Abstract
Sesame (Sesamum indicum L) is a high-value oilseed cash crop grown across different geographies. Although sesame is traditionally considered drought-tolerant, early seedling and vegetative stage plants are sensitive to drought, causing substantial yield losses. Roots are the sensors of water deficit and hence are determinants of drought stress-induced responses. Limited information is available about the molecular responses induced in sesame roots during osmotic stress. We performed RNA-sequencing to understand transcriptional changes in the roots of drought-tolerant (TEX-1) and drought-sensitive (VEN-1) sesame genotypes using Polyethylene glycol (PEG)-induced osmotic stress. The photosynthetic measurements and proline accumulation confirmed contrasting drought stress reactions between the two genotypes. A total of 1251 and 541 unique genes were differentially expressed in PEG-treated and untreated roots of TEX-1 and VEN-1 roots, respectively. Gene ontology (GO) function enrichment analysis revealed that the differentially expressed genes related to catalytic activity, ion binding, transferase activity, and cation binding activities were over-represented. Transcription-factor enrichment analysis showed that multiple members of WRKY, bZIP, MYB, and NAC families were over-represented in the roots of drought-tolerant genotype during osmatic stress. The study identified several genes involved in the primary metabolism, phenylpropanoid, and hormonal pathways induced in the roots of drought-tolerant genotype. The study provided a comparative transcriptome resource for understanding molecular responses underlying osmotic stress and identified target genes for enhancing abiotic stress tolerance in sesame.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sesame (Sesamum indicum L) is a high-value oilseed cash crop grown across diverse geographies in tropical and subtropical environments. Sesame has offered a viable alternative and profitable rotation crop in the Southwestern United States. Sesame oil is referred to as ‘queen of the oils’ due to its high unsaturated fatty acid content and resistance to oxidative deterioration (Bedigian and Harlan 1986). Sesame is traditionally grown in drought-prone and marginal lands, and hence often receives sub-optimal inputs such as water and nutrients. Although its deep root system helps in conserving water, early seedling and vegetative stage sesame plants are sensitive to drought stress (Boureima et al. 2011; Sun et al. 2010), causing significant yield and quality losses. There is increasing interest in breeding drought-tolerant varieties by utilizing available genetic resources adapted to various environments. High genetic variability and relatively small genome size (Zhang et al. 2013) make sesame a good model for identifying traits associated with abiotic stress tolerance in oilseed crops.
Drought stress tolerance is a complex process involving orchestrated responses at physiological, biochemical, and molecular levels across different plant tissues. Several approaches have been used to investigate the drought-induced responses in plants such as genomic (Cushman and Bohnert 2000; Langridge and Reynolds 2015; Tuberosa and Salvi 2006), transcriptomic (Bashir et al. 2019; Deyholos 2010; Takahashi et al. 2018), proteomic (Ghatak et al. 2017; Ghosh and Xu 2014; Johnová et al. 2016; Wang 2019), or metabolic (Fàbregas and Fernie 2019; Jorge et al. 2016; Krasensky and Jonak 2012). Drought tolerance is a polygenic trait determined by physiological adjustments regulated by an interplay of signaling cascades and integrated responses from molecular to whole-plant level (Fleury et al. 2010; Kebede et al. 2019). Roots are the sensors of water deficit and hence studied as determinants of drought stress-induced responses (Goodger and Schachtman 2010; Steudle 2000). During drought stress, the expression of a large number of genes is altered, with no single biological, cellular, or molecular mechanism providing a full level of protection (Golldack et al. 2014; Nakashima et al. 2014; Takahashi et al. 2018). The stress-induced genes can be grouped into two classes (Yang et al. 2017)—(1) the effector proteins that include genes involved in the biosynthesis of osmolytes, secondary metabolites, photosynthesis, and growth-related genes, and (2) the regulatory proteins that indirectly respond to stress through regulation of the expression of downstream target genes such as transcription factors (TFs), signal transduction pathways.
Studies to understand drought tolerance in sesame have predominantly been focused on evaluating above-ground (shoot and leaves) traits using morphological (Boureima et al. 2016) and physiological (Boureima et al. 2012) changes during the stress period. Transcriptome analysis and Next-Generation Sequencing (NGS) have been extensively used in several crop species to study changes in the molecular responses during different abiotic stresses (Abdel-Ghany et al. 2020; Formentin et al. 2018; Kang et al. 2020). Except for a few studies (Dossa et al. 2016, 2017, 2019b), limited information is available about molecular responses in sesame roots during drought stress. Polyethylene glycol (PEG) is exclusively used to mimic drought stress conditions as it effectively decreases water potential (Ψw) and disrupts the absorption of water by roots (Verslues et al. 1998). To understand the molecular responses associated with drought tolerance in sesame roots, we carried out a comparative transcriptome analysis of two sesame genotypes with contrasting responses using PEG-induced osmotic stress. The transcriptomic analysis identified 770 and 243 genes uniquely up-regulated in osmotically stressed roots of TEX-1 and VEN-1, respectively. Osmotic stress treatment showed strong up-regulation of genes such as several members of WRKY and NAC TF families, and genes involved in the phenylpropanoid and hormonal pathways in TEX-1 roots. Outcomes of this comparative analysis of sesame root transcriptome can be utilized as a reference to identify molecular markers and screen suitable breeding populations or germplasm to enhance drought tolerance.
Materials and Methods
Plant Material, Osmotic Stress Management, and Physiological Trait Measurements
Seeds of drought-tolerant sesame (Sesamum indicum L) genotype ‘TEX-1′ and drought-sensitive genotype ‘VEN-1′ proprietary experimental lines were made available by Dr. Eric Votava, Sesaco Corporation (Austin, Texas, USA) for this study. Seeds were grown in a greenhouse at the AgriLife Research and Extension Center in Uvalde in a Turface® (Quick Dry Infield Conditioner) and fertilized with standard Hoagland solution (Caisson Laboratories, UT). Four-week-old seedlings of both genotypes were carefully transferred to water containing 20% (m/v) polyethylene glycol—6000 (PEG-6000). Seedlings placed in water alone were used as control. Roots samples from three independent biological replicates for both genotypes were harvested 12 h after initiation of treatment, immediately frozen in liquid nitrogen, and stored at − 80 °C for further analyses.
Further, to validate contrasting drought responses in TEX-1 and VEN-1 genotypes, an experiment was carried out under managed stress conditions in a controlled greenhouse at the Uvalde AgriLife Research and Extension Center. Plants were grown in 10-gallon pots containing soil and sand mix (80:20), adopting standard agronomic practices. The drought stress was imposed by withholding water (30% RWC) when plants were 4 weeks old. Leaf samples from both sesame genotypes were collected for metabolic analysis seven days after the initiation of stress treatment. Well-watered plants were treated as controls. Leaf photosynthetic rate (Pn), transpiration rate (E), stomatal conductance (Gs), and intercellular CO2 concentration (Ci) were measured from two uppermost fully expanded leaves using the LI-COR-6400 XT Photosynthetic system (Lincoln, USA) from three independent plants for both genotypes.
Amino Acid Extraction and Quantification
Amino acid analysis of roots from PEG-stress experiments and leaves from the soil-based experiment was carried out using Waters UPLC-ESI-MS/MS platform comprising Waters Acquity H-class UPLC system coupled to Waters Xevo TQs mass spectrometer with electrospray ionization (ESI) probe following an established protocol. l-Norvaline (TCI AMERICA, USA) was used as an internal control. Amino acid calibrators were obtained from Amino Acid Kit (Waters Corporation, Milford, MA, USA). Lyophilized powder of the mixture of amino acids was reconstituted in 0.1 M HCl before derivatization. Multiple Reaction Monitoring (MRM) transitions and collision energy values and cone voltage were optimized for each amino acid using the Water’s IntelliStart software. Water's MassLynx™ software was used for instrument monitoring and data acquisition. The TargetLynxTM Application Manager (Waters Corporation, Milford, MA, USA) was used to perform data integration, preparation of calibration curves, and quantitation of amino acids.
RNA Extraction, cDNA Library Preparation, and Sequencing
The frozen root samples from the PEG-stress experiment were homogenized using a mortar and pestle, and total RNA was extracted using the RNeasy Plant Mini kit (QIAGEN Sciences, Germantown, MD, USA). The purity of RNA was analyzed using a Nanodrop (DS-11 + spectrophotometer, DeNovix Inc.), and quality was verified using Agilent 2100 Bioanalyzer (Texas A&M AgriLife Genomics & Bioinformatics Service, College Station, TX) before further processing.
Illumina RNA-Sequencing (RNA-Seq)
The construction of the cDNA library and sequencing was carried out using an Illumina HiSeq-4000 sequencing platform at the Genomics and Bioinformatics Services, Texas A&M University, College Station, TX, USA. A total of 12 RNA samples were prepared for library preparation using the TruSeq RNA Library Prep Kit v2 (Illumina, San Diego, CA). A set of three root samples of TEX-1 and VEN-1 genotypes collected from control (water alone), and PEG solution each were used for library construction. The sequencing was performed using 75 bp pair-end reads on one lane of the Illumina HiSeq-4000 platform.
Read Filtration, Annotation, and Functional Enrichment Analysis of Differentially Expressed Genes (DEGs)
The removal of sequence adapters, mapping to reference genome, and normalization of gene expression were performed using CLC Genomics Workbench (V4.5, CLC Bio.). The RNA-Seq reads were first assessed for quality with FastQC; adapters were removed with Cutadapt (Martin 2011), and the low-quality bases were trimmed (q < 20) with FastX-toolkit (Gordon and Hannon 2010). Quality-filtered reads were then mapped to the reference database Sinbase 2.0 ver 1.0 (Wang et al. 2014, 2016a) using TopHat2 (Kim et al. 2013) with default parameters. Novel transcripts were assembled by Cufflink (Trapnell et al. 2010), and then combined with available gene annotation using CuffMerge. Normalized expression levels of genes were expressed in FPKM values as generated by Cuffquant and Cuffnorm running with the option “–frag-bias-correct” and “–multi-read-correct” (Trapnell et al. 2012). The differential expressed genes (DEGs) of pooled samples from each condition were determined using Cuffdiff, using the FPKM values. The unigenes were recognized as differentially expressed when FDR-adjusted P-value ≤ 0.05, and the absolute value of Log2 ratio ≥ 1. The Blast2GO (Conesa and Götz 2008; Conesa et al. 2005) program was employed to obtain Gene Ontology (GO) annotations for unigenes. mRNA functional categorization and pathway enrichment were performed by mapping all unigenes against the Kyoto Encyclopedia of Genes and Genomes (KEGG) Ontology (KO) database (www.genome.jp/dbget/) BLASTX to assign to known biological pathways. Gene ontology (GO) enrichment analysis was carried using NCBI blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for Nr (NCBI non-redundant protein sequences) and generating combined graphs of molecular function, cellular component, and biological process with default settings in Blast2GO. The interaction between TFs and their target genes was evaluated using the ‘TF enrichment tool’ from the PlantRegMap portal (Jin et al. 2016), and the number of TFs with significantly over-represented target number genes was retrieved (P-value < 0.01; Fisher’s exactly test). The portal identifies the transcriptional regulations from literature and ChIP-seq data or inferred by combining TF binding motifs and regulatory elements data. Online tool Venny v.2.0.2 (Oliveros 2007) was used to show distribution of differentially expressed genes or TFs between the genotypes using Venn diagrams.
Availability of Transcriptome Sequencing Data
The transcriptional sequence data have been submitted to the National Center for Biotechnology Information (NCBI) through NCBI Gene Expression Omnibus (GEO) repository accession number GSE148340 (https://www.ncbi.nlm.nih.gov/geo/), raw reads through SRA (SRP255755) and GTF annotation file (Sesamum_indicum_v1.0.gene) describing the location of genes and exons pairing with the reference fasta file to retrieve the transcript sequences as Online Resource (File S1).
Gene Expression Analyses
For validation of RNA-Seq results, real-time quantitative PCR was performed. Root tissues collected from PEG-stressed and control seedlings of TEX-1 and VEN-1 were harvested and frozen in liquid nitrogen. Total RNA was extracted using an RNeasy® Plant Mini Kit (QIAGEN Sciences, Germantown, MD, USA) as per the manufacturer’s protocol. Total RNA was treated with DNase1 (QIAGEN Sciences, Germantown, MD, USA), quantified and subjected to reverse transcription (cDNA synthesis) using iScript RT Supermix (Bio-Rad Laboratories, Inc, Hercules, USA). For qRT-PCR, specific primers were designed using the PrimerQuest® program (IDT, Coralville, Iowa, USA). Gene expression analysis via reverse transcription-qPCR was performed using a Bio-Rad CFX96 qPCR instrument and LightCycler480 SsoAdv Univer SYBR GRN Master Kit (Bio-Rad Laboratories, Inc, Hercules, USA). All samples were amplified in triplicate along with a negative control without cDNA. The efficiency of each primer pair used in RT-qPCR was calculated (Supplementary Material Table S1) using LinRegPCR (Ramakers et al. 2003). Sesame UBQ6 and CYP genes (Wei et al. 2013) were used as internal controls, and relative expression levels (Cq values) for each gene were normalized to the transcription of both reference genes by taking an average of three biological replicates. The relative expression levels were calculated using the ΔΔCq (quantitative cycle) method provided with Bio-Rad CFX software.
Results and Discussion
Drought Stress Responses in Sesame Genotypes
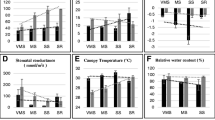
The differences in drought stress responses between two sesame genotypes were confirmed by quantifying the abundance of proline (Pro) and branched-chain amino acids in roots of PEG-stressed seedlings (Fig. 1) and leaves of drought-stressed plants (Fig. 2a) using LC–MS-based analysis. PEG-stressed roots of TEX-1 showed increases up to 11-fold in Pro, 16-fold in valine (Val), fourfold in isoleucine (Ile), and 14-fold in leucine (Leu). The increases in amino acids in VEN-1 were limited to fourfold for Pro and Val, and ninefold for Leu. These results were comparable to drought-stressed leaves of soil-grown plants, where TEX-1 leaves accumulated significantly higher amino acids than VEN-1 (Fig. 2a). Increased accumulation of Pro in drought-tolerant genotype is consistent with previous studies in sesame (Dossa et al. 2017; Hussein et al. 2015; Kadkhodaie et al. 2014). Additionally, we measured photosynthesis, stomatal conductance, intercellular CO2 concentration, and transpiration rates in drought-stressed sesame plants grown in pots before flowering (Fig. 2b). The net photosynthetic rate under stress was 54% lower than under well water conditions in VEN-1. The decreased photosynthesis was accompanied by reduced stomatal conductance, intercellular CO2 concentration, and transpiration rate, which was 79%, 37%, and 67% lower under stress compared to well-watered conditions. In contrast, a less profound impact on photosynthetic machinery was observed in TEX-1 as the same physiological parameters under drought stress were only 26%, 30%, 14%, and 27% lower than the control condition. The results demonstrated the presence of relatively higher levels of osmolytes and photosynthetic capacity of TEX-1 than VEN-1 under osmotic or drought conditions.
Validation of drought responses in the vegetative tissues of sesame genotypes. a Proline and branched-chain amino acid accumulation in drought-stressed leaves. Differences between varieties within treatments are shown as *Significant at P < 0.05. b Responses of gas exchange parameters to water stress in two sesame varieties. Pn—net photosynthetic rate; E—transpiration rate, Gs—stomatal conductance; Ci—intercellular CO2 concentration. Different letters within the same column denote the differences between the treatments (P < 0.05) within the genotype. NS no significant at the 0.05 level; *Significant at P < 0.05
Transcriptome Sequencing and Assembly of Sequencing Data
To understand transcriptional differences between roots of sesame genotypes under normal and control conditions, we carried out RNA-Seq analysis. A total of six cDNA libraries were constructed for PEG-stressed roots of drought-tolerant TEX-1 and drought-sensitive VEN-1 genotypes using the Illumina HiSeq-4000 sequencing platform. After a stringent quality assessment, on an average, 61 million (TEX-1) and 59 million (VEN-1) raw reads, and approximately 57 million (TEX-1) and 56 million (VEN-1) clean reads were generated (Supplementary Material Table S2) for control and PEG-stressed roots, respectively. A total of 48 million (TEX-1) and 51 million (VEN-1) reads in the PEG-stressed roots were mapped to the reference transcriptome (Sinbase V 1.0). Out of the total clean reads, 94% and 7% reads were uniquely and multi-mapped to the reference genome, respectively. The plot showing the combined assembly of transcripts in PEG-stressed and control roots for the two sesame genotypes is presented in Supplementary Material (Supplementary Material Fig. S1). A total of 18,923 and 19,045 transcripts in the control treatment and 19,019 and 19,244 transcripts in osmotically stressed roots were generated in TEX-1 and VEN-1, respectively. The numbers of raw reads expressed in roots and unigenes identified are comparable to other transcriptomic studies in sesame (Dossa et al. 2016; Su et al. 2019; Wang et al. 2014; Wei et al. 2015; Zhang et al. 2020).
Identification of Differentially Expressed Genes (DEGs)
Approximately 95% of clean reads were mapped to the 27,148 unigenes from the reference sesame genome, with more than 66% among them being uniquely mapped (Supplementary Material Table S2). We used normalized FPKM values to quantify transcript levels in reads, which facilitated a comparison of mRNA levels both within and between sesame genotypes. A graphical sketch showing a total 1251 and 542 unique genes differentially expressed between PEG-treated and untreated roots of TEX-1 and VEN-1, respectively, is presented as a Venn diagram (Fig. 3a) and the total distribution of all genes as a volcano plot (Supplementary Material Fig. S2). Together, 849 DEGs were expressed in both genotypes. Among DEGs, 770 and 243 genes were uniquely up-regulated due to osmotic stress in TEX-1 and VEN-1 roots, respectively (Fig. 3b). In total, 481 and 299 unique DEGs were down-regulated in TEX-1 and VEN-1 roots, respectively. A total of 440 up-regulated and 403 down-regulated genes were common to both genotypes. The DEGs from both genotypes comprised many diverse classes suggesting the presence of a complex regulatory mechanism in sesame roots in response to osmotic stress. The numbers of DEGs with Log2 fold change of 2 or above in roots of TEX-1 and VEN-1 genotypes are shown in a separate Venn diagram (Supplementary Material Fig. S3), and a list of genes is presented as a Supplementary Material Table S3. The numbers of genes showing either up- or down-regulation in roots of osmotically stressed roots were much higher in TEX-1 than VEN-1.
Differentially expressed gene profiles. a Comparison between the differential expressed genes (DEGs) in the TEX-1 and VEN-1 in PEG-stressed roots. The numbers of DEGs exclusively expressed in one variety are shown in each circle of the Venn diagram. The numbers of DEGs with common expression changes during osmotic stress are shown in the overlapping regions. b The total numbers of DEGs showing either up-regulation or down-regulation in both varieties are shown in the petal Venn diagram. The number of genes unique to variety is shown in the clear zones, while genes common to both varieties are shown in the overlapping parts. Genes with a fold change ≥ 1.5 and an FDR (q-value) < 0.05 were considered DEGs
Functional Annotation and Gene Ontology (GO) Classification
The gene ontology (GO) enrichment analysis was carried out to understand the biological relevance of DEGs in drought-sensitive VEN-1 and drought-tolerant TEX-1. We used Blast2GO (Conesa and Götz 2008) to retrieve GO functional categorization of DEGs in TEX-1 (Supplementary Material Fig. S4) and VEN-1 (Supplementary Material Fig. S5), which includes three categories: biological process, molecular function, and cellular component. The biological processes mediated by these DEGs mainly represented categories like “response to the stimulus,” “metabolic processes,” and “biological regulation” in both genotypes. The molecular functions of DEGs were primarily related to “catalytic activity,” “transporter activity,” and “transcription regulator activity,” while the cellular component mostly encompassed “cell organelle” and “membranes.”
GO term enrichment analysis of DEGs in TEX-1 and VEN-1 was conducted using the R package TopGO (Alexa et al. 2006), and significance was calculated based on Fisher’s exact test with a cut-off threshold of p < 0.01. All sesame genes were used as a background. GO annotation and statistical analyses demonstrated that 1507 DEGs (some contigs had more than one GO annotation) in TEX-1 and 997 DEGs in VEN-1 had GO annotation, of which 149 GO terms in TEX-1 (Fig. 4) and 119 GO terms in VEN-1 (Fig. 5) were enriched. In the case of cellular component, 325 and 211 DEGs for membrane (GO:0016020), 184 and 130 DEGs for cell periphery (GO:0071944,) and 142 and 87 DEGs for plasma membrane (GO:0005886,) were significantly enriched in TEX-1 and VEN-1 roots, respectively, compared to whole sesame transcriptome (Supplementary Material Table S4 for TEX-1 and Supplementary Material Table S5 for VEN-1). In the molecular function category, the majority of genes were related to “catalytic activity” (GO: 0003824, 662 DEGs in TEX-1 and 468 DEGs in VEN-1) followed by ion binding (GO:0043167, 474 DEGs in TEX-1 and 312 in VEN-1), transferase activity (GO:0016740, 278 DEGs in TEX-1 and 180 in VEN-1), and cation binding (GO:0043167, 254 DEGs in TEX-1 and 178 in VEN-1) activities. In the case of biological processes, DEGs over-represented terms like “response to stimulus” (GO0050896, 359 DEGs in TEX-1 and 232 in VEN-1) and “response to stress” (GO:0006950, 219 DEGs in TEX-1 and 148 in VEN-1).
Enrichment Analysis of Transcription Factors (TFs) in Response to Osmotic Stress
To identify possible regulatory connections between TFs and their putative target genes, we used TF enrichment analysis available through a web portal PlantRegMap (Jin et al. 2016). The portal allowed us to find the TFs possessing significantly over-represented targets in the DEGs of both the genotypes. We identified a total of 129 TFs in TEX-1, and 110 TFs in VEN-1 roots that possessed significantly over-represented target DEGs (Supplementary Material Table S6 for TEX-1 and Table S7 for VEN-1).
The functions of the putative targets of the TFs spans several categories, such as responses to stresses, including oxygen-containing compound, chemical, oxidative stress with the TFs mentioned above in both varieties regulating the expression of genes of a broad spectrum of biological processes (Supplementary Material Table S8 for TEX-1 and Table S9 for VEN-1). The transcription-factor classification (Fig. 6) revealed that 129 TFs identified in TEX-1 belonged to 23 transcription-factor families, of which 108 were up-regulated and 21 down-regulated. In VEN-1 roots, 110 TFs were grouped into 19 transcription-factor families, of which 51 were up-regulated and 59 down-regulated. The numbers of uniquely over-represented TFs in osmotically stressed TEX-1 and VEN-1 roots are shown as a Venn diagram (Supplemental Material Fig. S6), and a complete list provided as Supplemental Material Table S10. Among the enriched TFs, the members of bZIP (5), WRKY (4), and TCP (3) families were uniquely up-regulated in TEX-1 roots, suggesting their utility as potential markers for breeding drought-tolerant sesame varieties.
Transcription-factor enrichment analysis. The number of genes from transcription-factor families represented in the differentially expressed genes in TEX-1 and VEN-1 roots after osmotic stress treatment (P-value ≤ 0.0001). The induced genes are shown as green bars and the repressed as red bars (Color figure online)
Among the total TFome, the enriched TFs showing predicted interactions with the up-regulated TF DEGs were more comprehensive and broadly distributed into TF families in TEX-1 than those based on the down-regulated TF DEGs, suggesting an increased demand for specific TFs in response to osmotic stress in roots of stress-tolerant genotype. On the contrary, enriched TFs were widely distributed among the down-regulated TF in VEN-1, implying suppression or inactivation of such stress-induced responses.
The TFs enrichment analysis suggested over-representation of TFs belonging to the WRKY, bZIP, MYB, and NAC families in roots of osmotically stressed TEX-1 plants. Induction of the number of members of WRKY and MYB TF families in response to abiotic stresses is in agreement with the previous studies in drought-stressed and waterlogged sesame plants (Li et al. 2017; Mmadi et al. 2017). The WRKY family represents one of the most abundant TF family that activates a network of signaling cascades in response to different abiotic stresses in plants (Jiang et al. 2017; Phukan et al. 2016; Rushton et al. 2012; Schluttenhofer and Yuan 2015). The root architectural traits, such as root length and spread, contribute to maintaining plant productivity under water stress (Comas et al. 2013). In Arabidopsis, WRKY TF facilitated the growth of lateral roots in response to osmotic stress through the activation of ABA signaling and auxin homeostasis (Ding et al. 2015). Likewise, heterologous expression of sorghum WRKY30 enhanced drought tolerance in Arabidopsis and rice by influencing the root architecture, proline contents, and superoxide dismutase (SOD), polyphenol oxidase (POD), and catalase (CAT) activities (Yang et al. 2020). Recently, an ectopic over-expression of sesame MYB75 in Arabidopsis also demonstrated an increase in root growth, endogenous ABA level, and tolerance to drought and salt stresses (Dossa et al. 2019a).
Consistent with our data, the up-regulation of bZIP and NAC TFs has been demonstrated in roots during abiotic stress. The functional role of bZIP TFs in roots has been validated in several crops such as maize (Ma et al. 2018; Seeve et al. 2017), rice (Yang et al. 2019), and wheat (Agarwal et al. 2019) during abiotic stress. Similarly, the expression and role of NAC TFs in root development and drought tolerance have been demonstrated in rice (Jeong et al. 2010, 2013; Redillas et al. 2012) and wheat (Mao et al. 2020; Xue et al. 2011).
Drought-induced responses in roots are complex and are influenced by genotype, the environment, and their interactions. Our analysis suggests that the osmotic stress-induced responses were accompanied by changes in the expression of several genes in the roots of drought-tolerant TEX-1 than the susceptible genotype VEN-1. The changes in the expression of genes are probably directed towards the maintenance of root architecture, which may allow a better acclimation to the osmotic stress conditions. In part, the drought response may be regulated by activation of the members of WRKY, MYB, bZIP, and NAC TF families that could regulate root architecture as well as hormonal signaling pathways. The TFs identified in this study may serve as targets for the enhancement of drought tolerance in sesame.
Quantitative Real‑Time PCR (qRT-PCR) Validation
Next, to validate gene expression levels obtained from RNA-Seq transcriptome, we selected 9 DEGs that showed at least twofold changes (Log2) and determined their relative expression by qRT-PCR analysis in the osmotically stressed and control roots of TEX-1 and VEN-1. A set of nine unigenes selected for expression analysis represented different functional classes such as transcription factors, membrane proteins, and primary metabolism. Genes that were induced strongly in PEG-stressed roots selected for qRT-PCR assays included (1) 1-Amino-cyclopropane-1-carboxylate synthase (ACS): XLOC_005397 (SIN_1024771), (2) Phosphoenolpyruvate carboxylase kinase1 (PEPC): XLOC_016295 (SIN_1001672), (3) Cinnamyl alcohol dehydrogenase 7 (CAD7): XLOC_021096 (SIN_1005800), (4) Branched-chain amino acid transaminase 2 (BCAT2): XLOC_017622 (SIN_1006354), (5) NAC Transcription factor (NAC): XLOC_021943 (SIN_1007268), (6) Transmembrane protein (TMEM): XLOC_012774 (SIN_1015912), (7) BTB and TAZ domain protein (BTB): XLOC_013444 (SIN_1020232), (8) F-Box protein (FBXO): XLOC_022774 (SIN_1026951), and (9) Glutathione S-transferase (GST): XLOC_018225. The qRT-PCR analysis validated the relative expression of genes selected from the RNA-seq analysis in both the genotypes (Fig. 7). Furthermore, the linear regression y = 1.3357 × − 1.0936 and high correlation (R2 = 0.84) revealed a substantial similarity between both the analytical techniques (Supplementary Material Fig. S7), confirming the reliability of RNA-Seq data.
Validation of transcriptome sequencing by RT-qPCR. The vertical axis shows the ratios of 9 differentially expressed genes in osmotically stressed (PG) and control (CT) roots of TEX-1 and VEN-1. The horizontal axis shows genes in the following order: (1) 1-Amino-cyclopropane-1-carboxylate synthase (ACS), XLOC_005397 (SIN_1024771), (2) Phosphoenolpyruvate carboxylase kinase1 (PEPC), XLOC_016295 (SIN_1001672), (3) Cinnamyl alcohol dehydrogenase 7 (CAD7), XLOC_021096 (SIN_1005800), (4) Branched-chain amino acid transaminase 2 (BCAT2), XLOC_017622 (SIN_1006354), (5) NAC Transcription factor (NAC), XLOC_021943 (SIN_1007268), (6) Transmembrane protein (TMEM), XLOC_012774 (SIN_1015912), (7) BTB and TAZ domain protein (BTB), XLOC_013444 (SIN_1020232), (8) F-Box protein (FBXO), XLOC_022774 (SIN_1026951), and (9) Glutathione S-transferase (GST), XLOC_018225
Key Genes Associated with Osmotic Stress Tolerance
The accumulation of compatible solutes such as proline, branched-chain amino acids, or polyamines plays a critical role during osmotic stress. Consistent with the amino acid changes (Fig. 1) in the PEG-treated roots, expression of Δ1- pyrroline-5-carboxylate synthase XLOC_015307 (SIN_1012216) was significantly up-regulated in TEX-1 but not in VEN-1 roots. The bifunctional enzyme Δ1-pyrroline-5-carboxylate synthase (P5CS) catalyzes the rate-limiting reaction in proline biosynthesis (Delauney and Verma 1993). In plants, its transcription is induced by environmental stresses such as drought, salinity, or temperature in several species (Kavi Kishor and Sreenivasulu 2014; Strizhov et al. 1997). The early activation of the gene involved its biosynthesis, and its accumulation in roots may partly explain the osmoprotective role of Pro in the adaption of sesame to drought stress environment.
Besides Pro, the levels of other amino acids, mainly three types of branched-chain amino acids (BCAAs), could elevate during abiotic stress to levels comparable to proline or even higher (Joshi et al. 2010) due to decreased protein synthesis or increased protein degradation (Hildebrandt 2018; Huang and Jander 2017). The increased accumulation of Val, Leu, and Ile in TEX-1 roots during osmotic stress was in agreement with a strong up-regulation in the expression of branched-chain amino acid transaminase 2, XLOC_017622 (SIN_1006354), which catalyzes both the synthesis and catabolism of branched-chain amino acids (Hildebrandt et al. 2015).
Among the selectively activated genes in TEX-1, XLOC_016295 (SIN_1001672) and XLOC_012893 (SIN_1011314) coding for a phosphoenolpyruvate carboxykinase (PEPCK; 4.1.1.49) were strongly up-regulated (Log2 fold change 4.7 and 2.9, respectively). The PEPCK enzymes are ubiquitous across the plant kingdom, playing a pivotal role in carbon metabolism and physicochemical processes in photosynthesis. Although the role of phosphoenolpyruvate carboxylase (PEPCase) and PEPCK is well understood in C4 and CAM photosynthesis, it also plays a wide range of roles in non-photosynthetic and photosynthetic tissues of C3 plants in energy and biosynthetic metabolism (Izui et al. 2004). Several studies have demonstrated the induction of PEPCK genes and their functional role in tolerance to abiotic and biotic stresses (Feria et al. 2016; García-Mauriño et al. 2003; González et al. 2003; Hýsková et al. 2014; O'Leary et al. 2011; Wang et al. 2016b; Waseem and Ahmad 2019). Increased PEPC activity in roots initiates the production of metabolic intermediates required in several metabolic processes such as pyruvate, organic acids (malate or citrate) that chelate cations in nutrient deprivation, and synthesis of osmolytes like proline during osmotic stress (Doubnerová and Ryšlavá 2011; González et al. 2003; O'Leary et al. 2011). Induction of in the gene expression or increased enzyme activities of PEPC and PEPCK in responses to abiotic stresses, especially in roots, has been demonstrated in Arabidopsis (Feria et al. 2016), wheat (González et al. 2003; Zang et al. 2018), and tomato (Gong et al. 2014).
Pyruvate and pyruvate-derived amino acids accumulate in roots under oxidative conditions (Lehmann et al. 2009, 2012) and in stressed seedlings (Obata et al. 2011). Pyruvate synthesis occurs in the cytoplasm directly by the phosphorylation of phosphoenolpyruvate catalyzed by kinase enzyme or by the action of PEPCase. In addition to PEPCK in our study, up-regulation of pyruvate kinase family protein (XLOC_027450; SIN_1001467) and phosphoenolpyruvate hydratase (XLOC_005608; SIN_1012968)) along with down-regulation of both putative pyruvate dehydrogenases (XLOC_019412; SIN_1022485 and XLOC_010651; SIN_1018146) implicates an orchestrated induction of biosynthetic genes and suppression of catabolism to promote pyruvate accumulation in osmotically stressed TEX-1 roots (Supplementary Material Fig. S8). Pyruvate is the primary substrate for the tricarboxylic acid (TCA) cycle and synthesis of several amino acids, fatty acid biosynthesis, and anaerobic fermentation. Several studies based on expression analysis and transgenic approaches support the role of PEP-derived pyruvate formation in roots during environmental stresses (Bouthour et al. 2015; Chen et al. 2010; Feria et al. 2016; Wang et al. 2016b; Zhao et al. 2019).
In plants, the phenylpropanoid pathway comprises a complex network of metabolic processes related to the synthesis of powerful antioxidants such as phenolics (flavonoids, isoflavonoids, hydroxycinnamic acids, and lignin) and secondary metabolites. The enzymes involved in the phenylpropanoid pathway have been shown to play a direct role in abiotic and biotic stresses in a different plant species (Dixon and Paiva 1995; Sharma et al. 2019). In TEX-1, expression of at least four members of cinnamyl alcohol dehydrogenase family (XLOC_021096; SIN_1005800, XLOC_011357; SIN_1021093, XLOC_021100; SIN_1005796, XLOC_021101; SIN_1005795), 4-Coumarate–CoA ligase (XLOC_023104; SIN_1006083), Cinnamoyl-CoA reductase (XLOC_009932; SIN_1007428), and peroxidase (XLOC_017799; SIN_1023583) were induced at least fourfold higher in osmotically stressed roots (Supplementary Material Fig. S9). Increased expression of genes involved in flavonoid and lignin biosynthesis (such as phenylalanine ammonia-lyase (PAL), 4-coumarate 3-hydroxylase (C3H), 4-coumarate: coenzyme A ligase (4-CL), caffeoyl coenzyme A O-methyltransferase, cinnamyl alcohol dehydrogenase (CAD), peroxidase) during water stress has been demonstrated in several plant species (Li et al. 2017; Liu et al. 2018; Preisner et al. 2018; Yan et al. 2012; Yoshimura et al. 2008). Further, lignin modification is also known to be associated with the activation of genes involved in oxidative stress responses aligning with the up-regulation of expression of glutathione S-transferase (XLOC_018225; 4.5-fold change at Log2) in TEX-1 roots. Several transcriptomic and metabolomic studies in plants have confirmed that enhanced flavonoid accumulation positively impacts drought stress tolerance (Ballizany et al. 2012; Fleck et al. 2011; Nakabayashi et al. 2014; Nichols et al. 2015).
Plant hormones mediate responses to adapt to environment changes (Burgess and Huang 2016; Peleg and Blumwald 2011). We assessed the DEGs associated with hormonal pathways such as ethylene, auxin, cytokinin, gibberellic acid (GA), abscisic acid (ABA), and jasmonic acid (JA). The phytohormone ethylene is well known for its role in various biotic and abiotic stresses (Ju and Chang 2015) and in promoting root growth (Stepanova et al. 2007; Swarup et al. 2007). Ethylene is synthesized from methionine through 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) and ACC oxidases (ACO). In the roots of drought-tolerant TEX-1, we found that the expression of both ACC (XLOC_005397; SIN_1024771) and ACS (XLOC_010050; SIN_1013258) were strongly up-regulated (> threefold change at Log2). Consistent with these observations, an increase in the expression of ACS and ACO genes in response to water stress has also been reported in soybean (Arraes et al. 2015) and barley (Montilla-Bascón et al. 2017). Several independent studies have confirmed increased ethylene production in roots during water deficit (da Costa et al. 2013; Munné-Bosch and Müller 2013; Sharp 2002). Ethylene acts as a signaling molecule during abiotic and biotic stresses (Johnson and Ecker 1998; Ju and Chang 2015) and promotes adventitious root development predominantly by alternating auxin biosynthesis, transport, and signaling (Negi et al. 2010; Qin and Huang 2018). In line with this theory, in TEX-1 roots, we also observed induction of expression of auxin response factor (XLOC_000963; SIN_1010841) and SAUR-like auxin-responsive proteins (XLOC_006093; SIN_1014231, XLOC_001224; SIN_1013943). Drought stress inhibits the synthesis and transport of cytokinin (Prerostova et al. 2018), causing a decline in stress tolerance. Our results demonstrated that the expression of isopentenyl transferase (XLOC_019702; SIN_1012440), a key rate-limiting step in the biosynthesis of cytokinin (Sakakibara 2006), was strongly up-regulated in TEX-1 roots in response to osmotic stress. The role of cytokinin production in abiotic stress-induced responses has been demonstrated in a dicot (Rivero et al. 2007) and monocot plants (Peleg et al. 2011). Moreover, down-regulation of cytokinin oxidase (XLOC_004591; SIN_1024740), which promotes root growth, also indicates possible crosstalk between ethylene, auxin, and cytokinin hormonal pathways in alternating root growth in TEX-1 during osmotic stress. JA is involved in the regulation of many physiological and stress-related processes in plants (Wasternack and Hause 2013). Jasmonate ZIM-domain (JAZ) proteins regulate JA signaling pathways and the crosstalk between various phytohormones (Zhu et al. 2011). Unlike VEN-1, at least four of the putative JAZ proteins (XLOC_006351; SIN_1006670, XLOC_012304; SIN_1010464, XLOC_016643; SIN_1007561, XLOC_010484; SIN_1021313) were up-regulated in osmotically stressed TEX-1 roots. Similarly, up-regulation of gibberellin 2-oxidase (XLOC_020788; SIN_1022458), which plays an important role in the GA catabolism, in TEX-1 roots is consistent with its role in root development during abiotic stress (Shan et al. 2014). On the other hand, the expression of ABI5 binding protein 2 (AFP, XLOC_010602), which interacts with transcription factor ABA-Insensitive 5 (ABI5), a key regulator in ABA signaling and stress response (Garcia et al. 2008) was up-regulated in both TEX-1 and VEN-1 roots suggesting its conserved role in roots in response to osmotic stress. ABA modifies root hydraulic properties and regulates water balance during drought stress (Aroca et al. 2012), promoting root growth to access water in deeper soil layers (Skirycz and Inzé 2010). ABI5 acts as a negative regulator of lateral root development during stress (Shkolnik-Inbar and Bar-Zvi 2010), while AFP attenuates ABA signals by targeting ABI5 for ubiquitin-mediated degradation (Lopez-Molina et al. 2003).
Taken together, it is evident that the processes associated with osmotic stress responses in sesame roots are regulated in a complex way not by a few selected hormones but by the crosstalk between several hormonal networks and the regulatory elements. The complexity of the osmotic stress-induced responses observed in the current study suggesting multiple levels of metabolic and molecular regulations is in agreement with transcriptomic studies in other oilseed crops such as sunflower (Liang et al. 2017), safflower (Wei et al. 2020), peanut (Zhao et al. 2020), soybean (Ha et al. 2015).
Conclusions
In conclusion, comparative analysis of sesame root transcriptomes indicated a coordinated regulation of multiple genes and pathways (effector and regulatory genes) and provided a catalog of transcriptomic variation between genotypes, with contrasting responses to osmotic stress. The RNA-Seq data generated in this study would be a valuable resource for the identification of genes related to drought tolerance across systems. Furthermore, genes involved in amino acid metabolism, phenylpropanoid, and hormonal pathways and TFs induced in TEX-1 roots would serve as potential biomarkers for identifying drought-tolerant genotypes. The use of systems biology approaches incorporating transcriptomic, proteomic, and metabolomic information would help in understanding the functional role and biological relevance of these genes in roots during drought stress in plants. Further, the RNA-Seq data made available in the public repository would serve the interests of researchers characterizing the drought tolerance mechanisms and accelerate the efforts to breed crops for sustainable production.
Data Availability
The RNA-Seq dataset is accessible through NCBI Gene Expression Omnibus (GEO) repository accession number GSE148340 (https://www.ncbi.nlm.nih.gov/geo/) and the raw data through SRA (SRP255755).
References
Abdel-Ghany SE, Ullah F, Ben-Hur A, Reddy ASN (2020) Transcriptome analysis of drought-resistant and drought-sensitive sorghum (sorghum bicolor) genotypes in response to PEG-induced drought stress. Int J Mol Sci. https://doi.org/10.3390/ijms21030772
Agarwal P, Baranwal VK, Khurana P (2019) Genome-wide analysis of bZIP transcription factors in wheat and functional characterization of a TabZIP under abiotic stress. Sci Rep 9:4608. https://doi.org/10.1038/s41598-019-40659-7
Alexa A, Rahnenführer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22:1600–1607
Aroca R, Porcel R, Ruiz-Lozano JM (2012) Regulation of root water uptake under abiotic stress conditions. J Exp Bot 63:43–57
Arraes FBM et al (2015) Implications of ethylene biosynthesis and signaling in soybean drought stress tolerance. BMC Plant Biol 15:213. https://doi.org/10.1186/s12870-015-0597-z
Ballizany WL, Hofmann RW, Jahufer MZ, Barrett BA (2012) Multivariate associations of flavonoid and biomass accumulation in white clover (Trifolium repens) under drought. Funct Plant Biol 39:167–177
Bashir K, Matsui A, Rasheed S, Seki M (2019) Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress F1000Res 8:F1000. Faculty Rev. https://doi.org/10.12688/f1000research.18424.1
Bedigian D, Harlan JR (1986) Evidence for cultivation of sesame in the ancient world. Econ Bot 40:137–154
Boureima S, Eyletters M, Diouf M, Diop T, Van Damme P (2011) Sensitivity of seed germination and seedling radicle growth to drought stress in sesame (Sesamum indicum L.). Res J Environ Sci 5:557–564
Boureima S, Oukarroum A, Diouf M, Cisse N, Van Damme P (2012) Screening for drought tolerance in mutant germplasm of sesame (Sesamum indicum) probing by chlorophyll a fluorescence. Environ Exp Bot 81:37–43
Boureima S, Diouf S, Amoukou M, Van Damme P (2016) Screening for sources of tolerance to drought in sesame induced mutants: assessment of indirect selection criteria for seed yield. Int J Pure Appl Biosci 4:45–60
Bouthour D, Kalai T, Chaffei HC, Gouia H, Corpas FJ (2015) Differential response of NADP-dehydrogenases and carbon metabolism in leaves and roots of two durum wheat (Triticum durum Desf.) cultivars (Karim and Azizi) with different sensitivities to salt stress. J Plant Physiol 179:56–63. https://doi.org/10.1016/j.jplph.2015.02.009
Burgess P, Huang B (2016) Mechanisms of hormone regulation for drought tolerance in plants. In: Hossain M, Wani S, Bhattacharjee S, Burritt D, Tran LS (eds) Drought stress tolerance in plants, vol 1. Springer, Cham, pp 45–75
Chen M, Tang Y, Zhang J, Yang M, Xu Y (2010) RNA Interference-based suppression of phosphoenolpyruvate carboxylase results in susceptibility of rapeseed to osmotic stress. J Integr Plant Biol 52:585–592. https://doi.org/10.1111/j.1744-7909.2010.00948.x
Comas L, Becker S, Cruz VMV, Byrne PF, Dierig DA (2013) Root traits contributing to plant productivity under drought. Front Plant Sci 4:442
Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008:619832
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Cushman JC, Bohnert HJ (2000) Genomic approaches to plant stress tolerance. Curr Opin Plant Biol 3:117–124
da Costa CT, de Almeida MR, Ruedell CM, Schwambach J, Maraschin FS, Fett-Neto AG (2013) When stress and development go hand in hand: main hormonal controls of adventitious rooting in cuttings. Front Plant Sci 4:133–133. https://doi.org/10.3389/fpls.2013.00133
Delauney AJ, Verma DPS (1993) Proline biosynthesis and osmoregulation in plants. Plant J 4:215–223. https://doi.org/10.1046/j.1365-313X.1993.04020215.x
Deyholos MK (2010) Making the most of drought and salinity transcriptomics. Plant Cell Environ 33:648–654. https://doi.org/10.1111/j.1365-3040.2009.02092.x
Ding ZJ, Yan JY, Li CX, Li GX, Wu YR, Zheng SJ (2015) Transcription factor WRKY 46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J 84:56–69
Dixon RA, Paiva NL (1995) Stress-induced phenylpropanoid metabolism. Plant Cell 7:1085–1097. https://doi.org/10.1105/tpc.7.7.1085
Dossa K, Diouf D, Cissé N (2016) Genome-wide investigation of Hsf genes in sesame reveals their segmental duplication expansion and their active role in drought stress response. Front Plant Sci 7:1522
Dossa K et al (2017) Dynamic transcriptome landscape of sesame (Sesamum indicum L.) under progressive drought and after rewatering. Genomics Data 11:122–124
Dossa K et al (2019a) Ectopic expression of the sesame MYB transcription factor SiMYB305 promotes root growth and modulates ABA-mediated tolerance to drought and salt stresses in Arabidopsis. AoB Plants. https://doi.org/10.1093/aobpla/plz081
Dossa K et al (2019b) Depicting the core transcriptome modulating multiple abiotic stresses responses in sesame (Sesamum indicum L.). Int J Mol Sci 20:3930
Doubnerová V, Ryšlavá H (2011) What can enzymes of C4 photosynthesis do for C3 plants under stress? Plant Sci 180:575–583
Fàbregas N, Fernie AR (2019) The metabolic response to drought. J Exp Bot 70:1077–1085. https://doi.org/10.1093/jxb/ery437
Feria AB et al (2016) Phosphoenolpyruvate carboxylase (PEPC) and PEPC-kinase (PEPC-k) isoenzymes in Arabidopsis thaliana: role in control and abiotic stress conditions. Planta 244:901–913. https://doi.org/10.1007/s00425-016-2556-9
Fleck AT, Nye T, Repenning C, Stahl F, Zahn M, Schenk MK (2011) Silicon enhances suberization and lignification in roots of rice (Oryza sativa). J Exp Bot 62:2001–2011. https://doi.org/10.1093/jxb/erq392
Fleury D, Jefferies S, Kuchel H, Langridge P (2010) Genetic and genomic tools to improve drought tolerance in wheat. J Exp Bot 61:3211–3222. https://doi.org/10.1093/jxb/erq152
Formentin E et al (2018) Transcriptome and cell physiological analyses in different rice cultivars provide new insights into adaptive and salinity stress responses. Front Plant Sci. https://doi.org/10.3389/fpls.2018.00204
García-Mauriño S, Monreal J, Alvarez R, Vidal J, Echevarría C (2003) Characterization of salt stress-enhanced phosphoenolpyruvate carboxylase kinase activity in leaves of Sorghumvulgare: independence from osmotic stress, involvement of ion toxicity and significance of dark phosphorylation. Planta 216:648–655
Garcia ME, Lynch T, Peeters J, Snowden C, Finkelstein R (2008) A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol Biol 67:643–658. https://doi.org/10.1007/s11103-008-9344-2
Ghatak A, Chaturvedi P, Weckwerth W (2017) Cereal crop proteomics: systemic analysis of crop drought stress responses towards marker-assisted selection breeding. Front Plant Sci 8:757. https://doi.org/10.3389/fpls.2017.00757
Ghosh D, Xu J (2014) Abiotic stress responses in plant roots: a proteomics perspective. Front Plant Sci 5:6. https://doi.org/10.3389/fpls.2014.00006
Golldack D, Li C, Mohan H, Probst N (2014) Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci 5:151. https://doi.org/10.3389/fpls.2014.00151
Gong B, Zhang C, Li X, Wen D, Wang S, Shi Q, Wang X (2014) Identification of NaCl and NaHCO3 stress responsive proteins in tomato roots using iTRAQ-based analysis. Biochem Biophys Res Commun 446:417–422. https://doi.org/10.1016/j.bbrc.2014.03.005
González M-C, Sanchez R, Cejudo FJ (2003) Abiotic stresses affecting water balance induce phosphoenolpyruvate carboxylase expression in roots of wheat seedlings. Planta 216:985–992
Goodger JQD, Schachtman DP (2010) Re-examining the role of ABA as the primary long-distance signal produced by water-stressed roots. Plant Signal Behav 5:1298–1301. https://doi.org/10.4161/psb.5.10.13101
Gordon A, Hannon G (2010) Fastx-toolkit. FASTQ/A short-reads pre-processing tools Unpublished. https://www.hannonlabcshledu/fastx_toolkit5. Accessed 1 Sep 2019
Ha C et al (2015) Comparative analysis of root transcriptomes from two contrasting drought-responsive Williams 82 and DT2008 soybean cultivars under normal and dehydration conditions. Front Plant Sci. https://doi.org/10.3389/fpls.2015.00551
Hildebrandt TM (2018) Synthesis versus degradation: directions of amino acid metabolism during Arabidopsis abiotic stress response. Plant Mol Biol 98:121–135. https://doi.org/10.1007/s11103-018-0767-0
Hildebrandt TM, Nesi AN, Araújo WL, Braun H-P (2015) Amino acid catabolism in plants. Mol Plant 8:1563–1579
Huang T, Jander G (2017) Abscisic acid-regulated protein degradation causes osmotic stress-induced accumulation of branched-chain amino acids in Arabidopsis thaliana. Planta 246:737–747. https://doi.org/10.1007/s00425-017-2727-3
Hussein Y, Amin G, Azab A, Gahin H (2015) Induction of drought stress resistance in sesame (Sesamum indicum L.) plant by salicylic acid and kinetin. J Plant Sci 10:128
Hýsková VD, Miedzińska L, Dobrá J, Vankova R, Ryšlavá H (2014) Phosphoenolpyruvate carboxylase, NADP-malic enzyme, and pyruvate, phosphate dikinase are involved in the acclimation of Nicotiana tabacum L. to drought stress. J Plant Physiol 171:19–25
Izui K, Matsumura H, Furumoto T, Kai Y (2004) Phospho enol pyruvate carboxylase: a new era of structural biology. Annu Rev Plant Biol 55:69–84
Jeong JS et al (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153:185–197
Jeong JS et al (2013) OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol J 11:101–114
Jiang J, Ma S, Ye N, Jiang M, Cao J, Zhang J (2017) WRKY transcription factors in plant responses to stresses Journal of Integrative. Plant Biol 59:86–101. https://doi.org/10.1111/jipb.12513
Jin J, Tian F, Yang D-C, Meng Y-Q, Kong L, Luo J, Gao G (2016) PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res 45:D1040
Johnová P, Skalák J, Saiz-Fernández I, Brzobohatý B (2016) Plant responses to ambient temperature fluctuations and water-limiting conditions: a proteome-wide perspective. Biochim Biophys Acta 1864:916–931. https://doi.org/10.1016/j.bbapap.2016.02.007
Johnson PR, Ecker JR (1998) The ethylene gas signal transduction pathway: a molecular perspective. Annu Rev Genet 32:227–254
Jorge TF, Rodrigues JA, Caldana C, Schmidt R, van Dongen JT, Thomas-Oates J, António C (2016) Mass spectrometry-based plant metabolomics: metabolite responses to abiotic stress. Mass Spectrom Rev 35:620–649. https://doi.org/10.1002/mas.21449
Joshi V, Joung J-G, Fei Z, Jander G (2010) Interdependence of threonine, methionine and isoleucine metabolism in plants: accumulation and transcriptional regulation under abiotic stress. Amino Acids 39:933–947
Ju C, Chang C (2015) Mechanistic insights in ethylene perception and signal transduction. Plant Physiol 169:85–95. https://doi.org/10.1104/pp.15.00845
Kadkhodaie A, Razmjoo J, Zahedi M, Pessarakli M (2014) Selecting sesame genotypes for drought tolerance based on some physiochemical traits. Agron J 106:111–118. https://doi.org/10.2134/agronj2013.0260
Kang W-H et al (2020) Transcriptome profiling of abiotic responses to heat, cold, salt, and osmotic stress of Capsicum annuum L. Sci Data 7:17. https://doi.org/10.1038/s41597-020-0352-7
Kavi Kishor PB, Sreenivasulu N (2014) Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ 37:300–311. https://doi.org/10.1111/pce.12157
Kebede A, Kang M, Bekele E (2019) Advances in mechanisms of drought tolerance in crops, with emphasis on barley. Adv Agron. https://doi.org/10.1016/bs.agron.2019.01.008
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36
Krasensky J, Jonak C (2012) Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot 63:1593–1608. https://doi.org/10.1093/jxb/err460
Langridge P, Reynolds MP (2015) Genomic tools to assist breeding for drought tolerance. Curr Opin Biotechnol 32:130–135. https://doi.org/10.1016/j.copbio.2014.11.027
Lehmann M et al (2009) The metabolic response of Arabidopsis roots to oxidative stress is distinct from that of heterotrophic cells in culture and highlights a complex relationship between the levels of transcripts, metabolites, and flux. Mol Plant 2:390–406. https://doi.org/10.1093/mp/ssn080
Lehmann M, Laxa M, Sweetlove LJ, Fernie AR, Obata T (2012) Metabolic recovery of Arabidopsis thaliana roots following cessation of oxidative stress. Metabolomics 8:143–153. https://doi.org/10.1007/s11306-011-0296-1
Li D et al (2017) Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biol 17:152
Liang C et al (2017) Identification of differentially expressed genes in sunflower (Helianthus annuus) leaves and roots under drought stress by RNA sequencing. Bot Stud. https://doi.org/10.1186/s40529-017-0197-3
Liu W, Jin Y, Li M, Dong L, Guo D, Lu C, Qi H (2018) Analysis of CmCADs and three lignifying enzymes in oriental melon (‘CaiHong7’) seedlings in response to three abiotic stresses. Sci Hortic 237:257–268. https://doi.org/10.1016/j.scienta.2018.04.024
Lopez-Molina L, Mongrand S, Kinoshita N, Chua N-H (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17:410–418. https://doi.org/10.1101/gad.1055803
Ma H et al (2018) ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development. Plant Physiol 178:753–770
Mao H et al (2020) Regulatory changes in TaSNAC8-6A are associated with drought tolerance in wheat seedlings. Plant Biotechnol J 18:1078–1092. https://doi.org/10.1111/pbi.13277
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12
Mmadi M et al (2017) Functional characterization of the versatile MYB gene family uncovered their important roles in plant development and responses to drought and waterlogging in sesame. Genes 8:362
Montilla-Bascón G et al (2017) Reduced nitric oxide levels during drought stress promote drought tolerance in barley and is associated with elevated polyamine biosynthesis. Sci Rep 7:13311. https://doi.org/10.1038/s41598-017-13458-1
Munné-Bosch S, Müller M (2013) Hormonal cross-talk in plant development and stress responses. Front Plant Sci 4:529–529. https://doi.org/10.3389/fpls.2013.00529
Nakabayashi R et al (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J 77:367–379
Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci 5:170. https://doi.org/10.3389/fpls.2014.00170
Negi S, Sukumar P, Liu X, Cohen JD, Muday GK (2010) Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J 61:3–15. https://doi.org/10.1111/j.1365-313X.2009.04027.x
Nichols SN, Hofmann RW, Williams WM (2015) Physiological drought resistance and accumulation of leaf phenolics in white clover interspecific hybrids. Environ Exp Bot 119:40–47
O'Leary B, Park J, Plaxton WC (2011) The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem J 436:15–34
Obata T et al (2011) Alteration of mitochondrial protein complexes in relation to metabolic regulation under short-term oxidative stress in Arabidopsis seedlings. Phytochemistry 72:1081–1091. https://doi.org/10.1016/j.phytochem.2010.11.003
Oliveros JC (2007) VENNY. An interactive tool for comparing lists with Venn Diagrams. https://www.bioinfogpcnbcsices/tools/venny/indexhtml. Accessed 5 Sep 2019
Peleg Z, Blumwald E (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 14:290–295
Peleg Z, Reguera M, Tumimbang E, Walia H, Blumwald E (2011) Cytokinin-mediated source/sink modifications improve drought tolerance and increase grain yield in rice under water-stress. Plant Biotechnol J 9:747–758
Phukan UJ, Jeena GS, Shukla RK (2016) WRKY transcription factors: molecular regulation and stress responses in plants. Front Plant Sci 7:760–760. https://doi.org/10.3389/fpls.2016.00760
Preisner M, Wojtasik W, Kostyn K, Boba A, Czuj T, Szopa J, Kulma A (2018) The cinnamyl alcohol dehydrogenase family in flax: differentiation during plant growth and under stress conditions. J Plant Physiol 221:132–143. https://doi.org/10.1016/j.jplph.2017.11.015
Prerostova S et al (2018) Cytokinins: their impact on molecular and growth responses to drought stress and recovery in arabidopsis. Front Plant Sci 9:655–655. https://doi.org/10.3389/fpls.2018.00655
Qin H, Huang R (2018) Auxin controlled by ethylene steers root development. Int J Mol Sci 19:3656. https://doi.org/10.3390/ijms19113656
Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66. https://doi.org/10.1016/s0304-3940(02)01423-4
Redillas MCFR et al (2012) The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol J 10:792–805. https://doi.org/10.1111/j.1467-7652.2012.00697.x
Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc Natl Acad Sci 104:19631–19636
Rushton DL et al (2012) WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol J 10:2–11
Sakakibara H (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57:431–449
Schluttenhofer C, Yuan L (2015) Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol 167:295–306
Seeve CM et al (2017) Water-deficit-induced changes in transcription factor expression in maize seedlings. Plant Cell Environ 40:686–701
Shan C, Mei Z, Duan J, Chen H, Feng H, Cai W (2014) OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS ONE 9:e87110. https://doi.org/10.1371/journal.pone.0087110
Sharma A, Shahzad B, Rehman A, Bhardwaj R, Landi M, Zheng B (2019) Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 24:2452. https://doi.org/10.3390/molecules24132452
Sharp R (2002) Interaction with ethylene: changing views on the role of abscisic acid in root and shoot growth responses to water stress. Plant Cell Environ 25:211–222. https://doi.org/10.1046/j.1365-3040.2002.00798.x
Shkolnik-Inbar D, Bar-Zvi D (2010) ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 22:3560–3573. https://doi.org/10.1105/tpc.110.074641
Skirycz A, Inzé D (2010) More from less: plant growth under limited water. Curr Opin Biotechnol 21:197–203. https://doi.org/10.1016/j.copbio.2010.03.002
Stepanova AN, Yun J, Likhacheva AV, Alonso JM (2007) Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19:2169–2185. https://doi.org/10.1105/tpc.107.052068
Steudle E (2000) Water uptake by roots: effects of water deficit. J Exp Bot 51:1531–1542. https://doi.org/10.1093/jexbot/51.350.1531
Strizhov N et al (1997) Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J 12:557–569. https://doi.org/10.1046/j.1365-313x.1997.00557.x
Su R et al (2019) Root diversity in sesame (Sesamum indicum L.): insights into the morphological, anatomical and gene expression profiles. Planta 250:1461–1474. https://doi.org/10.1007/s00425-019-03242-y
Sun J, Rao Y, Yan T, Yan X, Zhou H (2010) Effects of drought stress on sesame growth and yield characteristics and comprehensive evaluation of drought tolerance. Chin J Oil Crop Sci 32:525–533
Swarup R et al (2007) Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19:2186–2196. https://doi.org/10.1105/tpc.107.052100
Takahashi F, Kuromori T, Sato H, Shinozaki K (2018) Regulatory gene networks in drought stress responses and resistance in plants. Adv Exp Med Biol 1081:189–214. https://doi.org/10.1007/978-981-13-1244-1_11
Trapnell C et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. https://doi.org/10.1038/nbt.1621
Trapnell C et al (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562
Tuberosa R, Salvi S (2006) Genomics-based approaches to improve drought tolerance of crops. Trends Plant Sci 11:405–412
Verslues PE, Ober ES, Sharp RE (1998) Root growth and oxygen relations at low water potentials. Impact of oxygen availability in polyethylene glycol solutions. Plant Physiol 116:1403–1412. https://doi.org/10.1104/pp.116.4.1403
Wang X (2019) Protein and proteome atlas for plants under stresses: new highlights and ways for integrated omics in post-genomics era. Int J Mol Sci. https://doi.org/10.3390/ijms20205222
Wang L et al (2014) Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol 15:R39. https://doi.org/10.1186/gb-2014-15-2-r39
Wang L et al (2016a) Updated sesame genome assembly and fine mapping of plant height and seed coat color QTLs using a new high-density genetic map. BMC Genomics 17:31–31. https://doi.org/10.1186/s12864-015-2316-4
Wang N, Zhong X, Cong Y, Wang T, Yang S, Li Y, Gai J (2016b) Genome-wide analysis of phosphoenolpyruvate carboxylase gene family and their response to abiotic stresses in soybean. Sci Rep 6:38448
Waseem M, Ahmad F (2019) The phosphoenolpyruvate carboxylase gene family identification and expression analysis under abiotic and phytohormone stresses in Solanum lycopersicum L. Gene 690:11–20
Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111:1021–1058
Wei L, Miao H, Zhao R, Han X, Zhang T, Zhang H (2013) Identification and testing of reference genes for Sesame gene expression analysis by quantitative real-time PCR. Planta 237:873–889. https://doi.org/10.1007/s00425-012-1805-9
Wei X, Wang L, Yu J, Zhang Y, Li D, Zhang X (2015) Genome-wide identification and analysis of the MADS-box gene family in sesame. Gene 569:66–76. https://doi.org/10.1016/j.gene.2015.05.018
Wei B, Hou K, Zhang H, Wang X, Wu W (2020) Integrating transcriptomics and metabolomics to studies key metabolism, pathways and candidate genes associated with drought-tolerance in Carthamus tinctorius L. Under drought stress. Ind Crops Prod 151:112465. https://doi.org/10.1016/j.indcrop.2020.112465
Xue G-P, Way HM, Richardson T, Drenth J, Joyce PA, McIntyre CL (2011) Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant 4:697–712
Yan L et al (2012) Identification of differentially expressed genes in sorghum (Sorghum bicolor) brown midrib mutants. Physiol Plant 146:375–387. https://doi.org/10.1111/j.1399-3054.2012.01646.x
Yang Z et al (2017) Transcriptome analysis of two species of jute in response to polyethylene glycol (PEG)-induced drought stress. Sci Rep 7:16565
Yang S et al (2019) A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice. BMC Plant Biol 19:260. https://doi.org/10.1186/s12870-019-1872-1
Yang Z et al (2020) SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J Plant Physiol 246–247:153142. https://doi.org/10.1016/j.jplph.2020.153142
Yoshimura K, Masuda A, Kuwano M, Yokota A, Akashi K (2008) Programmed proteome response for drought avoidance/tolerance in the root of a C3 xerophyte (wild watermelon) under water deficits. Plant Cell Physiol 49:226–241
Zang X et al (2018) Overexpression of the wheat (Triticum aestivum L.) TaPEPKR2 gene enhances heat and dehydration tolerance in both wheat and arabidopsis. Front Plant Sci. https://doi.org/10.3389/fpls.2018.01710
Zhang H, Miao H, Wang L, Qu L, Liu H, Wang Q, Yue M (2013) Genome sequencing of the important oilseed crop Sesamum indicum L. Genome Biol 14:401
Zhang Y et al (2020) A collection of transcriptomic and proteomic datasets from sesame in response to salt stress. Data Brief 32:106096. https://doi.org/10.1016/j.dib.2020.106096
Zhao X, Bai X, Jiang C, Li Z (2019) Phosphoproteomic analysis of two contrasting maize inbred lines provides insights into the mechanism of salt-stress tolerance. Int J Mol Sci 20:1886. https://doi.org/10.3390/ijms20081886
Zhao N et al (2020) Identification and expression analysis of WRKY gene family under drought stress in peanut (Arachis hypogaea L.). PLoS ONE 15:e0231396. https://doi.org/10.1371/journal.pone.0231396
Zhu Z et al (2011) Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci 108:12539–12544
Acknowledgements
We thank Dr. Eric Votava, Sesaco Corporation (Austin, Texas USA), for kindly providing seed material for this study. We thank Diwas Silwal for his help in plant sampling. This work was partly supported by ‘Crops of Texas Genomics of Plant Water Use Seed Grant Program’ Texas A&M AgriLife Research.
Author information
Authors and Affiliations
Contributions
QS and MJ carried out the experiment. SW, QS, CJ, and VJ carried out sequencing and analyzed the data. VJ conceived the idea, supervised the project, and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Song, Q., Joshi, M., Wang, S. et al. Comparative Analysis of Root Transcriptome Profiles of Sesame (Sesamum indicum L.) in Response to Osmotic Stress. J Plant Growth Regul 40, 1787–1801 (2021). https://doi.org/10.1007/s00344-020-10230-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-020-10230-0