Abstract
Nitrogen uptake by plants is a key step for efficient nitrogen use, which affects plant growth and yield. Arabidopsis thaliana gene NRT1.1 was identified as a transporter related to nitrate (NO3−) signaling and uptake. In rice, three orthologs of NRT1.1, named OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C, have been identified. This study evaluated the potential of OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C in NO3− signaling and uptake through overexpression in the Arabidopsis chl1-5 mutant. The expression of OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C was evaluated in the roots and shoots of rice cultivated with NO3− or NH4+. OsNRT1.1A was expressed in the roots and shoots cultivated with NO3− and NH4+. OsNRT1.1B was expressed predominantly in roots of rice cultivated with NO3−, while the expression of OsNRT1.1C was low in roots and shoots. Arabidopsis chl1-5 plants were transformed by the floral dip method using Agrobacterium tumefaciens to overexpress OsNRT1.1A and the alternative splicing product named OsNRT1.1As, OsNRT1.1B, and OsNRT1.1C. The chlorate test showed the ability of OsNRT1.1A, OsNRT1.1B or OsNRT1.1C to take up chlorate, as evidenced by the decrease in fresh weight. The OsNRT1.1B lineages presented higher toxicity to chlorate. Gene expression analyses showed that the insertion of OsNRT1.1A and OsNRT1.1B into Arabidopsis chl1-5 induced the expression of NRT2.1 and NAR2.1. OsNRT1.1As overexpression did not significantly affect the expression of NRT2.1 and NAR2.1. The results show the differential ability of NRT1.1 orthologs in rice to take up chlorate and signal the expression of other nitrate transporters, which may affect the efficiency of nitrogen utilization and its uptake.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice is one of the most consumed cereals worldwide and is part of the daily and staple diet of more than half the world's population; it is one of the main crops for the maintenance of food security of various countries. The majority of crops remove significant amounts of N from the soil, and significant applications of nitrogen fertilizers are necessary to maintain the productivity of these crops (O'Brien et al. 2016). Nitrogen is usually the element required the most by the plants and often limits crop yield, and the preferentially absorbed forms are inorganic NO3− and NH4+ (Glass, 2003). NO3− is absorbed by transporters of the NRT (Nitrate Transporter) and NPF (Nitrate Peptide Transporter Family) gene families, which may belong to the group of HATS (High-Affinity transport system) or LATS (Low-Affinity transport system), allowing the uptake of NO3− and H+ ions via symport (2H+/1NO3−) over a wide range of concentration (Glass et al. 1992; Glass 2003).
Characterization of different NRT1 transporters indicated the transport of several molecules besides NO3−, for example, the AtNRT1.1 transporter of Arabidopsis thaliana, besides transporting NO3− can also facilitate auxin transport (Krouk et al. 2010). Thus, Leran et al. (2014) and von Wittgenstein et al. (2014) performed a new classification of the NRT1 family and renamed it NPF family (Nitrate Peptide Transporter Family). In Arabidopsis, the NRT1/NPF family genes encode for LATS transporters that operate at high concentrations of nitrate, usually above 1 mM (Glass 2003). The transporter AtNRT1.1 (AtNPF6.3) has the ability to absorb NO3− in the high-affinity and low-affinity range and is regulated by phosphorylation and dephosphorylation of threonine 101 (T101) (Ho et al. 2009). This dual affinity transporter needs the protein kinase CIPK23 when NO3− is present at low concentrations (Ho et al. 2009; Sun and Zheng 2015). This regulatory mechanism describes how the plants adapt to varying concentrations of nitrogen in the environment (Liu and Tsay 2003; Fredes et al.2019). Phosphorylation of T101 of NRT1.1 is essential for the regulation of the transporter in response to NO3−, and for controlling the growth of lateral roots mediating the flow of auxin (Bouguyon et al. 2015, 2016). The deletion of the NRT1.1 gene gave rise to the Arabidopsis mutant chl1-5, which confers chlorate tolerance and impairs signaling for the AtNRT2.1 gene expression (Ho et al.2009).
In rice, three potential orthologs of the AtNRT1.1 gene were identified, named OsNRT1.1A (OsNPF6.3), OsNRT1.1B (OsNPF6.5) and OsNRT1.1C (OsNPF6.4) (Plett et al. 2010). OsNRT1.1B is associated with uptake of NO3− through the roots and transport to the shoot, demonstrated by osnrt1.1b mutant rice plants that also exhibited decreased expression of OsNIA1 and OsNIA2 but no change in OsNRT2.1 expression (Hu et al.2015). OsNRT2.1 is induced by NO3−, and osnrt2.1 mutant rice plants have decreased uptake of NO3− (Feng et al. 2011). In Arabidopsis, the expression of NRT2.1 is controlled by NRT1.1 and depends on the phosphorylation of T101 (Ho et al. 2009; Bouguyon et al. 2015). Interestingly, OsNRT1.1B shares a common ancestor with NRT1.1 from Arabidopsis (Hu et al. 2015).
It is essential to properly understand the regulation of the expression of NO3− transporters (NRT) to increase the efficiency of NO3− uptake (Plett et al. 2018). The uptake capacity of NO3− promoted by NRT2.1 depends on the NAR2.1 in Arabidopsis and rice (Orsel et al. 2006; Yan et al. 2011). It is difficult to study the individual function of OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C in rice because of a possible countervailing effect by another isoform of OsNRT1.1 (Plett et al. 2010). Therefore, we used chl1-5 mutant Arabidopsis plants which do not have the NRT1.1 gene and thus could express OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C individually.
In the present study, the rice genes OsNRT1.1A, OsNRT1.1As (OsNRT1.1A alternative splicing), OsNRT1.1B and OsNRT1.1C were overexpressed to verify which genes are capable of reversing the Arabidopsis chl1-5 mutant phenotype. Thus, the objectives of this study were to: (i) study the expression of the OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C genes in rice roots and shoots grown with NO3− or NH4+, (ii) evaluate the survival of Arabidopsis chl1-5 mutant plants overexpressing the OsNRT1.1A, OsNRT1.1As, OsNRT1.1B, and OsNRT1.1C genes in the presence of chlorate, and (iii) assess whether the chl1-5 mutant plants with overexpressed OsNRT1.1A, OsNRT1.1As, OsNRT1.1B and OsNRT1.1C had restored the lost nitrate transport and signaling capacity in the chl1-5 mutant. This is the first report showing the capacity of these genes to reverse the Arabidopsis chl1-5 mutant for the NRT1.1 transceptor.
Material and Methods
Expression of OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C Genes in Roots and Shoots of Rice Cultivated with NO3 − and NH4 +
Nipponbare rice cultivar seeds were disinfected in 2% sodium hypochlorite, followed by washes with distilled water, and germinated on gauze in containers containing distilled water. Six days after germination (DAG), four seedlings were transferred to 700 mL pots containing Hoagland's solution (Hoagland and Arnon 1950) modified to its 1∕4 ionic strength (1.5 mM K, 0.25 mM P, 0.5 mM Mg, 1.0 mM Ca, 0.5 mM S, 2.72 µM Cl, 11.57 µM B, 2.25 µM Fe, 0.075 µM Cu, 0.025 µM Mo, 2.72 µM Mn, and 0.2 µM Zn) and containing 2 mM NO3−-N or 2 mM NH4+-N and pH 5.8. After three days the two groups (NO3− and NH4+) of plants received a modified solution of 1∕2 ionic strength, which was changed every three days and with 2 mM NO3−-N or 2 mM NH4+-N. 18 DAG, 2/3 of the pots of each group received a modified solution without N while the rest of the pots continued receiving solution with 2 mM of NO3−-N or 2 mM of NH4+-N. At 21 DAG plants with 2 mM N solution received the solution with the same earlier composition, while half of the plants grown in N-free solution for three days received 2 mM of N (NO3− or NH4+), and the other half received new solution without N (starvation). Sampling occurred on the 21 DAG three hours after the application of the solutions (Online Resource 1). Thus, the treatments were: (i) constant supply with NO3−-N, (ii) resupply with 2.0 mM NO3−-N after 72 h without NO3−, (iii) deficiency of NO3− for 72 h, (iv) constant supply with 2.0 mM of NH4+-N, (v) resupply with 2.0 mM of NH4+-N after 72 h without NH4+, and (vi) deficiency of NH4+ for 72 h.
Extraction of Total RNA
Total RNA was extracted as per the procedure described by Gao et al. (2001) with modifications cited in Sperandio et al. (2011). Samples of each plant were macerated in liquid nitrogen, and approximately 100 mg of macerated tissue was transferred to microtubes containing 500 µL of extraction buffer (200 mM Tris–HCl pH 7.5, 100 mM LiCl, 5 mM EDTA, and 1% SDS). After mixing samples and the extraction buffer, 250 µL of phenol and 250 µL of chloroform were added, mixed on a vortex mixer for one minute and then centrifuged for 10 min at 20000 × g. The aqueous phase was placed in fresh tubes containing 250 µL each of phenol and chloroform. This mixture was again mixed on a vortex mixer for one minute, centrifuged for 10 min at 20000 × g, and the aqueous phase transferred to a fresh tube. The RNA was precipitated with one volume of 6 M LiCl for 16 h at 4 °C.
The samples were centrifuged for 10 min at 20000 × g, the supernatant discarded and the precipitate was resuspended in 1 mL of 3 M LiCl for a second precipitation for 10 min at 20000 × g. The supernatant was discarded and the precipitate resuspended in 250 µL of ultrapure H2O treated with diethylpyrocarbonate (Milli-Q-DEPC). After complete dissolution of the precipitate, 0.1 volume of 3 M sodium acetate (25 µL) and two volumes of absolute ethanol (550 µL) were added. This mixture was maintained in the freezer −80 °C for 2 h and then centrifuged for 15 min at 20,000 × g. The supernatant was discarded and the precipitate was washed with 300 µL of 70% ethanol. After drying the precipitate, the RNA was resuspended in 50 µL of milliQ-DEPC water and stored in a freezer at −80 °C.
Treatment with DNase and Synthesis of cDNA
After quantification of total RNA using a Nanodrop 2000C (Thermo Scientific), 500 ng of total RNA was treated with DNase I using a kit (Thermo Scientific) according to the manufacturer's instructions. The synthesis of cDNA was performed using the GoScript™ Reverse Transcription System kit (Promega) according to the manufacturer's instructions.
Real-Time PCR
PCR reactions were performed in a StepOne Plus system (Applied Biosystems) using cDNA, primers (Online Resource 2) and EvaGreen (Solis Biodyne) according to the manufacturer's specifications. The gene sequences were obtained from the TAIR site (https://www.arabidopsis.org/) for Arabidopsis and TIGR (https://rice.plantbiology.msu.edu/) for rice. The specificity of the primers was verified using the primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). After the PCR reaction, the specificity of the reactions was verified by analyzing the dissociation curve. All primers used showed high specificity for the target gene. The expression was calculated according to Livak and Schmittgen (2001) using the Ct values (cycle threshold) obtained after real-time PCR. The sequences of the primers used for real-time PCR are listed in Online Resource 2.
Construction of Vectors Overexpressing OsNRT1.1A, OsNRT1.1A Splicing, OsNRT1.1B and OsNRT1.1C in Arabidopsis chl1-5
Total RNA from the Nipponbare rice variety was used to synthesize cDNA using the SuperScript™ III Kit (Thermo Fisher Scientific) according to the manufacturer's recommendations. Genes were cloned using the Gateway cloning system (Thermo Scientific) according to the manufacturer's recommendations. The primers for OsNRT1.1A, OsNRT1.1As, OsNRT1.1B, and OsNRT1.1C were synthesized containing the Gateway attB recombination sites (Online Resource 3). Two PCR reactions were performed, where a part of the attB site was placed in the primer containing the annealing site in the target gene, and the remainder of the attB site was placed in a second PCR reaction. PCR reactions were performed using the Phusion kit (Finnzymes) according to the manufacturer's instructions. The OsNRT1.1A gene undergoes alternative splicing, one called OsNRT1.1A complete (or only OsNRT1.1A) containing all exons, while the second part, OsNRT1.1As does not contain the first exon (alternative splicing).
After isolating the genes with the attB sites, they were cloned in the vector pDONR™221 using the Gateway BP Clonase™ II Enzyme mix kit (Thermo Scientific) according to the manufacturer's recommendations to obtain the entry vectors containing the OsNRT1.1A, OsNRT1.1As, OsNRT1.1B and OsNRT1.1C sequences. The reaction product was transferred in Escherichia coli strain DH5α by electroporation and plated on solid LB medium containing the antibiotic kanamycin (50 µg mL−1). The vector was extracted using the PureYield™ Plasmid Miniprep system (Promega) according to the manufacturer's recommendations. The expression vectors were obtained by carrying out a triple LR reaction containing the pK7m34GW target vector and the entry vectors containing the 35S promoter (pENTR-35S), the genes of interest (OsNRT1.1A, OsNRT1.1As, OsNRT1.1B, and OsNRT1.1C) and a HA tag (pENTR-3xHA), using the Gateway™ LR Clonase™ Enzyme mix Kit (ThermoScientific) according to the manufacturer's recommendations. The reaction product was electroporated into E. coli strain DH5α, plated on solid LB medium containing the antibiotics spectinomycin/streptomycin (Sm/Sp) (50 µg mL−1 and 20 µg mL−1, respectively). Vectors were isolated with the PureYield™ Plasmid Miniprep System Kit (Promega) and were used to transform Agrobacterium tumefaciens strain LBA4404 by heat shock for further transformation of Arabidopsis. A schematic model of expression vector assembly is presented in Online Resource 4.
Transformation of Arabidopsis chl1-5 Mediated by Agrobacterium tumefaciens
Agrobacterium tumefaciens strain LBA4404 was used to transform the Arabidopsis chl1-5 mutant with the expression vectors containing the genes OsNRT1.1A, OsNRT1.1As, OsNRT1.1B, and OsNRT1.1C. After the transformation of Agrobacterium by heat shock and confirmation by colony PCR, pre-inoculum was grown in 1 mL of YEB medium with Sm/Sp antibiotics (50 µg mL−1 and 20 µg mL−1, respectively). Subsequently, the volume was raised to 10 mL with YEB medium and stirred overnight to reach an OD (optical density) close to 2. Forty milliliters of a solution containing 10% sucrose and 0.05% silwet was added to the 10 mL culture and floral dip was performed by dipping the inflorescence of Arabidopsis chl1-5 mutant plants in the pre-flowering phenological phase (Clough and Bent 1998; Mara et al. 2010). Eight chl1-5 plants, grown under the same previously reported conditions were used for each construct.
Selection of Homozygous Arabidopsis Plants
Seeds of Arabidopsis chl1-5 submitted to the floral dip transformation process were transferred to Petri dishes of 12 mm diameter containing MS culture medium (Murashige and Skoog, 1962) at half of the ionic strength with 1% sucrose, 0.5 g L−1 of MES, 50 mg L−1 of kanamycin, solidified with 0.7% bacto-agar, pH 5.7. The dishes were closed and sealed with microporous tape, and placed in the refrigerator at 4 °C for three days. The dishes were transferred to a BOD at 25 °C with approximately 80 µmol photons m−2 s−1 and 14 h photoperiod. The plants with the kanamycin-resistance phenotype were selected for propagation on the substrate to obtain homozygous inbred lineages (T3) (Clough and Bent 1998; Zhang et al.2006; Mara et al. 2010). For the subsequent analytical steps, two homozygous inbred lineages (T3) were chosen for each transformation: OsNRT1.1A lineages 1.2 and 3.1, OsNRT1.1As lineages 2.2 and 1.1, OsNRT1.1B lineages 1.8 and 2.2, and OsNRT1.1C lineages 1.1 and 2.1. The confirmation of the insertion and expression levels of Arabidopsis chl1-5 overexpressing OsNRT1.1A, OsNRT1.1As, OsNRT1.1B and OsNRT1.1C genes was done by Real-Time PCR as described above with the primers listed in Online Resource five. The expression of NRT1.1 in Arabidopsis WT was used as control.
Analysis of Gene Expression, Nitrate and Amino Acid Content in Arabidopsis chl1-5 Containing the Isoforms OsNRT1.1A, OsNRT1.1As, OsNRT1.1B, and OsNRT1.1C
Seeds of WT Arabidopsis Columbia ecotype (Col 0), chl1-5 mutant and homozygous plants transformed with the OsNRT1.1A, OsNRT1.1As, OsNRT1.1B, and OsNRT1.1C genes were propagated on substrate mixed with vermiculite (2:1) in 150 mL pots and placed in plastic trays covered with PVC film. The trays were placed in a plant growth chamber with an average luminous flux of 140 µmol photons m−2 s−1, an average temperature of 23 °C and 65% humidity. Twenty-three days after planting, eight applications of a 15 mL solution containing 12 mM sodium chlorate, 3.75 mM ammonium sulfate and 8.25 mM potassium nitrate per pot (Wang et al. 1988) were initiated. On the 39th day after planting, the plants were photographed whit a camera Canon model SX400, after they harvested to obtain comparative parameters of the effect of chlorate on their metabolism. The control treatment consisted of seeds of all transformed, WT and chl1-5 mutant treated with the same nutrient concentrations without chlorate.
The plants cultivated without chlorate were used for the analysis of gene expression by real-time PCR. The extraction of total RNA, treatment with DNase, synthesis of cDNA and real-time PCR were performed as previously described. The expression of the NRT2.1 and NAR2.1 (Santos et al.2012) were assessed. The primers used are listed in Online Resource 6. The content of NO3−-N and amino acid were performed according to the methods of Miranda et al. (2001) and Yemm and Cocking (1955), respectively.
Alignment of Rice OsNRT1.1 Isoforms and Arabidopsis NRT1.1
Multiple sequence alignment of the amino acids sequences between rice OsNRT1.1 isoforms (OsNRT1.1A, OsNRT1.1As, OsNRT1.1B and OsNRT1.1C) and Arabidopsis NRT1.1 was performed using Clustalw2 with default parameters (https://www.genome.jp/tools-bin/clustalw).
Statistical Analysis
The experiments were conducted using a completely randomized design. The results were subjected to analysis of variance using the program Sisvar (Ferreira 2014). Comparisons of the means were made by the LSD test (p < 0.05).
Results
Expression of OsNRT1.1A, OsNRT1.1B and OsNRT1.1C Genes in Roots and Shoots of Rice
The expression profile of the isoforms OsNRT1.1A, OsNRT1.1B and OsNRT1.1C varied with treatments in NO3− and NH4+ (Fig. 1). The isoform OsNRT1.1A displayed higher expression in the roots of plants grown with NO3− and NH4+, in particular with a constant supply of 2 mM N (Fig. 1). The expression of OsNRT1.1B was higher in the roots with a constant supply of 2 mM of NO3−-N, while it was downregulated in plants grown with NH4+ (Fig. 1a, b). Resupply of 2 mM of NO3−-N did not induce the expression of OsNRT1.1A and OsNRT1.1B in roots (Fig. 1a, b). Expression of OsNRT1.1A and OsNRT1.1B are lower in shoots, meanwhile OsNRT1.1C displayed low expression in roots and shoots in plants grown with NO3− or NH4+ (Fig. 1). Thus, the expression of OsNRT1.1A in roots occurred regardless of the form of N applied, while OsNRT1.1B was expressed mainly in the roots of plants grown in a constant supply of NO3− and downregulated by NH4+ provision. The gene expression analysis suggests the higher importance of OsNRT1.1A and OsNRT1.1B in rice compared to OsNRT1.1C.
Expression of OsNRT1.1A, OsNRT1.1B and OsNRT1.1C in the roots a and b and shoots c and d of rice cultivated with NO3− a and c and NH4+ b and d. The treatments used were constant supply with 2 mM of NO3−-N or NH4+-N, resupply with 2 mM of NO3−-N or NH4+-N after 72 h of N-deprivation and NO3− and NH4+ deficit for 72 h. *Significantly different from OsNRT1.1A with constant NO3− treatment in the roots according to a Least Significant Difference test (p < 0.05)
Effect of Overexpression of OsNRT1.1A, OsNRT1.1As, OsNRT1.1B, and OsNRT1.1C in Arabidopsis chl1-5
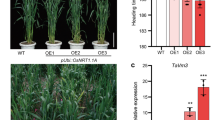
The chl1-5 lineages overexpressing OsNRT1.1A, OsNRT1.1As, OsNRT1.1B, and OsNRT1.1C presented different expression levels (Fig. 2). The lineages OsNRT1.1A L3.1, OsNRT1.1B L2.2 and OsNRT1.1C L2.1 presented higher expression levels compared to OsNRT1.1As L2.2, OsNRT1.1B L1.8 and OsNRT1.1C L1.1 (Fig. 2).
Expression analysis of OsNRT1.1 in lineages of Arabidopsis chl1-5 overexpressing the rice genes OsNRT1.1A splicing (OsNRT1.1As), OsNRT1.1A, OsNRT1.1B and OsNRT1.1C. Two lineages were used for each mutant overexpressing OsNRT1.1 family. The WT expression of NRT1.1 was assigned value 1. *Significantly different from WT according to a Least Significant Difference test (p < 0.05)
As expected, the application of chlorate to WT plants resulted in a strong reduction in growth, while no toxic effect of chlorate was observed in the chl1-5 mutant (Fig. 3). On comparing the lineages overexpressing OsNRT1.1A and OsNRT1.1As, a variation in growth compared to chl1-5 was observed (Fig. 3). The chlorate-sensitive phenotype by the OsNRT1.1B transporter overexpression for the OsNRT1.1B L1.8 and L2.2 was more characteristic and approached the WT plant (Fig. 4), indicating that OsNRT1.1B may be the predominant isoform in low-affinity nitrate absorption in rice, as its expression was able to revert the mutant chl1-5 phenotype as that of WT. The overexpression of OsNRT1.1C L1.1 and L2.1 also showed evidence of damage due to the chlorate treatment (Fig. 4).
When 12 mM of chlorate was applied there was no significant reduction of fresh mass in the mutant chl1-5 (Fig. 5). However, in the case of the WT plant and all the transformed lineages, a significant reduction of fresh weight was observed with chlorate application (Fig. 5).
Plant fresh weight of mutant Arabidopsis chl1-5, WT and lineages of Arabidopsis chl1-5 with overexpressed rice genes OsNRT1.1A splicing (OsNRT1.1As), OsNRT1.1A, OsNRT1.1B and OsNRT1.1C, with and without chlorate. Two lineages were used for each mutant overexpressing OsNRT1.1. *Significantly different from control (without chlorate) according to a Least Significant Difference test (p < 0.05)
The importance of the chlorate test and the function of NRT1.1 were characterized and the effect of comparison of the effect of chlorate on plants with and without chlorate treatment, are shown in Fig. 6. Arabidopsis plants overexpressing OsNRT1.1B showed growth reduction similar to that of WT plants when treated with chlorate (90% fresh mass reduction), while plants overexpressing OsNRT1.1A (complete and splicing) and OsNRT1.1C plants displayed more than 40% fresh mass reduction when treated with chlorate (Fig. 6).
Effect of chlorate on the reduction of fresh weight (%) in Arabidopsis mutant chl1-5, WT and lineages of Arabidopsis chl1-5 overexpressing the rice genes OsNRT1.1A splicing (OsNRT1.1As), OsNRT1.1A, OsNRT1.1B and OsNRT1.1C. Two lineages were used for each mutant overexpressing OsNRT1.1. *Different letter represents statistically significantly different according to a Least Significant Difference test (p < 0.05)
The overexpression of OsNRT1.1A and OsNRT1.1B increased the expression of transporter NRT2.1, although it was not affected by the overexpression of OsNRT1.1C when compared with chl1-5 plants (Fig. 7a). Curiously, the overexpression of the alternative splicing product of OsNRT1.1A (OsNRT1.1As) did not increase the expression of NRT2.1 when compared with chl1-5 plants, and unlike plants overexpressing the complete sequence of OsNRT1.1A (Fig. 7a). The lineage OsNRT1.1A L3.1 displayed higher expression of OsNRT1.1A (Fig. 2) and NRT2.1 (Fig. 7a), suggesting the differences of NRT2.1 expression was partially caused by different levels of OsNRT1.1A expression in Arabidopsis chl1-5.
Expression of the NO3− transporter NRT2.1 a and NAR2.1 b in Arabidopsis mutant chl1-5, WT and lineages of Arabidopsis chl1-5 overexpressing the rice genes OsNRT1.1A splicing (OsNRT1.1As), OsNRT1.1A, OsNRT1.1B and OsNRT1.1C. Two lineages were used for each mutant overexpressing OsNRT1.1. The expression in the mutant chl1-5 was assigned value 1. *Significantly different from chl1-5 according to a least significant difference test (p < 0.05)
WT Arabidopsis plants displayed a higher expression of NAR2.1 compared to chl1-5, as well as OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C lineages, while OsNRT1.1As overexpression did not affect the expression of NAR2.1 (Fig. 7b). WT Arabidopsis induced NAR2.1 by twofold compared to chl1-5, meanwhile overexpression of OsNRT1.1A L1.2 and L3.1 induced NAR2.1 by 77 and 26 times compared to chl1-5, respectively. Overexpression of OsNRT1.1B L1.8 and L2.2 induced NAR2.1 by 53 and 14 times compared to chl1-5, respectively (Fig. 7b). Analysis of the amino acid sequences of OsNRT1.1A, OsNRT1.1As, OsNRT1.1B, OsNRT1.1C, and Arabidopsis NRT1.1 shows that OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C have a threonine (T) residue in a position similar to the T101 on the Arabidopsis NRT1.1 transporter (positions T107, T104 and T118 in OsNRT1.1A, OsNRT1.1B and OsNRT1.1C, respectively), while OsNRT1.1As does not present the threonine residue due to loss of the first exon with alternative splicing (Fig. 8). Altogether, the results indicate the first exon in OsNRT1.1A containing the T107 is essential to NRT2.1 and NAR2.1 induction.
Alignment of partial amino acid sequences of OsNRT1.1A, OsNRT1.1As (alternative splicing of OsNRT1.1A without the first exon), OsNRT1.1B, OsNRT1.1C and NRT1.1 (Arabidopsis) in the region where residue T101 is found in Arabidopsis NRT1.1. Threonine residues in a similar position to T101 of Arabidopsis NRT1.1 characterized by Ho et al. (2009) are in red and underlined. The position of threonine residues in rice are T107, T104 and T118 in OsNRT1.1A, OsNRT1.1B and OsNRT1.1C, respectively (Color figure online)
WT Arabidopsis presented increased NO3−-N and amino acid content compared to chl1-5 (Fig. 9a, b). According to WT plants, Arabidopsis chl1-5 overexpressing OsNRT1.1A and OsNRT1.1B increased NO3−-N and amino acid content compared to chl1-5 as well, however, chl1-5 overexpressing OsNRT1.1As and OsNRT1.1C did not increase NO3−-N and amino acid content (Fig. 9a, b).
Plant NO3−-N content a and amino acid content b of mutant Arabidopsis chl1-5, WT and lineages of Arabidopsis chl1-5 with overexpressed rice genes OsNRT1.1A splicing (OsNRT1.1As), OsNRT1.1A, OsNRT1.1B and OsNRT1.1C. Two lineages were used for each mutant overexpressing OsNRT1.1. *Significantly different from chl1-5 according to a Least Significant Difference test (p < 0.05)
Discussion
Plants need to regulate the expression of NO3− transporters to grow in environments with varying levels of N. The expression of transporters essential for the absorption of NO3− in Arabidopsis is regulated by the NRT1.1 transporter (Ho et al. 2009; Bouguyon et al. 2015, 2016). Rice has three NRT1.1 orthologs, namely OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C (Plett et al. 2010). The importance of OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C in N uptake and expression of NRT2.1 and NAR2.1 promoted by NO3− was assessed by evaluating their expression in rice plants and subsequently by overexpressing OsNRT1.1A, OsNRT1.1As, OsNRT1.1B, and OsNRT1.1C in Arabidopsis chl1-5.
The expression results indicate a complex regulation of the OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C genes in rice (Fig. 1). While OsNRT1.1A expression in roots and shoots displayed less variation, OsNRT1.1B seems to be expressed mainly in the roots and with a constant supply of NO3−, and OsNRT1.1C exhibited lower expression between OsNRT1.1 orthologs. These results indicate a greater involvement of OsNRT1.1A and OsNRT1.1B orthologs in rice, and the participation of OsNRT1.1A in the movement of NO3− in the shoots. NH4+ and its assimilation products, especially glutamine, are powerful inhibitors of several NO3− transporters (Glass et al. 2003). Here, NH4+ was used as a source of N in the nutrient solution to verify the expression of OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C in repressive conditions. The expression of OsNRT1.1A presented little inhibitory effect on the roots of plants grown in the presence of NH4+, indicating low sensitivity to variations in the N status in the plant. On the other hand, the expression of OsNRT1.1B was only induced by NO3− in the roots of rice and repressed by NH4+, suggesting that it can contribute to the uptake of NO3− in the roots and was less significant in the flow of NO3− in the shoots. A higher expression of OsNRT1.1B is related to higher NO3− uptake and transport to the shoots in rice (Hu et al. 2015), while, high expression of OsNRT1.1A in the shoots (Fig. 1) suggests that this transporter can also participate in the internal flow of NO3−.
Rice plants overexpressing OsNRT1.1A reveal its regulation by NH4+, suggesting that this transporter is important in flooded areas for rice production (Wang et al. 2018). The results reported by Wang et al. (2018) are similar to the results of our study (Fig. 1), where NH4+ is responsible for inducing the expression of OsNRT1.1A. Wang et al. (2018) also demonstrated that under field conditions the mutant osnrt1.1a has decreased plant size, panicle size, and weight of 1000 seeds, on the other hand, the overexpression of OsNRT1.1A in Arabidopsis resulted in an increase of the size of the plant and shorter flowering with a reduction of 6 to 15 days (Wang et al. 2018).
The Arabidopsis chl1-5 plants overexpressing OsNRT1.1A, OsNRT1.1B and OsNRT1.1C and OsNRT1.1As were assessed by applying chlorate. Fig. 3, 4 show the toxicity promoted by the chlorate (molecule analogous to nitrate). After being absorbed, chlorate is reduced within the cells to chlorite by nitrate reductase, which is toxic to the plant, resulting in the reduction of fresh mass for all transformed isoforms as well as the WT plant. The decrease of fresh weights with the overexpression of OsNRT1.1A, OsNRT1.1B, OsNRT1.1C, and OsNRT1.1As indicates the individually isoform function in the transport of nitrate, however, overexpression of OsNRT1.1B led to a greater reduction of fresh weight between the lineages and similar to that exhibited by WT after the application of chlorate (Fig. 6), which could be confirmed visually (Fig. 4). The fresh weight reduction of Arabidopsis overexpressing OsNRT1.1A and OsNRT1.1As treated with chlorate clearly indicates that the first exon is not necessary for the absorption function of NO3− (Fig. 3). On the other hand, even a low expression of OsNRT1.1C in rice (Fig. 1) may enable the transport of NO3− (Fig. 4).
The NRT1.1 transporter is essential for the regulation of NRT2.1 expression in Arabidopsis (Ho et al. 2009; Bouguyon et al. 2015, 2016). The analysis of expression in Arabidopsis chl1-5 plants overexpressing OsNRT1.1A, OsNRT1.1B, OsNRT1.1C, and OsNRT1.1As revealed that only OsNRT1.1A and OsNRT1.1B could increase the expression of NRT2.1 compared to the Arabidopsis chl1-5 (Fig. 7a). The nitrate-induced high-affinity transport system requires NAR2.1 for its functionality (Orsel et al. 2006; Yan et al. 2011), although the NAR2.1 does not transport NO3− (Orsel et al. 2006). Arabidopsis chl1-5 lineages overexpressing OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C exhibited high NAR2.1 induction compared to chl1-5, while Arabidopsis WT displayed low induction (Fig. 7b).
The results obtained show the ability of OsNRT1.1A and OsNRT1.1B to induce the expression of NRT2.1 and NAR2.1 in Arabidopsis chl1-5 and may also have signaling functions in rice, as the expression of OsNRT2.1 and OsNAR2.1 is induced by NO3− in a manner similar to that in Arabidopsis (Araki and Hasegawa 2006). The T101 residue in the NRT1.1 transporter of Arabidopsis is essential for signaling activity (Ho et al. 2009; Bouguyon et al. 2015, 2016). The overexpression of OsNRT1.1As in Arabidopsis chl1-5 was not able to induce the expression NRT2.1 and NAR2.1 as observed in plants overexpressing OsNRT1.1A and OsNRT1.1B which possess the threonine residue in a position similar to the T101 of Arabidopsis NRT1.1 (Fig. 8). The gene expression analysis indicates that the first exon present in OsNRT1.1A, OsNRT1.1B, and OsNRT1.1C containing the T107, T104, and T118 residue, respectively (Fig. 8), may present an essential function for the signaling promoted by NO3− in rice. However, the first exon in OsNRT1.1A does not seem to be essential for the transport capacity of NO3−, given that Arabidopsis chl1-5 overexpressing OsNRT1.1As displayed effects of toxicity in the presence of chlorate (Fig. 6). The importance of OsNRT1.1A and OsNRT1.1B to induce the expression of NRT2.1 and NAR2.1 was also supported by the increased NO3−-N and amino acid content in chl1-5 lineages overexpressing OsNRT1.1A and OsNRT1.1B compared to chl1-5 plants, similar to those presented in WT plants (Fig. 9). The protein CPSF30-L regulates NRT1.1 expression in Arabidopsis and the cpsf30 mutant decreases NRT1.1 expression and NO3− accumulation compared to WT (Li et al. 2017). Compared to chl1-5 mutants, chl1-5 overexpressing OsNRT1.1As and OsNRT1.1C did not increase NRT2.1 expression, NO3− content and amino acid, indicating the importance of OsNRT1.1A and OsNRT1.1B in NO3− signaling.
In rice, the OsNRT1.1A and OsNRT1.1B transporters are highly expressed and can signal for the expression of OsNAR2.1, since OsNRT2.1 depends also on OsNAR2.1 to transport NO3− (Yan et al. 2011). The failure of OsNRT1.1A splicing overexpression in inducing the expression of NAR2.1 and NRT2.1 (Fig. 7) clearly indicates the importance of the first exon for signaling mediated by OsNRT1.1A. The osnrt1.1b mutant rice plant did not present a lower expression of OsNRT2.1 (Hu et al. 2015), however, the results obtained in this study suggest that other homologs to Arabidopsis NRT1.1, as OsNRT1.1A can control the expression of NO3− transporters. In rice, OsNRT1.1B promotes the ubiquitination and degradation of SPX4, by activating NO3− and phosphorus (P) response genes (Hu et al. 2019). The results showed differences in intensity of chlorate sensitivity and the ability to induce NRT2.1/NAR2.1 expression in Arabidopsis chl1-5 plants overexpressing OsNRT1.1A and OsNRT1.1B, indicating that they may have distinct functions.
Altogether, using chl1-5 mutant overexpression system we were able to study the individual function of OsNRT1.1A, OsNRT1.1As, OsNRT1.1B and OsNRT1.1C, avoiding potential redundancy of functions among the OsNRT1.1 in rice. In this study, OsNRT1.1A and OsNRT1.1B genes were overexpressed in Arabidopsis chl1-5 mutant and their NO3− transport function was revealed, as well as the NRT2.1/NAR2.1 induction in the Arabidopsis chl1-5 overexpression system. The overexpression of OsNRT1.1A transporter showed a strong effect on the expression of NRT2.1/NAR2.1 promoted by NO3−, while the overexpression of OsNRT1.1B displayed a strong sensitivity towards chlorate, showing its likely role in the absorption of NO3. Although OsNRT1.1A has higher protein sequence identity to Arabidopsis NRT1.1 (Wang et al. 2018), the present study indicates OsNRT1.1A and OsNRT1.1B contribute to NO3− uptake and NRT2.1 and NAR2.1 expression. Additionally, OsNRT1.1A and OsNRT1.1B display different expression among rice root and shoot as well as N source (NO3− or NH4+ ), indicating different functions of NRT1.1 family in rice. Rice is NH4+ tolerant and responsive to NO3− and NH4+ provision (Kronzucker et al. 2000). Although chl1-5 lineages overexpressing OsNRT1.1C presented chlorate sensitivity, those lineages did not induce NRT2.1 expression compared to chl1-5 mutants. Further, the low expression of OsNRT1.1C in rice indicates the importance of the expression of OsNRT1.1A and OsNRT1.1B in rice. The T101 residue in NRT1.1 is essential to mediate NO3− signaling in Arabidopsis (Ho et al. 2009). The analysis of alternative splicing of OsNRT1.1A revealed the importance of the first exon in signaling and the likely importance of T107 of OsNRT1.1A in signaling promoted by NO3−.
References
Araki R, Hasegawa H (2006) Expression of rice (Oryza sativa L.) genes involved in high- affinity nitrate transport during the period of nitrate induction. Breed Sci 56:295–302. https://doi.org/10.1270/jsbbs.56.295
Bouguyon E, Brun F, Meynard D, Kubeš M, Pervent M, Leran S, Lacombe B, Krouk G, Guiderdoni E, Zažímalová E, Hoyerová K, Nacry P, Gojon A (2015) A Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nature Plants 1:1–8. https://doi.org/10.1038/nplants.2015.15
Bouguyon E, Perrine-Walker F, Pervent M, Rochette J, Cuesta C, Benkova E, Martinière A, Bach L, Krouk G, Gojon A, Nacry P (2016) Nitrate controls root development through post transcriptional regulation of the NRT1.1/NPF6.3transporter/sensor. Plant Physiol 172:1237–1248. https://doi.org/10.1104/pp.16.01047
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. https://doi.org/10.1046/j.1365-313x.1998.00343.x
Feng H, Yan M, Fan X, Li B, Shen Q, Miller AJ, Xu G (2011) Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J Exp Bot 62:2319–2332. https://doi.org/10.1093/jxb/erq403
Ferreira DF (2014) Sisvar: a guide for its bootstrap procedures in multiple comparisons. Ciência e Agrotecnologia 38:109–112. https://doi.org/10.1590/S1413-70542014000200001
Fredes I, Moreno S, Díaz FP, Gutiérrez RA (2019) Nitrate signaling and the control of Arabidopsis growth and development. Curr Opin Plant Biol 47:112–118. https://doi.org/10.1016/j.pbi.2018.10.004
Gao J, Liu J, Li B, Li Z (2001) Isolation and purification of functional total RNA from blue-grained whet endosperm tissues contaning high levels of starches and flavonoids. Plant Mol Biol Rep 19:185–186. https://doi.org/10.1007/BF02772163
Glass ADM (2003) Nitrogen use efficiency of crop plants: physiological constraints upon nitrogen absorption. Crit Rev Plant Sci 22:453–470. https://doi.org/10.1080/07352680390243512
Glass ADM, Shaff JE, Kochian LV (1992) Studies of nitrate uptake in barley IV Electrophysiology. Plant Cell 99:456–463. https://doi.org/10.1104/pp.99.2.456
Ho CH, Lin SH, Hu HC, Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell 138:1184–1194. https://doi.org/10.1016/j.cell.2009.07.004
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Exp Stn Bull 347:1–32
Hu B, Wang W, Ou S, Tang J, Li H, Che R, Zhang Z, Chai X, Wang H, Wang Y, Liang C, Liu L, Piao Z, Deng Q, Deng K, Xu C, Liang Y, Zhang L, Li L, Chu C (2015) Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet 47:834–838. https://doi.org/10.1038/ng.3337
Hu B, Jiang Z, Wang W, Qiu Y, Zhang Z, Liu Y, Li A, Gao X, Liu L, Qian Y, Huang X, Yu F, Kang S, Wang Y, Xie J, Cao S, Zhang L, Wang Y, Xie Q, Kopriva S, Chu C (2019) Nitrate–NRT1.1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nature Plants 5:401–413. https://doi.org/10.1038/s41477-019-0384-1
Kronzucker HJ, Glass ADM, Siddiqi MY, Kirk GJD (2000) Comparative kinetic analysis of ammonium and nitrate acquisition by tropical lowland rice: implications for rice cultivation and yield potential. New Phytol 145:471–476. https://doi.org/10.1046/j.1469-8137.2000.00606.x
Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, Zazimalova E, Benkova E, Nacry P, Gojon A (2010) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18:927–937. https://doi.org/10.1016/j.devcel.2010.05.008
Leran S, Varala K, Boyer J-C, Chiurazzi M, Crawford N, Daniel-Vedele F, David L, Dickstein R, Fernandez E, Forde B, Gassmann W, Geiger D, Gojon A, Gong J-M, Halkier BA, Harris JM, Hedrich R, Limami AM, Rentsch D, Seo M, Tsay Y-F, Zhang M, Coruzzi G, Lacombe B (2014) A unified nomenclature of nitrate transporter 1/peptide transporter family members in plants. Trends Plant Sci 19:5–9. https://doi.org/10.1016/j.tplants.2013.08.008
Li Z, Wang R, Gao Y, Wang C, Zhao L, Xu N, Chen K-E, Qi S, Zhang M, Tsay Y-F, Crawford NM, Wang Y (2017) The Arabidopsis CPSF30-L gene plays an essential role in nitrate signaling and regulates the nitrate transceptor gene NRT1.1. New Phytol 216:1205–1222. https://doi.org/10.1111/nph.14743
Liu KH, Tsay Y-F (2003) Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. The EMBO Journal 22:1005–1013. https://doi.org/10.1093/emboj/cdg118
Livak KJ, Schmittgen TD (2001) Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCт Method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Mara C, Grigorova B, Liu Z (2010) Floral-dip Transformation of Arabidopsis thaliana to Examine pTSO2:β-glucuronidase Reporter Gene Expression. J Vis Exp 40:e1952. https://doi.org/10.3791/1952
Miranda KM, Espey MG, Wink DA (2001) A rapid simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5:62–71. https://doi.org/10.1006/niox.2000.0319
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
O’Brien JA, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez RA (2016) Nitrate transport, sensing and responses in plants. Mol Plant 9:837–856. https://doi.org/10.1016/j.molp.2016.05
Orsel M, Chopin F, Leleu O, Smith SJ, Krapp A, Daniel-Vedele F, Miller AJ (2006) Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiol Protein-Protein Interact Plant Physiol 142:1304–1317. https://doi.org/10.1104/pp.106.085209
Plett D, Toubia J, Garnett T, Tester M, Kaiser BN, Baumann U (2010) Dichotomy in the NRT gene families of dicots and grass species. PLoS ONE 5:e15289. https://doi.org/10.1371/journal.pone.0015289
Plett DC, Holtham LR, Okamoto M, Garnett TP (2018) Nitrate uptake and its regulation in relation to improving nitrogen use efficiency in cereals. Semin Cell Dev Biol 74:97–104. https://doi.org/10.1016/j.semcdb.2017.08.027
Santos LA, Souza SR, Fernandes MS (2012) Osdof25 expression alters carbon and nitrogen metabolism in Arabidopsis under high N-supply. Plant Biotechnol rep 6:327–337. https://doi.org/10.1007/s11816-012-0227-2
Sperandio MVL, Santos LA, Bucher CA, Fernandes MS, Souza SR (2011) Isoforms of plasma membrane H+-ATPase in rice root and shoot are differentially induced by starvation and resupply of NO3− or NH4+. Plant Sci 180:251–258. https://doi.org/10.1016/j.plantsci.2010.08.018
Sun J, Zheng N (2015) Molecular mechanism underlying the plant NRT1.1 dual-affinity nitrate transporter. Front Physiol. 6:386. https://doi.org/10.3389/fphys.2015.00386
von Wittgenstein NJ, Le CH, Hawkins BJ, Ehlting J (2014) Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC Evol Biol 14:11. https://doi.org/10.1186/1471-2148-14-11
Wang X, Feldmann KA, Scholl RL (1988) A chlorate-hypersensitive, high nitrate/chlorate uptake mutant of Arabidopsis thaliana. Physiol Plant 73:305–310. https://doi.org/10.1111/j.1399-3054.1988.tb00602.x
Wang W, Hu B, Yuan D, Liu Y, Che R, Hu Y, Ou S, Liu Y, Zhang Z, Wang H, Li H, Jiang Z, Zhang Z, Gao X, Qiu Y, Meng X, Liu Y, Bai Y, Liang Y, Wang Y, Zhang L, Li L, Sodmergen JH, Li J, Chu C (2018) Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. Plant Cell 30:638–651. https://doi.org/10.1105/tpc.17.00809
Yan M, Fan X, Feng H, Miller AJ, Shen Q, Xu G (2011) Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell & Enviroment 34:1360–1372. https://doi.org/10.1111/j.1365-3040.2011.02335.x
Yemm EW, Cocking EC (1955) The determination of amino-acid with ninhydrin. Anal Biochem 80:209–213. https://doi.org/10.1039/AN9558000209
Zhang X, Henriques R, Lin S-S, Niu Q-W, Chua N-H (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1:641–646. https://doi.org/10.1038/nprot.2006.97
Acknowledgments
We would like to thank the National council for scientific and technological development (CNPq), Brazilian Federal Agency for the support and evaluation of graduate education (CAPES) and foundation Carlos Chagas Filho research support of the state of Rio de Janeiro (FAPERJ) for financial support. We would like to thank editage (www.editage.com) for english language editing.
Author information
Authors and Affiliations
Contributions
VJS: conceptualization, plant transformation, performing the experiments, experiment analysis, data analysis, and writing the manuscript. SMVL: conceptualization, gene expression analysis, vectors construction, data analysis, and writing the manuscript. FMS: conceptualization, data analysis, and writing the manuscript. SLA: conceptualization, supervision, project administration, vectors construction, and writing the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vera, J.S., Sperandio, M.V.L., Fernandes, M.S. et al. Overexpression of Rice Genes OsNRT1.1A and OsNRT1.1B Restores Chlorate Uptake and NRT2.1/NAR2.1 Expression in Arabidopsis thaliana chl1-5 Mutant. J Plant Growth Regul 40, 1701–1713 (2021). https://doi.org/10.1007/s00344-020-10219-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-020-10219-9