOR Y, SUBUNIT A2 (NF-YA2), and NF-YB3. Additionally, the ROS generated as secondary stress responses during the HS reaction in Arabidopsis are sensed by HSFA4a and HSFA8 (Qu et al. 2013). HSFBs are transcriptional repressors that control the expression of certain heat-inducible HSFs (HSFA2, HSFA7s, and HSPs) in class B HSFs (e.g., HSP101, HSP70). The regulatory network that controls the expression of HS-responsive genes is formed by HSFBs, which are also downstream target genes of HSFA1s in plants. Through their effect and interaction, they also operate as a component of this network. The functions and roles of Class C HSFs are not well understood. Overexpression of FaHSFC1b (cloned from tall fescue) in Arabidopsis improves heat tolerance by activating or upregulating the expression of HSPs. In addition, HS also upregulates the expression of HSFC genes in wheat, cabbage, and soybean (Zhao et al. 2021).
TF families such as MBF1C, NAC, WRKY, bZIP, and MYB play a role in the regulation of heat-responsive genes. MBF1C, a highly conserved transcriptional co-activator, plays a crucial role in thermotolerance (Suzuki et al. 2011). The expression of HSFB and DREB2A during heat stress has been observed to decrease in an mbf1c mutant, and the expression of MBF1C is regulated by HSFA1s during heat stress. NAC transcription factors, which are a main group of transcription factors in plants, modulate the response to heat stress (Zhao et al. 2021). NACs bind to the promoters of HSFs (including HSFA1b, HSFA6b, HSFA7a, and HSFC1) to enhance their expression, thereby improving thermotolerance. Additionally, TaNAC2L improves heat resistance by controlling the expression of wheat heat-stress response genes, including AtHSFA3 and AtDREB2A (Guo et al. 2015). Altering ROS homeostasis, OsNAC3 overexpression in rice improves tolerance to heat stress. Under heat stress, the NAC transcription factor JUNGBRUNNEN1 (JUB1) controls DREB2A expression. Under non-stressful conditions, the association of heat shock protein (HSP)70/90 and heat shock transcription factor (HSF)A1s suppresses the activity of HSFA1s, whereas heat stress causes HSFA1 to disassociate from HSP70 and HSP90, resulting in HSFA1 activation (Zhao et al. 2021). Plant thermotolerance heavily relies on the bZIP transcription factors and the unfolded protein response (UPR). The endoplasmic reticulum (ER) membrane proteins bZIP28 and bZIP60 are transported to the nucleus, where they trigger the expression of genes that are responsive to stress (Zhao et al. 2021). Under heat stress, the RNA-splicing factor IRE1 (INOSITOL-REQUIRING ENZYME 1), which is anchored on the ER membrane, splices the bZIP60 mRNA to produce a spliced variant of the protein (sbZIP60), which proceeds into the nucleus (Ding et al. 2020). Under non-stressful contexts, the ER-localized chaperone BiP (BINDING PROTEIN) binds to bZIP28 and suppresses its activation. The control of the heat-stress response in Arabidopsis is influenced by the interaction between bZIP28 and HSFA2. In Arabidopsis, MYB30 controls heat-stress responses via ANNEXIN (ANN)-mediated cytosolic calcium signaling. MYB30 binds to the ANN1 and ANN4 promoters and suppresses protein expression. Decreased heat-stress responses are then elicited as a result of ANNs modulating heat-induced Ca2+flux. Furthermore, OsMYB55 overexpression in rice contributes to the increase in amino acid accumulation, such as glutamic acid, gamma-aminobutyric acid, arginine, and proline, further enhancing rice’s capacity to endure heat stress at the vegetative stage (Zhao et al. 2021). These amino acids are known to play a role in osmotic stress tolerance and may also act as antioxidants, protecting the plant from heat-induced damage. Overall, the regulation of heat-stress response in plants is a complex process involving the coordination of various transcription factors, signaling pathways, and stress-responsive genes.
Low-Temperature Stress
Many plant species are limited in their growing seasons due to cold stress. Many agriculturally significant crops, such as maize, rice, tomatoes, and fruits like bananas, mangoes, and papaya, which originate from tropical or subtropical climates, are highly susceptible to chilling (Hong et al. 2017). Cold stress can be broadly categorized into two types: freezing and chilling stressors. Common symptoms of cold stress include surface damage, discoloration, drying out, accelerated aging, reduced shelf life, increased ethylene production, and other indications (Imran et al. 2021). Alterations in plant physiology include increase in cellular Ca2+ concentrations and accumulation of reactive oxygen species (ROS) (Cramer et al. 2011), a reduction in membrane fluidity caused by the unsaturation of fatty acids in the membrane, and changes in the ratio of lipids to protein in the membrane (Wang et al. 2006). Low temperatures can also cause dehydration, primarily due to a reduced ability to absorb water. Freezing, similar to chilling, can damage membranes by causing severe dehydration and ice accumulation. Cells and tissues may also be physically disrupted when ice forms in intracellular areas. Protein denaturation and solute precipitation are additional effects of freezing. Both freezing and chilling can cause the generation of ROS, similar to other abiotic stressors, which disrupt the redox balance of cells.
Plant low-temperature stress tolerance is a complex trait influenced by genes and QTLs. Key genes include cold-responsive transcription factors like CBF/DREB1, Antioxidant enzymes such as SOD and CAT, Chaperones like HSPs, ABA metabolism genes such as ABA biosynthesis and ABA receptor genes. QTLs for low-temperature stress tolerance can be identified through genetic mapping and association studies and provide valuable information for improving stress tolerance in crops.
Salinity Stress
Salinity, either in water or soil, can be a limiting factor for crop growth and production, particularly in arid or semi-arid regions (Chourasia et al. 2022). According to an estimate published in 2011, over 932 million hectares of world land are affected by high levels of salinity (IAEA 2021). About 17 million hectares of agricultural land will be affected by salinity by 2050. The effects of salinity on plants include reducing growth rate, smaller and fewer leaves, and a reduction in root length (Munns and Termaat 1986). Akin to other abiotic stresses, the effects of salinity on plants and plant responses depend on the duration and severity of the stress. In general, salinity has short-term effects (such as ion-independent growth reduction) that take place within minutes to hours or days after perception of the stimuli, such as closing of stomata and inhibition of cell expansion which is shoot specific. Long-term effects of salinity can occur over days or even weeks, such as building up cytotoxic ion levels, slowing down metabolic activities, causing early senescence and ultimately cell death (Imran et al. 2021). Salt stress in plants is caused by high salt concentrations in soil, leading to reduced growth and survival. Plant responses to salt stress include I. Physiological responses: 1. Osmotic adjustment to balance water and counteract salt-induced water loss by increasing solute concentration in cells. 2. Reduced water loss through stomatal closure. 3. Increased uptake of sodium ions and extrusion of potassium ions to reduce salt damage. II. Anatomical responses: 1. Root growth inhibition to reduce salt uptake. 2. Leaf margin necrosis due to salt damage. 3. Leaf abscission to reduce water loss. These responses help plants cope with salt stress, but also affect growth and productivity. III. Salt exclusion mechanisms include 1. Hydrogen ion secretion from roots to decrease soil pH and reduce salt uptake. 2. Selective ion uptake regulation. 3. Vacuole sequestration of excess ions. 4. Rhizosphere modification to reduce salt uptake. 5. Root adaptation in plants like mangroves, with specialized roots adapted to tolerate high salt levels. These mechanisms help plants mitigate salt stress and maintain growth and survival in salty environments (Raza et al. 2022).
A plant’s tolerance to salinity is achieved by a multitude of physio-molecular mechanisms, osmotic and ionic tolerance, and tissue tolerance (Fig. 5). Among these, osmotic tolerance is a rapid response associated with a quick decrease in stomatal conductance to store water which employs fast signaling mechanisms between roots and shoots. The ionic tolerance is achieved by the activation of several signaling cascades that restrict net Na+ influx and reduce net Na+ translocation. The tissue-specific tolerance is achieved by the translocation of toxic ions to the vacuole to avoid their detrimental effects (Yasin and Singh 2019) on cytoplasmic-based metabolic processes (Isayenkov and Maathuis 2019). The salt overly sensitive (SOS) pathway comprising different SOS genes, in this case, is the key to directing toxic ions to the vacuole.
A simulation of a plant's response to salt stress; Salt stress is primarily responsible for ionic stress as well as osmotic stress. After recognizing Na+ and hyperosmolality, plants gather Ca2+, activate ROS signalling, and alter the composition of their phospholipids. The plasma membrane calcium-permeable channel OSCA1 functions as a putative osmosensor in osmotic stress and is necessary for Ca2+ signalling brought on by this condition. The loss-of-function mutant osca1 exhibits reduced Ca2+ signal amplification under osmotic stress. In addition, by regulating the H2O2-triggered influx of Ca2+ , the leucine-rich-repeat receptor kinase, hydrogen-peroxide-induced Ca2+ increases 1 (HPCA1), recognizes the rise in H2O2 under stress stimuli for stomatal closure. The monovalent-cation sensor MOCA1 (monocation-induced Ca2+ increases 1) encodes a glucuronosyltransferase and participates in the manufacture of glycosyl inositol phosphorylceramide (GIPC), which senses Na + and controls salt stress responses by activating MOCA1. The cell wall's pectin component interacts with FER (the plasma membrane receptor-like kinase FERONIA), which can detect salt stress-related cell wall damage. Calmodulin regulates the calcium-permeable cyclic nucleotide-gated ion channels (CNGCs), which are blocked by cellular calcium concentrations (CaM). By phosphorylating CNGCs, FER controls calcium signaling alongside BAK1. These signals engage adaptive mechanisms that reduce salt stress by triggering phytohormone signaling, maintaining ion balance and osmotic equilibrium, controlling cytoskeleton dynamics, and controlling cell wall structure, ultimately inhibiting plant development and stimulating metabolism to become salt-tolerant (Zhao et al. 2021)
Plants respond to salinity stress at the molecular level through several pathways: 1. Osmolyte synthesis: plants produce osmolytes such as proline, trehalose, and glycine betaine to balance salt levels in cells. 2. Ion regulation: to maintain ion balance, plants regulate the transport of ions such as sodium and calcium. 3. Antioxidant defense: salinity triggers ROS production, causing oxidative stress, and plants respond by activating antioxidants such as SOD, catalase, and peroxidase. 4. Hormone adjustment: salinity affects hormone balance, for example increasing ABA levels to regulate stomatal closure and reduce water loss. 5. Gene expression changes: salinity leads to changes in gene expression, some stress response and ion uptake genes are upregulated while growth and development genes are downregulated. These molecular responses enable plants to adapt to salinity stress and survive.
Flooding Stress
Flooding can pose a significant challenge to plants, as the excess of water in the environment can deprive them of crucial necessities such as oxygen, carbon dioxide, and sunlight (Jackson et al. 2009). This deprivation can lead to difficulties in growth and survival for the plant and also cause issues with soil structure and nutrient availability, exacerbating the challenges even further (Voesenek and Bailey-Serres 2013). Several studies have shown that an overabundance of water causes intricate alterations, including hindered gas exchange, which can impact the soil microbiota and cause hypoxia and anoxia (Tewari and Arora 2013, 2016). Hypoxia during flooding can increase the redox potential between the waterlogged soil and plants, leading to the production of reactive oxygen species (ROS) (Sasidharan and Voesenek 2015). The rise of ethylene levels during a flood is also caused by oxygen-mediated conversion of 1-Aminocyclopropane-1-Carboxylate to ethylene, resulting in adaptations such as the production of shoot aerenchyma and leaf nastic motions and the role of endogenous plant growth regulators in flood stress (Fig. 6) (Sasidharan and Voesenek 2015). Plants can withstand oxidative stress by modifying their anatomy or metabolism, such as the control of adaptation to floods by low oxygen-sensing ethylene response factors (ERFs), ERF VII (Gibbs et al. 2011; Licausi et al. 2011). During flooding, when plants cannot use oxygen for aerobic respiration, they switch to anaerobic, glycolytic, and fermentative metabolism to sustain cell viability. However, glycolysis is less effective and limited, producing only two ATP molecules for every hexose molecule, compared to 36 ATP produced through oxidative phosphorylation. It is also limited by NADH oxidation.
Plant metabolism is an energy-intensive process that gets shindered in anoxic conditions, which are common during flooding. Proteins involved in anaerobic and ROS metabolism are encoded during the tightly regulated translation process, which also involves protein synthesis. Gibbs and Greenway have described various anaerobic proteins (ANPs) that are produced in response to flooding stress, including pyruvate decarboxylase, sucrose synthase, LDH, and ADH.NADPH oxidase is used by waterlogging to increase ROS. By explaining the overexpression of the enzymes ADH and malic dehydrogenase in plant roots when exposed to waterlogging, Mcmanmon and Crawford (1971) proposed the metabolic theory of flood tolerance. Similarly, numerous data indicate that flooding significantly boosts the crop’s antioxidant potential. Under flooding stress, Sairam et al. (2009) found that NADPH oxidase activity and NADPH oxidase mRNA expression increased in genotypes of pigeon peas. SOD activity increased 14-fold during hypoxia in the iris plant, according to Monk et al. (1987). H2O2 sensors in plants are also known as heat shock transcription factors (HSF). Different plant species all respond to anoxia by upregulating the heat shock protein (HSP) (Mustroph et al. 2014). According to Banti et al. (2010), the transgenic model plant Arabidopsis that expresses HSFA2 is more resistant to hypoxia than the wild type. Researchers have also discovered that plants’ HSFs are activated in response to floods, causing the transcription of genes that code for both high- and low-molecular-weight HSPs. Under the flooding condition, the ROS-related transcription factor and HSFs are controlled by the NADPH oxidase-dependent ROS pathway. Since the proteolysis of transcription factors controlled by HSPs and ROS is not N-end rule pathway-dependent, ROS signaling is a self-regulating mechanism operating under flood tolerance.
Flooding tolerance in plants is a complex trait, orchestrated by numerous genes. Key genes in this area include 1. Transcription factors, such as NAC (NAM, ATAF1/2, and CUC2), WRKY, and MYB, regulating gene expression in response to flooding stress. 2. ROS scavenging genes, such as catalase (CAT), superoxide dismutase (SOD), and ascorbate peroxidase (APX), protecting plants from oxidative damage caused by flooding. 3. Ethylene response genes, such as ERF VII, playing a role in regulating plant responses to flooding through the ethylene signaling pathway. 4. Aquaporin genes, controlling water transport and crucial for regulating water uptake and distribution in flooded conditions. 5. Metabolic genes, involved in glycolysis and fermentative metabolism, assisting plants in maintaining energy production and viability in the absence of oxygen. 6. Cell wall modification genes, involved in xyloglucans and pectins biosynthesis, playing a role in regulating cell expansion and water uptake in flooded conditions. Understanding these genes and their functions is essential to comprehend the mechanisms of flooding tolerance in plants and develop crops with improved flooding tolerance.
The use of osmoprotectants such as mannitol and glycine betaine can help reduce stress. Glycine betaine is widely present in chloroplasts, where it protects thylakoid membranes and preserves photosynthetic efficiency. Mannitol, as a free radical scavenger, stabilizes cellular and subcellular structures and significantly contributes to carbon storage during flooding stress (Arora et al. 2012). A rise in proline content was observed in rice plants subjected to flooding compared to the control (non-flooded circumstances) (Chanu et al. 2015). Proline accumulation, as a result of de novo synthesis, serves as an adaptive mechanism to maintain osmoregulation under stress (Chen et al. 1993). Proline acts as a compatible solute that modifies the osmotic potential in the cytoplasm during a flooding state (Caballero et al. 2005).
Heavy Metal Stress
Plants are frequently exposed to abiotic and biotic stressors. Anthropogenic activities have led to the accumulation of heavy metals like Fe, Mn, Cu, Ni, Co, Cd, Zn, Hg, and arsenic in soils (Ghori et al. 2019). The presence of heavy metals in the nucleus can result in promutagenic damage to DNA, causing strand breaks, base alterations, rearrangements, and depurination. These metals can produce reactive species that lead to harmful effects on DNA repair mechanisms, creating the promutagenic adduct 8-0xoG, which can cause conversion of C to T. When heavy metals enter plant cells and bind to the nucleus, they can cause DNA damage, such as strand breaks, base alterations, rearrangements, and depurination. This damage is often due to the reactive species produced by metal poisoning. In A. thaliana, exposure to cadmium resulted in DNA damage and reduced the expression of DNA repair genes. In addition, nickel accumulation has been observed to lower mitotic activity in maize roots, while copper, nickel, and cadmium exposure caused clastogenic effects in Helianthus annuus. Studies have shown that some plant species can use mechanisms like chelation, compartmentalization, and vacuolar sequestration to avoid the harmful effects of heavy metals on DNA. These findings suggest that heavy metal exposure can cause DNA damage and other harmful effects in plant models (Yang et al. 2020).
Plants have an innate immune system that is activated under stress to survive (Li et al. 2020). When heavy metal stress is detected, a signal transduction network is triggered, leading to the creation and activation of stress-related proteins and signaling molecules (Ghori et al. 2019; Jamla et al. 2021). The signal transduction pathway activates transcription factors, which then triggers the transcription of metal stress-responsive genes. Different signaling pathways, including the MAPK cascade, hormonal signaling, ROS signaling system, and Ca-Calmodulin pathway are activated in response to different metal stressors (Ghori et al. 2019).
Calcium (Ca2+) is a crucial player in mediating heavy metal toxicity avoidance in plants. Ca2+-mediated signaling pathways are critical in the defense mechanism of plants against heavy metal toxicity. By regulating gene expression and activating cellular defense mechanisms, plants can tolerate and mitigate the toxic effects of heavy metals, ensuring their survival in contaminated environments. Heavy metals like copper, cadmium, nickel, and lead can cause severe damage to plant cells by inducing oxidative stress, altering cellular signaling pathways, and disrupting metabolic processes. To mitigate the toxic effects of heavy metals, plants rely on the complex network of calcium signaling pathways, which can activate defense mechanisms and induce changes in gene expression that enhance the plant’s tolerance to heavy metal stress. Studies have shown that increased levels of Ca2+ inside the cell can activate the calmodulin pathway, which functions as a cellular target for various metals. Calmodulin serves as a crucial signaling component that mediates the response to metal stress by triggering the activation of Mitogen-Activated Protein Kinases (MAPKs) that are responsible for transmitting signals. Furthermore, calcium-dependent protein kinases (CDPKs) and phosphatidylinositol are also necessary for the activation of MAPKs under heavy metal stress conditions. Transcription factors, such as WRKY and zinc finger transcription factors, play a critical role in regulating gene expression in response to heavy metal stress. For example, the expression of WRKY25, WRKY33, and ZAT12 was found to be downregulated by MPK4 during oxidative stress caused by metal exposure, showing that these transcription factors are responsible for controlling the production of reactive oxygen species (ROS).
Metallothioneins (MTs) are small, cysteine-rich proteins that play a key role in heavy metal toxicity avoidance in plants. MTs have a high affinity for heavy metals such as zinc, cadmium, copper, and lead, and they can bind to and detoxify these metals and have role in other stress tolerance by preventing harmful effects on cellular components (Yasin 2015; Singh et al. 2016). MTs are synthesized in response to heavy metal stress and their expression is regulated by various signaling pathways, including the calcium-calmodulin pathway and the MAPK cascade. In addition to detoxifying heavy metals, MTs also act as antioxidants by scavenging reactive oxygen species (ROS) and preserving cellular redox homeostasis. Studies have shown that the upregulation of MTs is essential for the survival of plants under heavy metal stress conditions, and it is thought that this protein may play a crucial role in mitigating the negative effects of heavy metal toxicity in plants.
The accumulation of reactive oxygen species is accelerated in a heavy metal stress environment, putting the plant metabolic pathways at risk (Fig. 7). This leads to altered calcium channel function and increased calcium influx into cells. Studies of the Ca-calmodulin pathway suggest that calmodulin serves as a key target for various metals, including Pb, Cd, and Ni, with over 90% enhancement of calmodulin’s activity by Pb, which binds to all four of its calcium-binding sites simultaneously (Ouyang and Vogel 1998). Most metals function as calcium analogs, causing calmodulin to initiate signal transduction (Snedden and Fromm 2001). The transmission of signals is crucial for the activation of Mitogen-Activated Protein Kinases (MAPKs), which are composed of three modules: MAPKK, MAPK, and MAPKKK. They are activated by phosphorylation of serine/threonine residues. Copper and cadmium stress requires phosphatidylinositol and a Ca2+−dependent protein kinase (CDPK) for MAPK activation (Thapa et al. 2012). The transcription factor WRKY and zinc finger transcription factors are implicated in plant heavy metal stress, with WRKY25, WRKY33, and ZAT12 expression being downregulated by MPK4 during oxidative stress caused by metal exposure (Pitzschke et al. 2009), demonstrating their role in controlling ROS production.
Heavymetals generate free radicals through single-step electron transfer reactions. The heavy metals severely disrupt the thylakoid membrane pathways, hampering the detoxification of free radicals. They inactivate antioxidants like superoxide dismutase and catalase, and deplete low molecular weight antioxidants like glutathione that chelate heavy metals
During metal stress, salicylic acid concentration has also been reported to rise. It has been observed that pre-treating plants with salicylic acid increases glutathione levels and increases resistance to nickel toxicity. Salicylic acid’s major function in plant cells is to control the levels of ROS and antioxidants and to induce the expression of a variety of genes (Hossain et al. 2012). In addition, nitrilase protein and the SAMT gene, which enhance the manufacture of salicylic acid and ABA, respectively, were found to be activated in Arabidopsis and pea plants that had been exposed to mercury using proteomic analysis (Shao et al. 2010). When Cd stimulates the creation of ethylene and ABA, these hormones launch a sequence of reactions in the cell (Polle and Schutzendubel 2004). Studies on the transcriptome have shown that heavy metals affect various transcription factors.
Signaling and Crosstalk
Signaling and crosstalk in response to abiotic stresses, constitute a complex and dynamic interplay of various phytohormones and molecular components (Ku et al. 2018). Phytochromes, pivotal for light perception, interact with other major phytohormones in response to environmental stressors, fine-tuning adaptive strategies crucial for survival under adverse conditions. Signaling crosstalk of pathways affects ROS production manipulated by stress-responsive gene expression, thereby facilitating plant adaptation to challenging conditions. This process, being abscisic acid (ABA) centered, assumes a role in regulating the plant’s response to diverse stressors. ABA’s interactions with hormones like auxin and other signaling molecules contribute significantly to the plant’s capacity to effectively modulate its stress responses. When a plant faces drought stress, it promptly activates the ABA signaling pathway, leading to stomatal closure and water conservation. Notably, ABA-induced stomatal closure, while conserving water, indirectly influences ROS production, with ROS molecules serving as secondary messengers that activate additional signaling pathways. To highlight, signaling crosstalk mediated by ABA intricately impacts the expression of stress-responsive genes modulated by a set of non-coding RNA to enhance stress tolerance (Yasin et al. 2020).
Signaling pathway research has uncovered intricate connections between phytochromes and hormones (Gavassi et al. 2023; Singh et al. 2023) emphasizing the dynamic and coordinated nature of signaling crosstalk at the molecular level, to fine tune a plant’s tolerance response. Based on the experimental results reported earlier and the databases, we have identified a set of genes having key roles in abiotic stress tolerance and having interactions and crosstalk in stress signaling (Fig. 8). These genes can further be explored to validate the identified nodes by genome editing in crop improvement.
Auxin is an important regulator of growth and development, orchestrating key roles in mediating the abiotic stress signals and control of the downstream stress pathways (Jing et al. 2023). H2A-Z histone variant plays a dynamic role in regulating SMALL AUXIN UP RNAs (SAURs) in the crosstalk between ABA and auxin signaling pathways in Arabidopsis (Yin et al. 2023). This study unravels the molecular intricacies of how histone modifications and small RNAs contribute to the precise regulation of ABA-auxin crosstalk, which holds paramount importance in optimizing plant responses to abiotic stresses.
The initiation of signaling pathways in plant cells is a multifaceted process, primarily governed by specialized receptor proteins that discern specific ligands. These ligands encompass a wide array, including hormones such as ABA, auxins, gibberellins, cytokinins, and signaling molecules like calcium ions (Ca2+) (Koenig and Hoffmann-Benning 2020). Ligand binding sets off conformational changes in receptors pivotal for activation. Once activated, receptors initiate kinase cascades or influence gene expression through transcription factors. For instance, the activation of the BRI1 receptor sets in phosphorylation cascade with far-reaching effects on gene expression. Signals are amplified via secondary messengers and phosphorylation cascades. Plant receptors can be either membrane-bound (receptor kinases) or intracellular like (nuclear receptors). Phosphorylation commonly follows ligand binding, activating membrane-bound receptors or generating secondary messengers such as Ca2+ or IP3. Post-transcriptional modification of signalling molecules has been found to reduce protein production by up to 15% (Bockaert and Pin 1999). Temporal control mechanisms are integral, encompassing desensitization and negative feedback loops, culminating in the termination and regulation of signaling responses, respectively. The phenomenon of crosstalk between pathways allows plants to synthesize multiple signals effectively, enabling them to respond adeptly. For instance, ABA signaling crosstalks with jasmonic acid signaling to adapt to both abiotic and biotic stresses.
Role of ncRNAs in Abiotic Stress Tolerance
Regulatory non-coding RNA species hold pivotal roles in intricate plant regulatory networks governing responses to abiotic stress. Under abiotic stress conditions, specific ncRNAs exhibit dynamic upregulation or downregulation, exerting influence over the expression of stress-responsive genes. Recent advancements in plant genomics have highlighted the critical role of regulatory non-coding RNA (ncRNA). Yasin et al. (2020) conducted a comprehensive study on stress-responsive horsegram (Macrotyloma uniflorum) accessions, employing genome-wide in silico analysis to unveil miRNA and target networks. This research sheds light on the intricate regulatory networks underpinned by ncRNAs that assist plants in coping with environmental challenges. In another study, Neha et al. (2017) showed the regulation of MYB overexpression through ncRNA-mediated mechanisms. This work underscores the pivotal role of ncRNAs in finely calibrating gene expression, ensuring effective modulation of stress responses. These findings emphasize that ncRNAs function as critical gatekeepers, preventing gene overexpression and finely regulating abiotic stress tolerance mechanisms.
Building upon this growing body of knowledge, Yasin et al. (2021) presented a roadmap that charts the landscape of long non-coding RNAs (lncRNAs) and large intervening ncRNAs (lincRNAs). The stress-responsive microRNAs identified in horsegram, including mun-miR150, mun-miR1507a, mun-miR156k, mun-miR156r, mun-miR390, mun-miR390b-5p, mun-miR482, mun-miR482a, mun-miR482a-5p, and mun-miR482d-3p, offer promising avenues for further exploration in diverse crops to enhance the development of stress-tolerant varieties. These non-coding RNAs (ncRNAs) are intricately connected with stress-responsive microRNAs and gene networks in pigeon pea, (Yasin et al. 2020). This research provides valuable insights within the broader context of crop improvement, showcasing how ncRNAs can be harnessed to enhance stress tolerance and overall crop performance. This comprehensive study offers a panoramic view of the ncRNA terrain within pigeon pea, particularly under abiotic stress conditions. It highlights the complex network of ncRNAs that manipulate the plant’s responses to stress. These ncRNAs serve as masters of gene expression, preventing overexpression and weaving intricate networks that empower plants to adapt and thrive in challenging environmental conditions.
Conclusions and Perspectives
In the context of food security and climate change, the significance of abiotic stresses in plants and the impact on crop productivity has necessitated intense research on how a plant senses a stress factor to trigger series of sensing and signaling pathways, downstream activation of stress pathways, as well as several pathways of defense including ROS, stress-induced gene regulation by coding and non-coding RNAs, metabolites, and hormones (Yasin et al. 2020). Abiotic stress tolerance is known to be influenced by gene expression regulation through the action of non-coding RNA (ncRNA) networks. These ncRNAs, such as microRNAs, are known to play a significant role in regulating the expression of genes involved in various stress tolerance mechanisms in plants. Plants rely on the sensing process via which they trigger a downstream signaling cascade to prepare their reaction to offer a well-articulated response to stress. Modern understanding of cellular signaling, ROS signaling, regulation of gene expression, intercellular communication, and long-distance integration of stress signals during abiotic stress responses has been shaped by the developments in genomics and other genomics tools (Yasin et al. 2020; Imran et al. 2021). The exact mechanism by which plants detect drought stress conditions and communicate that information to the cell to control ABA accumulation for the development of drought stress resistance is not completely known. The migration of ABA from the synthesis tissues to the target tissues is complemented by the mobility of these mobile molecules, transport of miRNA complexes, AGO complexes, role of motor proteins, and pH flux of the cells which produces the environment for action which has induce regulatory mechanisms involved in tissue-to-tissue communication and long-distance signaling to get attention recently (Yasin 2015).
Model plants like Arabidopsis, rice, and tomatoes are used to study how plants respond to heat stress. Minimal research has focused on non-model plants like various crops and forestry trees (woody plants). In the course of evolution, certain genes may have undergone functional divergence, and homologous genes in various plants may have acquired distinct functions. Thus, additional research on non-model plants is required to better understand the gene regulatory networks underpinning the plant heat-stress response (Zhao et al. 2021). The advances in gene discovery, transformation protocols, and genome editing are now facilitating the genetic engineering in non-model plant studies (Andrew et al. 2021). Therefore, studies should focus on the way that plants react to heat stress that lasts for a long time or occurs across several generations, including transgenerational and multigenerational heat stress. Model plants’ transcriptional level responses to heat stress have been steadily described. On the other hand, it is unknown how crucial epigenetic regulatory mechanisms interact to affect how plants respond to heat stress. The majority of prior research on epigenetic changes brought on by heat stress concentrated on methylation and acetylation, while less is known about phosphorylation, ubiquitination, and SUMOylation (Small Ubiquitin-like Modifier, SUMO). Furthermore, RNA methylation in response to heat stress is poorly understood because the majority of methylation studies focused on DNA. Research on epigenetic alterations under heat stress will be aided by recently established technologies, such as single-cell RNAseq, histone modification, RNA modification (m6A/m1A/m5C), and ATAC-seq (assay for transposase-accessible chromatin sequencing). How heat is perceived by a plant is a key issue in the HS response. The missing link between the heat cue and the response could be uncovered by identifying plant thermosensors (Jung et al. 2020; Raza et al. 2023a, b, c, d). In the future, it will be possible to study the abiotic stress signaling pathways, intercellular and extracellular metabolites involved in the stress response and manipulate crops with desirable traits using genome editing, and these research gaps should augment development of stress-tolerant crops.
References
Akhiyarova G, Veselov D, Ivanov R, Sharipova G, Ivanov I, Dodd IC, Kudoyarova G (2023) Root ABA accumulation delays lateral root emergence in osmotically stressed barley plants by decreasing root primordial IAA accumulation. Int J Plant Biol 14(1):77–90
Andrew PL, Selvaraj R, Kumar KK, Muthamilarasan M, Yasin JK, Pillai MA (2021) Loss of function of OsFBX267 and OsGA20ox2 in rice promotes early maturing and semi-dwarfism in γ-irradiated IWP and genome-edited Pusa Basmati-1. Front Plant Sci 12:714066. https://doi.org/10.3389/fpls.2021.714066
Arora NK, Tewari S, Singh S, Lal N, Maheshwari DK (2012) PGPR the for protection of plant health under saline conditions. In: Maheshwari DK (ed) Bacteria in agrobiology: stress management. Springer, Berlin Heidelberg, pp 239–258
Aslam M, Waseem M, Jakada BH, Okal EJ, Lei Z, Saqib HS, Yuan W, Xu W, Zhang Q (2022) Mechanisms of abscisic acid-mediated drought stress responses in plants. Int J Mol Sci 23(3):1084
Banti V, Mafessoni F, Loreti E, Alpi A, Perata P (2010) The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol 152:1471–1483
Bockaert J, Pin JP (1999) Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J 18(7):1723–1729
Caballero JL, Verderzco CV, Galam J, Jimenez ESD (2005) Proline accumulation as a symptom of water stress in maize: a tissue differentiation requirement. J Exp Bot 39:889–897
Chanu WS, Sarangthem K (2015) Changes in proline accumulation, amino acid, sugar and chlorophyll content in leaf and culm of Phourel-amubi, a rice cultivar of Manipur in response to flash flood. Ind J Plant Physiol 20:10–13
Chaudhry S, Sidhu S (2022) Climate change regulated abiotic stress mechanisms in plants: a comprehensive review. Plant Cell Rep 41(1):1–31
Chen CT, Kao CH (1993) Osmotic stress and water stress have opposite effects on putrescine and proline production in excised rice leaves. Plant Growth Regul 13:197–202
Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12(10):444–451
Chirag M, Mawlong I, Tyagi A (2022) Role of apetela2 (AP2)/ERF family transcription factors in stress-responsive gene expression. Response of field crops to abiotic stress. CRC Press, Boca Raton, pp 191–209
Chourasia KN, More SJ, Kumar A, Kumar D, Singh B, Bhardwaj V, Kumar A, Das SK, Singh RK, Zinta G, Tiwari RK (2022) Salinity responses and tolerance mechanisms in underground vegetable crops: an integrative review. Planta 255:68. https://doi.org/10.1007/s00425-022-03845-y
Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K (2011) Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol 11:163
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61:651–679
Daryanto S, Wang L, Jacinthe PA (2016) Global synthesis of drought effects on maize and wheat production. PLoSONE 11:e0156362
Davletova S, Rizhsky L, Liang H, Shengqiang Z, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17(1):268–281
DeWald DB, Torabinejad J, Jones CA, Shope JC, Canjelosi AR, Thompson JE, Prestwich GD, Hama H (2001) Rapid accumulation of phosphatidylinositol 4, 5-biphosphate, and inositol 1,4,5-triphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol 126:159–172
Ding Y, Liu N, Virlouvet L, Riethoven JJ, Fromm M, Avramova Z (2013) Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol 18(1):13
Ding YL, Shi YT, Yang SH (2020) Molecular regulation of plant responses to environmental temperatures. Mol Plant 13:544–564
FAO. 2021. Statistical year book, Land use. Rome. http://www.fao.org/faostat/en/#data/RL.
Foyer CH, Noctor G (2016) Stress-triggered redox signalling: what’s in prospect ? Plant Cell Environ 39:951–964. https://doi.org/10.1111/pce.12621
Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462(7273):660–664
Gavassi MA, Alves FR, Carvalho RF (2023) Phytochrome and hormone signaling crosstalk in response to abiotic stresses in plants. Plant hormones and climate change. Springer, Singapore, pp 145–165
Ghori NH, Ghori T, Hayat MQ, Imadi SR, Gul A, Altay V, Ozturk M (2019) Heavy metal stress and responses in plants. Int J Environ Sci Technol 16:1807–1828
Ghosh S, Bheri M, Bisht D, Pandey GK (2022) Calcium signaling and transport machinery: potential for development of stress tolerance in plants. Curr Plant Biol 29:100235. https://doi.org/10.1016/j.cpb.2022.100235
Gibbs DJ, Lee SC, Isa NM, Gramuglia S, Fukao T, Bassel GW, Correia CS, Corbineau F, Theodoulou FL, Bailey-Serres J, Holdsworth MJ (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479:415–418
Gjindali A, Johnson GN (2023) Photosynthetic acclimation to changing environments. Biochem Soc Trans 51:473
Guo W, Zhang J, Zhang N, Xin M, Peng H, Hu Z, Ni Z, Du J (2015) The Wheat NAC transcription factor TaNAC2L is regulated at the transcriptional and post-translational levels and promotes heat stress tolerance in transgenic Arabidopsis. PLoSONE 10:e0135667
Hahn A, Bublak D, Schleiff E, Scharf KD (2011) Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 23:741–755
Hong JH, Savina M, Du J, Devendran A, Ramakanth KK, Tian X, Sim WS, Mironova VV, Xu J (2017) A sacrifice-for-survival mechanism protects root stem cell niche from chilling stress. Cell 170(1):102–113
Hossain MA, Piyatida P, Silva JAT, Fujita M (2012) Molecular mechanism of heavy metal toxicity and tolerance in plants: central role of glutathione in detoxification of reactive oxygen species and methylglyoxal and in heavy metal chelation. J Bot 2012:1–37
Hotamisligil GS, Davis RJ (2016) Cell signaling and stress responses. Cold Spring Harb Perspect Biol 8(10):a006072. https://doi.org/10.1101/cshperspect.a006072
Hsieh TH, Lee JT, Yang PT, Chiu LH, Charng YY, Wang YC, Chan MT (2002) Heterology expression of the arabidopsis C –repeat /dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol 129:1086–94
Hu Y, Chen X, Shen X (2022) Regulatory network established by transcription factors transmits drought stress signals in plant. Stress Biol 2:26. https://doi.org/10.1007/s44154-022-00048-z
Huang H, Ullah F, Zhou D-X, Yi M, Zhao Y (2019) Mechanisms of ROS regulation of plant development and stress responses. Front Plant Sci 10:800. https://doi.org/10.3389/fpls.2019.00800
IAEA (2021) www.iaea.org.newscentre/news/
Imran QM, Falak N, Hussain A, Mun BG, Yun BW (2021) Abiotic stress in plants; stress perception to molecular response and role of biotechnological tools in stress resistance. Agronomy 11:1579
Isayenkov SV, Maathuis FJM (2019) Plant salinity stress: many unanswered questions remain. Front Plant Sci 10:80
Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T et al (2001) Regulation of drought tolerance by gene manipulation of 9-cisepoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27(4):325–333
Jackson MB, Ishizawa K, Ito O (2009) Evolution and mechanisms of plant tolerance to flooding stress. Ann Bot 103:137–142
Jamla M, Khare T, Joshi S, Patil S, Suprasanna P, Kumar V (2021) Omics approaches for understanding heavy metal responses and tolerance in plants. Curr Plant Biol. https://doi.org/10.1016/j.cpb.2021.100213
Jing H, Wilkinson EG, Sageman-Furnas K, Strader LC (2023) Auxin and abiotic stress responses. J Exp Bot. https://doi.org/10.1093/jxb/erad325
Joshi R, Wani SH, Singh B, Bohra A, Dar ZA, Lone AA, Pareek A, Singla-Pareek SL (2010) Transcription factors and plants response to drought stress: current understanding and future directions. Front Plant Sci 1:7
Jung JH, Barbosa AD, Hutin S, Kumita JR, Gao M, Derwort D, Silva CS, Lai X, Pierre E, Geng F et al (2020) A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585:256–260
Kavi Kishor PB, Ganie SA, Wani SH, Rajasheker G, Karumanchi AR, Sujatha E, Jalaja N, Kumar V, Rathnagiri P, Suravajhala P, Suprasanna P (2022) Nuclear factor-Y (NF-Y): developmental and stress-responsive roles in the plant lineage. Jour Plant Growth Reg 42:2711–2735
Kim JS, Jeon BW, Kim J (2021) Signaling peptides regulating abiotic stress responses in plants. Front Plant Sci 12:704490. https://doi.org/10.3389/fpls.2021.704490
Kobrinsky E, Mirshani T, Zhang H, Jin T, Longothesis DE (2000) Receptor-mediated hydrolysis of plasma membrane messenger PIP2 leads to K+ -current desensitisation. Nat Cell Biol 2:507–514
Koenig AM, Hoffmann-Benning S (2020) The interplay of phloem-mobile signals in plant development and stress response. Biosci Rep. https://doi.org/10.1042/BSR20193329
Ku Y-S, Sintaha M, Cheung M-Y, Lam H-M (2018) Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int J Mol Sci 19(10):3206. https://doi.org/10.3390/ijms19103206
Kumar S, Jeevaraj T, Yunus MH, Chakraborty S, Chakraborty N (2023) The plant cytoskeleton takes center stage in abiotic stress responses and resilience. Plant, Cell Environ 46:5–22
Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING-finger protein that displays cold-regulated nucleo cytoplasmic partitioning. Genes Dev 15:912–924
Li XP, Tian AG, Luo GZ, Gong ZZ, Zhang JS, Chen SY (2005) Soybean DRE-binding transcription factors that are responsive to abiotic stresses. Theor Appl Genet 115(5):687–696
Li M, Berendzen KW, Schoffl F (2010) Promoter specificity and interactions between early and late Arabidopsis heat shock factors. Plant Mol Biol 73:559–567
Li X, Wang X, Cai Y, Wu J, Mo B, Yu E (2017) Arabidopsis heat stress transcription factors A2 (HSFA2) and A3 (HSFA3) function in the same heat regulation pathway. Acta Physiol Plantarum 39:1–9
Li P, Lu YJ, Chen H, Day B (2020) The lifecycle of the plant immune system. CRC Crit Rev Plant Sci 39(1):72–100
Li T, Xiao X, Liu Q, Li W, Li L, Zhang W, Munnik T, Wang X, Zhang Q (2023) Dynamic responses of PA to environmental stimuli imaged by a genetically encoded mobilizable fluorescent sensor. Plant Commun 4(3):100500. https://doi.org/10.1016/j.xplc.2022.100500
Liang Y, Huang Y, Liu C, Chen K, Li M (2023) Functions and interaction of plant lipid signalling under abiotic stresses. Plant Biol J 25:361–378. https://doi.org/10.1111/plb.13507
Licausi F, DA KosmaczM W, Giuntoli B, Giorgi FM, Voesenek LACJ, Perata P, Van Dongen JT (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479(7373):419–422
Liu Q, He S (2017) Transcription factor OsAP2-39 involved in the regulation of osmotic stress response of rice. Crop J 5(2):126–134
Liu Y, Wei H, Ma M, Li Q, Kong D, Sun J, Ma X, Wang B, Chen C, Xie Y, Wang H (2019) Arabidopsis FHY3 and FAR1 regulate the balance between growth and defense responses under shade conditions. Plant Cell 29(12):2829–2846
Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, Xiao J (2015) COLD1 confers chilling tolerance in rice. Cell 160(6):1209–1221
McManmon M, Crawford RM (1971) A metabolic theory of flooding tolerance: the significance of enzyme distribution and behavior. New Phytol 70:299–306
Mitra GN (2015) Calcium (Ca) uptake. Regulation of nutrient uptake by plants: a biochemical and molecular approach. Springer, India, pp 53–70
Mittler R (2017) ROS are good. Trends Plant Sci 22(1):11–19
Mittler R, Blumwald E (2010) Genetic engineering for modern agriculture: challenges and perspectives. Annu Rev Plant Biol 61:443–462
Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2018) AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Et Biophys Acta Gene Regul Mech 1:2–12
Monk LS, Fagerstedt KV, Crawford RM (1987) Superoxide dismutase as an anaerobic polypeptide is a key factor in recovery from oxygen deprivation in Iris pseudacorus. Plant Physiol 85:1016–1020
Munns R, Termaat A (1986) Whole-plant responses to salinity. Funct Plant Biol 13:143–160
Mustroph A, Barding GA, Kaiser KA, Larive CK, Bailey-Serres J (2014) Characterization of distinct root and shoot responses to low-oxygen stress in Arabidopsis with a focus on primary C- and N-metabolism. Plant Cell Environ 37:2366–2380
Nakagawa Y, Katagiri T, Shinozaki K, Qi Z, Tatsumi H, Furuichi T et al (2007) Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc Natl Acad Sci USA 104(9):3639–3644
Narendra T, Sarvajeet GS (2016) Abiotic stress signaling in plants—an overview. Wiley, Hoboken, pp 1–12
Neha S, Inderjeet B, Abhishek K, Punit T, Girija S, Sakshi C, Yasin JK (2017) Stop the new gene, the alien: breakdown of transgenes and introgressions by ncRNA mediated gene regulations. J AgriSearch 4(2):133–40
Nuruzzaman M, Sharoni AM, Kikuchi S (2013) Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol 4:248
Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K (2017) Transcriptional regulatory network of plant heat stress response. Trends Plant Sci 22:53–65
Ouyang H, Vogel HJ (1998) Metal ion binding to calmodulin: NMR and fluorescence studies. Biometals 11(3):213–222
Pareek A, Joshi R, Gupta KJ, Singla-Pareek SL, Foyer C (2020) Sensing and signalling in plant stress responses: ensuring sustainable food security in an era of climate change. New Phytol 228:823–827. https://doi.org/10.1111/nph.16893
Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324(5930):1068–1071
Pitzschke A, Djamei A, Bitton F, Hirt H (2009) A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signaling. Mol Plant 2:120–137
Polle A, Schutzendubel A (2004) Heavy metal signaling in plants: linking cellular and organismic responses. In: Hirt H, Shinozaki K (eds) Plant responses to abiotic stress, vol 4. Topics in Current Genetics. Springer, Berlin, pp 187–215
Qu AL, Ding YF, Jiang Q, Zhu C (2013) Molecular mechanisms of the plant heat stress response. Biochem Biophys Res Commun 432:203–207
Raza A, Tabassum J, Kudapa H, Varshney RK (2021) Can omics deliver temperature resilient ready-to-grow crops? Crit Rev Biotechnol 41(8):1209–1232
Raza A, Tabassum J, Fakhar AZ, Sharif R, Chen H, Zhang C, Ju L, Fotopoulos V, Siddique KH, Singh RK, Zhuang W (2022) Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit Rev Biotechnol 12:1–28
Raza A, Charagh S, Najafi-Kakavand S, Abbas S, Shoaib Y, Anwar S, Sharifi S, Lu G, Siddique KH (2023a) Role of phytohormones in regulating cold stress tolerance: physiological and molecular approaches for developing cold-smart crop plants. Plant Stress 23:100152
Raza A, Charagh S, Salehi H, Abbas S, Saeed F, Poinern GE, Siddique KH, Varshney RK (2023b) Nano-enabled stress-smart agriculture: can nanotechnology deliver drought and salinity-smart crops? J Sustain Agric Environ 2(3):189–214
Raza A, Mubarik MS, Sharif R, Habib M, Jabeen W, Zhang C, Chen H, Chen ZH, Siddique KH, Zhuang W, Varshney RK (2023c) Developing drought-smart, ready-to-grow future crops. The Plant Genome 16(1):e20279
Raza A, Wang D, Zou X, Prakash CS (2023d) Developing temperature-resilient plants: a matter of present and future concern for sustainable agriculture. Agronomy 13(4):1006. https://doi.org/10.3390/agronomy13041006
Ren CG, Chen Y, Dai CC (2014) Cross-talk between calcium-calmodulin and brassinolide for fungal endophyte-induced volatile oil accumulation of Atractylodes lancea plantlets. J Plant Growth Regul 33:285–294. https://doi.org/10.1007/s00344-013-9370-4
Ren S, Ma K, Lu Z, Chen G, Cui J, Tong P, Wang L, Teng N, Jin B (2019) Transcriptomic and metabolomic analysis of the heat-Stress response of Populus tomentosa Carr. Forests 10:383
Sairam RK, Kumutha D, Ezhilmathi K, Chinnusamy V, Meena RC (2009) Water-logging induced oxidative stress and antioxidant enzymes activity in pigeon pea. Biol Plant 53:493–504
Saini N, Nikalje GC, Zargar, SM, Penna S (2022) Molecular insights into sensing, regulation and improving of heat tolerance in plants. Plant Cell Rep 41:799–813
Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18(5):1292–1309
Sasidharan R, Voesenek LA (2015) Ethylene-mediated acclimations to flooding stress. Plant Physiol 169:3–12
Sato H, Mizoi J, Tanaka H, Maruyama K, Qin F, Osakabe Y, Morimoto K, Ohori T, Kusakabe K, Nagata M et al (2014) Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. Plant Cell 26:4954–4973
Sato H, Takasaki H, Takahashi F, Suzuki T, Iuchi S, Mitsuda N et al (2018) Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc Natl Acad Sci USA 115(47):E11178–E11187
Scharf KD, Berberich T, Ebersberger I, Nover L (2012) The plant heat stress transcription factor (Hsf) family: structure, function, and evolution. Biochim Biophys Acta 1819:104–119
Shao HB, Chu LY, Ni FT, Guo DG, Li H, Li WX (2010) Perspective on phytoremediation for improving heavy metal contaminated soils. In: Ashraf M, Ozturk M, Ahmad M (eds) Plant adaptation and phytoremediation. Springer, Dordrecht, pp 227–244
Singh A, Roychoudhury A (2023) Abscisic acid in plants under abiotic stress: crosstalk with major phytohormones. Plant Cell Rep 42:1–14
Singh S, Modi MK, Gill SS, Tuteja N (2012) Rice: genetic engineering approaches for abiotic stress tolerance–retrospects and prospects. Improving crop productivity in sustainable agriculture. Wiley, Hoboken, pp 201–236
Singh N, Bhogal I, Mishra BK, Yasin JK (2016) Carbonic anhydrase genes network: key role players in pH flux and abiotic stress tolerance. J Agric Search 3(4):8
Snedden WA, Fromm H (2001) Calmodulin as a versatile calcium signal transducer in plants. New Phytol 151:35–66
Stockinger EJ, Mao Y, Regier MK, Triezenberg SJ, Thomashow MF (2001) Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interaction with CBF1, a transcriptional activator involved in cold regulated gene expression. Nucleic Acid Res 29:1524–1533
Suzuki N, Sejima H, Tam R, Schlauch K, Mittler R (2011) Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J 66:844–851
Takahashi F, Suzuki T, Osakabe Y, Betsuyaku S, KondoY DN et al (2018) A small peptide modulates stomatal control via abscisic acid in long-distance signaling. Nature 556(7700):235–238
Takahashi F, Kuromori T, Urano K, Yamaguchi-Shinozaki K, Shinozaki K (2020) Drought stress responses and resistance in plants: from cellular responses to long distance intercellular communication. Front Plant Sci 11:556972
Tewari S, Arora NK (2013) Plant growth promoting rhizobacteria for ameliorating abiotic stresses triggered due to climatic variability. Clim Change Environ Sustain 1:95–103
Tewari S, Arora NK (2016) Soybean production under flooding stress and its mitigation using plant growth promoting microbes. Environ Stresses Soybean Prod 14:2–23
Thapa G, Sadhukhan A, Panda SK, Sahoo L (2012) A molecular mechanistic model of plant heavy metal tolerance. Biometals 25(3):489–505
Tuteja N (2007) Mechanisms of high salinity tolerance in plants. Methods Enzymol 428:419–438
Voesenek LACJ, Bailey-Serres J (2013) Flooding tolerance: O2 sensing and survival strategies. Curr Opin Plant Biol 16:647–653
Wahid A, Gelani S, Ashraf M, Foolad MR (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61:199–223
Wang X, Li W, Li M, Welti R (2006) Profiling lipid changes in plant response to low temperatures. Physiol Plant 126:90–96
Xiong L, Zhu JK (2002) Molecular and genetic aspect of plant responses to osmotic stress. Plant Cell Environ 25:131–139
Yamanaka T, Nakagawa Y, Mori K, Nakano M, Imamura T, Kataoka H, Terashima A, Iida K, Kojima I, Katagiri T, Shinozaki K (2010) MCA1 and MCA2 that mediate Ca2+ uptake have distinct and overlapping roles in Arabidopsis. Plant Physiol 152(3):1284–1296
Yang D, Liu Y, Liu S, Li C, Zhao Y, Li L, Lu S (2020) Exposure to heavy metals and its association with DNA oxidative damage in municipal waste incinerator workers in Shenzhen, China. Chemosphere 250:126289. https://doi.org/10.1016/j.chemosphere.2020.126289
Yao HY, Xue HW (2018) Phosphatidic acid plays key roles regulating plant development and stress responses. J Integr Plant Biol 60:851–863
Yasin JK (2015) Intra cellular pH flux and cyclosis in plant cells under abiotic stress. J Agric Search 2(2):150–151
Yasin JK, Singh AK (2019) Intracellular trafficking and cytoplasmic streaming under abiotic stress conditions. J Agric Search. https://doi.org/10.21921/jas.v6i04.16894
Yasin JK, Nizar MA, Rajkumar S, Verma M, Radhamani J, Verma N, Pandey S (2014) Alternate antioxidant defence system in moisture stress responsive accessions of horse gram. Legum Res 37(2):145–154
Yasin JK, Mishra BK, Pillai MA, Nidhi Verma SH, Wani HO, Elansary DO, El-Ansary PS, Pandey VC (2020) Genome wide in-silico miRNA and target network prediction from stress responsive Horsegram (Macrotyloma uniflorum) accessions. Sci Rep 10:17203
Yasin JK, Mishra BK, Pillai MA, Chinnusamy V (2021) Physical map of lncRNAs and lincRNAs linked with stress responsive miRs and genes network of pigeonpea (Cajanus cajan L.). J Plant Biochem Biotechnol. https://doi.org/10.1007/s13562-021-00674-0
Yin C, Sun A, Zhou Y, Liu K, Wang P, Ye W, Fang Y (2023) The dynamics of H2A.Z on SMALL AUXIN UP RNAs regulate abscisic acid–auxin signaling crosstalk in Arabidopsis. J Exp Botany 74:4158
Zhang X, Fowler SG, Cheng H, Lou Y, Rhee SY, Stockinger EJ, Thomashow MF (2015) Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J 82(2):1–13
Zhang L, Shi X, Zhang Y, Wang J, Yang J, Ishida T, Jiang W, Han X, Kang J, Wang X, Pan L (2019) CLE9 peptide-induced stomatal closure is mediated by abscisic acid, hydrogen peroxide, and nitric oxide in Arabidopsis thaliana. Plant Cell Environ 42(3):1033–1044
Zhang H, Zhu J, Gong Z et al (2022) Abiotic stress responses in plants. Nat Rev Genet 23:104–119
Zhao J, Lu Z, Wang L, Jin B (2021) Plant responses to heat stress: physiology, transcription, noncoding RNAs, and epigenetics. Int J Mol Sci 22:117
Acknowledgements
The authors are grateful to their respective institutions.
Funding
This work was partially supported by the Indian Council of Agricultural Research—National Bureau of Plant Genetic Resources [PGR/DFP-BUR-DEL-01.01].
Author information
Authors and Affiliations
Contributions
YJK conceived the idea, YJK, SS, MS, VK, MAP, SHW, and SP wrote the MS.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Abidur Rahman.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shakespear, S., Sivaji, M., Kumar, V. et al. Navigating Through Harsh Conditions: Coordinated Networks of Plant Adaptation to Abiotic Stress. J Plant Growth Regul (2024). https://doi.org/10.1007/s00344-023-11224-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00344-023-11224-4