Abstract
Flowers of the complex orchid hybrid Burrageara ‘Stefan Isler Lava Flow’ had been shown previously to react sensitively to ethylene. Via Agrobacterium tumefaciens, the mutant ethylene receptor ETHYLENE RESPONSE 1 (etr1-1) from Arabidopsis thaliana under the control of the flower-specific promoter FLOWER BINDING PROTEIN 1 (fbp1) from Petunia hybrida was transferred to Burrageara. One single-copy event was analyzed in this study aiming to investigate the expression of the fbp1::etr1-1 transgene in different plant and flower organs by quantitative RT-PCR and the reaction of flowers and inflorescences to ethylene. It was shown that the heterologous promoter led to high expression levels in the perianth of the orchid flowers compared to low levels in leaves and roots. The expression shift to the first whorl (sepals) described here corresponds to extended expression of endogenous B class MADS box homeotic genes in orchids in general. The transgenic plants grew and developed similar to the wild-type plants, except for slightly faster rooting in vitro and smaller flowers. Flower longevity was improved by 7 days in 10 ppm ethylene. Moreover, bud drop starting at day 5 of incubation of inflorescences in 10 ppm ethylene in the wild-type was efficiently prevented for at least 19 days in the fbp1::etr1-1 transgenic plants. The function of the tissue-specific promoter fbp1 and the mutant receptor etr1-1 was shown for the first time in a monocotyledonous plant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The gaseous plant hormone ethylene controls a wide range of morphological and developmental processes in plants including senescence and abscission of organs, fruit ripening, flower initiation, and germination of seeds (reviewed by Abeles and others 1992). Numerous studies have reported ethylene-induced floral bud drop and floral senescence in different orchids grown for cut flowers or as potted plants (Hew and Clifford 1993; Rattanawisalanon and others 2003; Huang and Paull 2009; Sun and others 2009; Raffeiner and others 2009b).
Ethylene perception and signaling are well resolved mainly based on investigations of mutants of Arabidopsis thaliana. Five ethylene receptors have been grouped into two subfamilies, the first one containing ETHYLENE RESPONSE 1 (ETR1) and ETHYLENE RESPONSE SENSOR 1 (ERS1), and the second with ETR2, ERS2, and ETHYLENE-INSENSITIVE 4 (EIN4) (Guo and Ecker 2004). The differences between both groups are that the subfamily two receptors have a lack of catalytic activity of the histidine kinase and an additional predicted transmembrane domain (Shakeel and others 2013). The members of both subfamilies work as negative regulators of the pathway, actively repressing the ethylene response in the absence of the hormone. All receptors function as homodimers but form higher order complexes in the ER membrane by interacting with different other receptors (Merchante and others 2013). Thereby, plants can probably modulate their reactions to a wide range of ethylene concentrations in a tissue-specific way (Liu and Wen 2012). ETR1 was the first identified receptor for ethylene and its mutants are characterized by ethylene insensitivity (Chang and others 1993). One mutant gene etr1-1 with a substitution of the amino acid cystein in position 65 by tyrosine confers dominant ethylene insensitivity (Chang and others 1993). ETR1 interacts with and activates CTR1, a negative regulator of the ethylene response, whereas the mutated receptor etr1-1 upon exposure to ethylene does not change the conformation of CTR1 resulting in a dominantly suppressed ethylene response by repressing activation of the downstream factors EIN2 and EIN3 (Gamble and others 2002; Hall and others 2007).
In transgenic approaches, the A. thaliana etr1-1 gene was transferred to several ornamental plant species and was shown to effectively confer dominant ethylene insensitivity in the heterologous background, such as Petunia (Wilkinson and others 1997), Dianthus caryophyllus (Bovy and others 1999), Campanula carpatica (Sriskandarajah and others 2007), or Kalanchoë blossfeldiana (Sanikhani and others 2008). To avoid unintended effects of the inhibited ethylene response in organs other than the flowers (Wilkinson and others 1997), the flower-specific promoter fbp1 from Petunia hybrida was used in several studies involving Dianthus, Campanula, and Kalanchoë. The FLORAL-BINDING PROTEIN 1 (fbp1) gene was found to be exclusively expressed in stamen and petals of petunia flowers (Angenent and others 1992; Immink and others 2003) and encodes a class B MADS box transcription factor (Angenent and others 1993). Following the subdivision into DEFICIENS (DEF)/APETALA3 (AP3)-like and GLOBOSA (GLO)/PISTILLATA(PI)-like class B MADS box factors, FBP1 belongs to the latter group (Zahn and others 2005).
The orchid cultivar Burrageara ‘Stefan Isler Lava Flow’ is a complex hybrid composed of species of four different genera (Cochlioda × Miltonia × Odontoglossum × Oncidium) (The International Plant Names Index, http://www.ipni.org). Orchids in the group of Odontoglossum and Oncidium hybrids are outstanding due to their particular flower colors, patterns, and flower shapes, but have not reached the commercial importance of hybrids from other genera like Phalaenopsis or Cymbidium. Reasons might be their long cultivation time and asynchronous flower induction, among others. Additionally, their sensitivity to ethylene has been shown previously (Raffeiner and others 2009b) resulting in senescence symptoms like leaf yellowing, wilting of florets, and dropping of buds and flowers.
Burrageara ‘Stefan Isler Lava Flow’ inflorescences were protected from senescence by treatment with the ethylene perception inhibitor 1-methylcyclopropene (1-MCP) (Raffeiner and others 2009b). However, the treatment with the gaseous chemical needs a closed chamber and lasts only a few days. Thus, a genetic approach to achieve ethylene insensitivity by Agrobacterium tumefaciens-mediated transformation of the etr1-1 gene under the control of the fbp1 promoter was used in this study. The main objectives were to test florets and inflorescences of an fbp1::etr1-1 transgenic line for their response to ethylene compared to the wild-type and to investigate the organ specificity of the P. hybrida promoter in this monocotyledonous orchid by qRT-PCR analyses of the etr1-1 expression.
Materials and Methods
Plant Material and Culture Conditions
In vitro shoot cultures of the cultivar Burrageara ‘Stefan Isler Lava Flow’ (Fig. 1a) provided by Gartenbau Valtl Raffeiner, Bolzano, Italy, were used to induce protocorm-like bodies (PLBs) as described in Raffeiner and others (2009a). Transformation of these PLBs following Raffeiner and others (2009a) used the A. tumefaciens strain LBA4404 carrying the binary vector pBEO210 (Bovy and others 1999) with the selectable marker gene for neophosphotransferase (35S::npt II) and the mutated ethylene receptor gene etr1-1 from A. thaliana under control of the P. hybrida floral-binding protein 1 (fbp1) promoter. Transgenic plantlets derived from a single-transformation event were multiplied by axillary shoots and maintained under in vitro conditions as described by Raffeiner and others (2010).
Propagation and growth were recorded for the comparison of wild-type and transgenic shoots in two repetitions with six replicates of five shoots each. After 9 weeks of culture on propagation medium (modified half-strength Murashige and Skoog (1962) after Raffeiner and others (2010)) containing 1 g/l polyvinylpyrrolidone 10, 0.25 g/l Na2HPO4 × 2 H2O, 0.89 µM benzyladenine, and 1.1 µM indole acetic acid, the propagation rate was determined by dividing the number of shoots at the end of the culture period by the initial number of shoots for each vessel.
Rooting was obtained on the same basal medium but without plant growth regulators for five and six replicates with five shoots each, in two repetitions of the experiment. The number of rooted shoots was recorded weekly, whereas after 8 weeks the number of roots per rooted shoot was taken in addition. Culture conditions for all in vitro experiments were 23 °C with a 16 h photoperiod by white fluorescent tubes (about 20 µmol m−2 s−1).
Rooted shoots were transferred to greenhouse conditions and cultivated at 20/18 °C day/night temperature until flowering. Length and width of 50 flowers from wild-type and fbp1::etr1-1 transgenic plants (five flowers from ten plants each) were measured.
PCR and Southern Blot Hybridization
DNA was extracted from 80 to 100 mg of leaf and root tissue of in vitro and greenhouse grown plants using the NucleoSpin® Plant II kit (Macherey and Nagel, Dueren, Germany). Integrated genes nptII and fbp::etr1-1 were amplified by PCR using the following conditions: 0.5 U Taq DNA polymerase (DNA Cloning Service, Hamburg, Germany), 10 mM Tris–HCl, 50 mM KCl, 2 mM MgCl2, and 0.25 µM gene-specific primers (nptII_for: 5′ AGGCTATTCGGCTATGACTG 3′ and nptII_rev: 5′ ATCGGGAGCGGCGATACCGTA 3′ (Han and others 2007); annealing temperature TA 58 °C, 699 bp or fbp1-etr1-1_for: 5′ TGTGGTGAAAGAAAAACTCGAC 3′ and fbp1-etr1-1_rev: 5′ CACGGCTGATTTCTTCACAA 3′ (Sanikhani and others 2008), annealing temperature TA 64 °C, 1029 bp). PCR amplification was conducted in a thermocycler (Biometra, Göttingen, Germany) under the following conditions: 3 min. 94 °C following with 35 cycles of 45 s 94 °C, 45 s TA, 60 s 72 °C and terminating with 7 min. 72 °C. For Southern blot hybridization, approximately 10 μg of genomic DNA from each sample was digested with BamHI for 24 h. The resulting DNA fragments were separated on agarose gels and transferred to a membrane as described in Sriskandarajah and others (2007). The probe of 1,029 bp was labeled according to the manufacturer’s protocol (Roche Applied Science Co. Mannheim, Germany) using plasmid DNA containing the etr1-1 gene and the fbp1-etr1-1 primers described above. The membrane hybridization, post-hybridization washing, and detection were performed as described in Sriskandarajah and others (2007).
RT-PCR and qRT-PCR
Total RNA was isolated with the InviTrap® Spin Plant RNA Mini Kit (Stratec Molecular, Berlin, Germany) from 60 to 80 mg fresh mass of different tissues and organs. RNA samples were treated with DNaseI as described in Mibus and others (2011). RNA (1.5 µg) from different organs was reversely transcribed according to Mibus and others (2011).
To evaluate the expression pattern of the FBP1 gene in different organs was analyzed from Petunia plants. Semi-quantitative RT-PCR was carried out in a 20 μl reaction containing 6 ng cDNA template of different Petunia organs applying the PCR conditions described above and with 0.25 µM primers fbp_s 5′TGCCGATTCCACAAGTACAG 3′ and fbp_as CAAGCTTGCCTTCCAGATTC 3′ TA 64 °C; amplicon size 176 bp.
To evaluate the expression pattern of the etr1-1 gene, qRT-PCR with 0.25 µM primer Atetr1-1_for 5′GGTTCCGCTTCTCCACCTTT3′ and Atetr1-1_rev: 5′TCAGCGACGACTTCAACGAG 3′, annealing temperature TA 64 °C, and amplicon size 150 bp was carried out in a 20 µl reaction mixture containing 10 ng of cDNA template from different organs as described in Mibus and others (2011). The qRT-PCR was performed with at least three independent biological replicates in the Rotor Gene 3000 (Corbett Qiagen, Hilden, Germany) with three technical replications per run and at least two replications of each run. To normalize all samples, expression levels of α-tubulin (primer: alphaTub_for 5′GGATTAGGCTCTCTGCTGTTG 3′ and alphaTub_rev: 5′GTGTGGATAAGACGCTGTTGTATG 3′ (Hou and Yang 2009); annealing temperature TA 64 °C; and amplicon size 128 bp) were assayed in each sample in parallel with the gene of interest. Constant expression of the internal control α-tubulin was tested and confirmed across all organs. A mixture composed of cDNAs from all investigated organs was used for standard curves for estimation of primer efficiency as recommended by Livak and Schmittgen (2001). Melting curves were analyzed immediately after finishing qRT-PCR to verify single-product amplification. Analysis of data was completed using version 6.1.81 Rotor Gene software. Relative quantification of transcript abundance of the target gene etr1-1 in individual samples was determined by the \( 2 ^{- \varDelta \varDelta {C_{\text{T} }}} \) method that enabled fold change values for various genes relative to calibrator gene α-tubulin to be calculated for each replicate of each sample (Livak and Schmittgen 2001).
Postharvest Quality Tests
Single flowers were incubated at 20 °C and 12 h photoperiod (about 30 µmol m−2 s−1) in 100 ml lab flasks (total volume 148 ml) with caps bearing a septum. Half of the flasks were filled with ambient air; in the other half, 1.8 µl of pure ethylene was injected. All flasks were opened and ventilated every 2–3 days, followed by a fresh injection of ethylene. Directly after injection and 2 days later, the ethylene concentration was checked using a Perkin–Elmer portable digital gas chromatograph (GC Voyager FFKG312, Ontario, Canada) equipped with a photoionization detector in 3–5 randomly chosen flasks. Ethylene concentration varied among flasks between 8 and 13 ppm directly after the injection and between 5 and 7 ppm 2 days later. The flowers were inspected daily for any changes in color, wilting, necrotic spots, or fungal infections. The number of days until discoloration that was best visible at the lip which turned yellowish and the number of days until the end of the display life, mainly due to fungal infections were analyzed statistically (Table 1). The experiment was finished after 25 days, because the remaining transgenic flowers were mostly wilted.
Cut inflorescences were incubated in a 54 l sealed aquarium, into which 540 µl pure ethylene was injected resulting in an atmosphere containing 10 ppm ethylene. The inflorescences were kept in 50 ml centrifuge tubes filled with tap water at room temperature and low light conditions (1 m distance from a window) and were taken out of the aquarium and evaluated for dropping of buds and flowers every 2–3 days, whereby the ethylene injection was repeated.
Experimental Design and Statistics
Experiments were conducted using at least three biological replications per sample for gene expression analyses with at least three technical replications. Data were subjected to a single-factor analysis of variance (ANOVA) using the general linear models (Proc GLM) of Statistical Package for the Social Sciences (SPSS 20) program (IBM Cooperations New York, USA). Multiple comparisons among treatment means were done using the Tukey test at p ≤ 0.05 or Welch t test, p ≤ 0.05.
Results
One transgenic event was obtained following the transformation protocol of Raffeiner and others (2009a) using PLBs induced on leaf tips. Selection of the transgenic PLBs on 300 mg/l kanamycin leads to one resistant PLB from which shoots were formed and propagated and rooted in vitro (Fig. 1a, Supplementary Fig. 1). Wild-type and fbp1::etr1-1 transgenic cultures did not differ in the formation of axillary shoots (Fig. S1A), and the appearance of the shoot cultures was indistinguishable. However, in the rooting phase, differences between wild-type and transgenic shoots were observed: rooting occurred faster in the fbp1::etr1-1 transgenics, and after 8 weeks they had formed a significantly higher number of 2.6 roots/shoot compared to 1.9 roots/shoot in the wild-type (Fig. S1B,C).
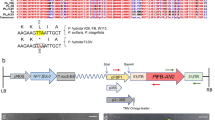
The presence of the transferred genes nptII and fbp1::etr1-1 (Fig. 2a) was proven by PCR for in vitro cultures as well as acclimatized plants in the greenhouse, both using DNA isolated from shoots and roots (Fig. 2b). Southern hybridization verified the integration of the fbp1::etr1-1 transgene into the genome of two clonally propagated transgenic plants (Fig. 2c). Due to the two BamHI restriction sites within the T-DNA of the pBEO210 plasmid (Fig. 2a), a 4.7 kb fragment can be expected in addition to the fragments that will include a part of plant genomic DNA. In both transgenic plants, one additional fragment of approximately 5.5 kb was detected, indicating a single-copy integration in the investigated transgenic event.
Confirmation of the integration of the transgenes. a T-DNA map of the vector pBEO210 (Bovy and others 1999) with indication of PCR amplification products and the probe used in Southern hybridization. b PCR amplification of fragments of the nptII and fbp1::etr1-1 genes from DNA isolated from leaves and roots of wild-type and fbp1::etr1-1 transgenic plants. NTC no template control. c Southern hybridization of DNA of a wild-type (WT) and an fbp1::etr1-1 transgenic plants (clones T11, T20). The DNA was digested with BamHI and probed with labeled fbp1::etr1-1 amplicon. M is a molecular weight marker
After acclimatization, both wild-type and fbp1::etr1-1 transgenic plants showed a high variability in growth, pseudo-tuber formation and flowering making a controlled culture with a prediction of the flowering time impossible. Flowers of fbp1::etr1-1 transgenic plants were significantly smaller (length: 45.8 ± 4 mm, width: 43.1 ± 5 mm) than those of wild-type plants (length: 53.2 ± 5 mm, width: 50.8 ± 5 mm), whereas the ratio of length to width remained unchanged (1.2).
Expression of the fbp1::etr1-1 Transgene
Because the integrated transgene was controlled by the fbp1 promoter from Petunia, it had to be tested if expression was flower-specific in the far related orchid hybrid Burrageara as well. Therefore, primers were designed for quantitative RT-PCR that amplified 150 and 128 bp of the etr1-1 and the reference α-tubulin gene, respectively. Expression of the etr1-1 gene was low in leaves and roots of in vitro cultivated plantlets and only slightly higher in roots (1.27-fold, no significant difference, Tukey test, p ≤ 0.05).
From flowering greenhouse grown plants, the leaves, three outer sepals, the petals, the labellum, the column, and the ovary with the pedicel were dissected. The expression of the fbp1 promotor controlled etr1-1 in these organs is presented in relation to the expression level in leaves in Fig. 3. The highest etr1-1 expression was detected in sepals (13.5-fold) followed by petals and labellum (5.6- and 4.6-fold, respectively), whereas the ovary/pedicel expression was similar to that in leaves. Thus, the fbp1 promoter gave rise to significantly increased expression levels of the etr1-1 gene in the outer two whorls of these orchid flowers (Fig. 3).
Relative normalized expression of the transgene etr1-1 in different flower organs compared to leaf tissue of transgenic plants (bars represent 1 cm). Data were analyzed by qRT-PCR according to the \( {\varDelta \varDelta {C_{\text{T} }}} \) method (Livak and Schmittgen 2001) and were based on the expression in the leaf. Given are means and standard deviations of three biological replicates which were based on three technical replicates each, values indicated by different letters are significantly different (Tukey Test, p ≤ 0.05)
Expression of fbp in Petunia
To evaluate the expression pattern of the fbp gene in Petunia, different organs were investigated by semi-quantitative RT-PCR. Expression of the fbp1 gene was detectable in leaves, shoots, and roots but much lower compared to the flowers (Fig. 4).
Postharvest Quality of Single Flowers and Inflorescences
When single flowers were harvested and incubated in room air, the first signs of discoloration that could be best observed on the lips showing a dull, more pale color were observed after 10.3 days for the wild-type, and significantly later (after 15.6 days) in the transgenic flowers (Table 1). This difference became even more striking when incubating the flowers in 10 ppm ethylene. Here, discoloration was recorded already after 4.3 days in wild-type, but after 11.6 days in transgenic flowers. The end of the display life in this experimental setup was either due to wilting or more often due to fungal infections. Under ambient air conditions, no difference between wild-type and fbp1::etr1-1 transgenics was found, whereas, under ethylene, the display life of wild-type flowers was only half that of transgenic ones (Table 1). These data clearly show a protection of fbp1::etr1-1 transgenic flowers from exogenous ethylene regarding senescence.
The second postharvest test involved inflorescences in ethylene. For wild-type inflorescences, buds on the positions directly above open flowers dropped starting after 5 days (Fig. 5). Only very young buds remained on the flower stalks after 12 days, and after 14 days the inflorescences had to be discarded because fungal mycelium was observed. In contrast, in the fbp1::etr1-1 transgenic inflorescences, all flower buds remained on the stalk and opened successively (Fig. 5a). Bud drop was completely prevented for 19 days, and only a single bud dropped at the end of the display life of one flower stalk of the fbp1::etr1-1 transgenic plants. Thus, the transgene effectively protected the single flowers as well as inflorescences from the senescing action of ethylene.
Postharvest quality of cut inflorescences of wild-type and fbp1::etr1-1 transgenic plants treated with 10 ppm ethylene. a Comparison of cut inflorescences of wild-type (left) and fbp1::etr1-1 (right) transgenic plants at days 0, 5, and 12 of the ethylene treatment. b Observation of bud drop during the ethylene treatment (means and standard deviation of eight inflorescences with 4–10 flower buds tested separately at different times)
Discussion
Transgenic Plants Show Comparable Growth and Development, but Accelerated Rooting and Smaller Flowers
One single-copy (Fig. 2) transgenic line of the genotype Burrageara ‘Stefan Isler Lava Flow’ was obtained and clonally multiplied in vitro in comparable propagation rates to wild-type shoots (Fig. S1A). In the rooting phase, fbp1::etr1-1 transgenic shoots rooted faster and produced more roots than the respective wild-types (Fig. S1B, C). The effect of ethylene on rooting is controversially reported in the literature (Clark and others 1999), probably due to differences between different species and most likely due to ethylene effects being strongly dependent on the phase of adventitious root formation. Clark and others (1999) analyzed wild-type and ethylene-insensitive tomato and petunia plants and found the perception of ethylene to be important for adventitious root formation. Similarly, Torenia fournieri transformed with a mutant ETR1 gene from carnation under the control of the 35S promoter formed less adventitious roots (Tanase and others 2011). In contrast, ethylene insensitivity of Atetr1-1 transgenic lettuce plants resulted in improved formation of adventitious roots (Kim and Botella 2004). In their study, the mutant ethylene receptor was driven by the senescence-specific promoter sag12 (senescence-associated gene 12) and the authors assumed it to be activated in cotyledon explants that showed yellowing during the regeneration process. They concluded that ethylene insensitivity promoted the root formation in lettuce.
In Campanula (Sriskandarajah and others 2007) and Kalanchoë (Sanikhani and others 2008), fbp1::etr1-1 transgenic plants did not differ from wild-type plants in rooting ability. In the present study, low expression of the transgene was detected in leaves and roots. Expression of fbp1::etr1-1 was also observed in roots of transgenic Kalanchoë (Sanikhani and others 2008) but without an effect on rooting. It remains to be shown in further studies, if in vitro rooting of Burrageara is influenced by ethylene.
Heterologous Function of the Flower-Specific fbp1 Promoter from Petunia Proven in Orchid
The functional evolutionary conservation of the ethylene receptor ETR1 enabled the successful operation of the mutant Arabidopsis receptor etr1-1 in several heterologous—but so far only dicotyledonous—plants, such as Petunia (Wilkinson and others 1997; Clark and others 1999), Dianthus (Bovy and others 1999), Campanula (Sriskandarajah and others 2007), and Kalanchoë (Sanikhani and others 2008). In monocots, like maize and rice, only two types of ethylene receptors are found (ETR2 and ERS1) (Chen and Gallie 2010). The introduction of the etr1-1 mutation at Cys65 into maize receptors Zmers1b and Zmetr2b resulted in dominant ethylene insensitivity. Both mutant maize alleles when transformed into Arabidopsis lead to insensitivity, if the subfamily I receptors were present (Chen and Gallie 2010). To our knowledge, no attempt to transfer the Arabidopsis etr1-1 gene to a monocot has been reported up to now. In our previous work on Oncidium and Odontoglossum hybrids, the integration of the fbp1::etr1-1 gene was proven, but no flowering plants were obtained. Thus, this study, for the first time shows the functionality of this gene in the monocotyledonous orchid Burrageara. Due to the time- and labor-intensive transformation protocol with efficiencies of about 1 % and due to the slow growth and development of wild-type and transgenic plants, positional effects of transgene integration at different locations in the plant genome, that would require independent transgenic events, could not be tested in this study.
An interesting and novel observation was the tissue-specific expression of the transgene in this orchid with the highest expression found in sepals, petals, and labellum (Fig. 3). The petunia fbp1 promoter belongs to a class B MADS box transcription factor of the GLO-like subtype. The exclusive expression of fbp1 in petals and stamen and not in sepals and carpels or in vegetative tissues (leaves and roots) was proven by RNA gel blot in Petunia (Angenent and others 1992). In contrast, expression of the etr1-1 gene controlled by the fbp1 promoter was detectable by qRT-PCR in leaves and roots of in vitro cultivated Burrageara and also in vegetative tissue harvested from plants grown in the greenhouse (Fig. 3). To investigate if the expression of etr1-1 (regulated by the fbp1 promotor) in vegetative tissue was unique in Burrageara, the fbp1 expression was evaluated in Petunia. The results demonstrated that the expression of the endogenous fbp1 was detectable in leaves and roots in Petunia by semi-quantitative RT-PCR (Fig. 4). This contradictory expression of fbp1 in vegetative tissue in Petunia could be explained by the use of the more sensitive RT-PCR compared to RNA gel blots in previous studies (Angenent and others 1992).
Orchid flower morphology is very diverse and special and was studied in detail regarding the homeotic genes leading to the postulation of the “orchid code” (Mondragon-Palomino and Theißen 2009). A set of four AP3/DEF-like and one PI/GLO-like class B MADS box genes seem to shape the orchid perianth organs, that is, the tepals (three sepals and two petals, see Fig. 3), and the labellum (Mondragon-Palomino and Theißen 2009). Like in tulips, class B MADS box gene expression expanded from whorls 2 and 3 (petals and stamens in the dicot models Antirrhinum and Arabidopsis) to whorls 1 and 4 (sepals and carpels) in orchid flowers (Aceto and Gaudio 2011). Thus, the expression of fbp1::etr1-1 in Burrageara flower organs of the first and second whorl which was shown in our expression analysis (Fig. 3) points to the functionality of this Petunia class B MADS box promoter in the heterologous orchid. Due to the limitations in materials, anthers and pollinia for which similar expression levels as in petals and lip can be expected were not investigated in this study. The column of orchid flowers is an organ that contains both the stigma and male flower parts and here also elevated fbp1::etr1-1 expression could be shown (Fig. 3). The low expression observed in the ovary probably can be explained by the sampling of the whole part below the sepals that is composed of the pedicel and the ovary. It will be important to study expression and ethylene sensitivity in the ovary in future studies, because ethylene may be the trigger for ovule differentiation as shown for Phalaenopsis, where a GLO-like gene was found to be involved in this process (Tsai and others 2008). Therefore, fertility of the transgenic plants needs to be analyzed.
Obviously, the promoter of the introduced gene was recognized by the endogenous homeotic transcription factors that are regulated in a complex network including dimerization, feedback and feedforward mechanisms, small RNAs and epigenetic control (Aceto and Gaudio 2011). For the other published fbp1::etr1-1 transgenic plants, no quantitative RT-PCR analyses were presented, but by Northern hybridization, Bovy and others (1999) detected expression in carnation petals, but not in ovaries, sepals, and leaves. Semi-quantitative RT-PCR revealed the strongest fbp1::etr1-1 expression in Campanula buds, followed by flowers and only weak expression in leaves (Sriskandarajah and others 2007), and expression of this transgene in roots and flowers, but not in leaves of Kalanchoë (Sanikhani and others 2008). These two studies point to activity in vegetative tissue of the promoter that was confirmed in our study.
Ethylene Insensitivity Delays Flower Senescence and Inhibits Bud Drop
Senescence of petals, normally observed after pollination, involves the remobilization of macromolecules, that is, nucleic acids, protein degradation and fatty acid breakdown, and autophagy in the vacuole, and can be regarded as a kind of programmed cell death (van Doorn and Woltering 2008). In ethylene-sensitive plants, this process is initiated or stimulated by ethylene, as shown for orchids in general (Woltering and van Doorn 1988) and also the species under investigation in this study (Raffeiner and others 2009b). The expression analyses showed that the introduced mutant ethylene receptor was transcribed in the floral organs (Fig. 3), and after exposure to room air or ethylene in relatively high concentrations, the transgenic flowers showed a significantly improved display life (Table 1). Thus, the dominant mutant receptor was functional also in the background of the endogenous orchid receptors. Previously, several ethylene receptors were cloned in orchids, but only one ethylene receptor from Oncidium ‘Grower Ramsey’ was investigated in detail (Huang and others 2007). The transcript accumulation of OgERS during ethylene-induced flower senescence gives an indication that the native sensitivity to ethylene is regulated by differential expression of the ethylene receptors in Oncidium (Huang and others 2007).
Similarly, in most other etr1-1 transgenic plants, postharvest quality of the flowers was improved (Wilkinson and others 1997; Bovy and others 1999; Sriskandarajah and others 2007; Sanikhani and others 2008) except for zonal Pelargonium for which no differences compared to the wild-type plants were observed (Boase and others 2012). More detailed analyses of the molecular and physiological effects of the etr1-1 transgene in Petunia revealed retarded DNA fragmentation in petals by a senescence-specific nuclease (Langston and others 2005) and a downregulation of transcription factors, genes involved in response to gibberellic acid and jasmonate, in GA biosynthesis, in cell wall modification, and ethylene biosynthesis (Wang and others 2013). Reduced autocatalytic ethylene production was also observed in the fbp1::etr1-1 transgenic carnations, and the corresponding analyses of ethylene biosynthesis in our transgenic Burrageara would be interesting. The increased ethylene production and ACC oxidase activity beginning at an early stage of senescence in Epidendrum ibaguense suggest the importance of autocatalytic production of ethylene during senescence in orchid flowers (Mapeli and others 2009).
From the economic point of view, the unexpected positive effect of the fbp1::etr1-1 transgene in preventing flower bud drop (Fig. 5) is most important and has not been described in other studies up to now. In Burrageara ‘Stefan Isler Lava Flow,’ the open flowers showed a response to ethylene in terms of discoloration and wilting, but remained on the flower stalks for 8–10 days, whereas flower buds, except the youngest two or three started to drop after 5 days incubation in ethylene (Fig. 5). Ethylene has a regulatory effect, at the transcriptional and the translational level, on expressed genes that are involved in organ abscission-related cell separation (Brown 1997). Bud and flower drop in orchids during transportation and dark storage is a serious problem, consequently, many farmers in South-East Asia have stopped growing Aranthera ‘Beatnice Eng’ and Dendrobium ‘Sri Siam’ because of this bud drop problem (Hew and Clifford 1993).
For Oncidium cut flowers, a higher ethylene sensitivity of buds compared to florets was reported (Huang and Paull 2009). In the case of Dendrobium, different cultivars reacted differently to ethylene treatments: whereas all genotypes started to drop buds after 5 days, in two genotypes open flowers remained attached to the inflorescences (Bunya-aticharta and others 2006). In one of these cultivars, ethylene sensitivity changed from very sensitive to insensitive with bud opening within 30 h. A follow-up study revealed a correlation of polygalacturonase activity in the abscission zone of buds and flowers and their ethylene sensitivity (Bunya-aticharta and others 2011). In Arabidopsis, floral organ abscission occurs within 2–3 days after anthesis, whereas in the ethylene response mutant etr1-1, this sequence is significantly delayed, and cells are not fully enlarged (Bleecker and Patterson 1997). However, ethylene is not necessary for the activation of organ abscission because floral abscission does still occur in the etr1-1 mutant of Arabidopsis. For that reason it can be assumed that both ethylene-dependent and ethylene-independent processes are involved in floral organ abscission in Arabidopsis (Patterson and Bleecker 2004). The expression analyses of the fbp1::etr1-1 transgene in the abscission zone of transgenic Burrageara would allow insights into the causes of the protection of flower buds from dropping.
The reduced cell enlargement that was reported for the Arabidopsis etr1-1 mutant (Bleecker and Patterson 1997) was also observed in the present study in which the transgenic Burrageara flowers were significantly smaller than the wild-type ones. The perianth organs, in which promoter activity was high (Fig. 3), were smaller probably due to reduced cell expansion. In microscopic sections, this hypothesis could be proven by measuring the cellular dimensions.
In conclusion, the promoter fpb1 was predominantly expressed in sepals, petals, labellum, and column indicating an operation in the heterologous plant Burrageara. For the first time, it could be shown that the mutant ethylene receptor from A. thaliana conferred ethylene insensitivity in a monocotyledonous orchid. Observations of male and female fertility as well as susceptibility to diseases are needed in the future, because in 35S::boers (mutant ethylene receptor from Brassica oleracea) transgenic Petunia plants, reduced disease resistance was observed (Shaw and others 2002).
Although transgenic approaches could be useful in reaching a number of breeding aims in ornamental plant breeding (Debener and Winkelmann 2010), their commercialization is lagging behind major agricultural crops. The costs for the production and testing of several stable transgenic lines and for deregulation of candidates for new cultivars are far too high for most ornamental species. Moreover, at least in Europe, the lack of public acceptance of transgenic plants, that also holds true for non-food species, is a severe restriction.
References
Abeles FB, Morgan PW, Saltveit ME (1992) Ethylene in plant biology. Academic Press, New York, p 414
Aceto S, Gaudio L (2011) The MADS and the beauty: genes involved in the development of orchid flowers. Curr Genom 12:342–356
Angenent GC, Busscher M, Franken J, Mol JNM, van Tunen AJ (1992) Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell 4:983–993
Angenent GC, Franken J, Busscher M, Colombo L, van Tunen AJ (1993) Petal and stamen formation in petunia is regulated by the homeotic gene fbp1. Plant J 4:101–112
Bleecker AB, Patterson SE (1997) Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9:1169–1179
Boase MR, Harriman RW, Smith FD, Deroles SC (2012) Herbicide-resistant, transgenic zonal pelargoniums produced by step-down glyphosate selection and plantlet recovery in the presence of aromatic amino acids. In Vitro Cell Dev Biol Plant 48:313–323
Bovy AG, Angenent GC, Dons HJM, van Altvorst AC (1999) Heterologous expression of the Arabidopsis etr1-1 allele inhibits the senescence of carnation flowers. Mol Breed 5:301–308
Brown KM (1997) Ethylene and abscission. Physiol Plant 100:567–576
Bunya-aticharta K, Ketsa S, van Doorn WG (2006) High floral bud abscission and lack of open flower abscission in Dendrobium cv. Miss Teen: rapid reduction of ethylene sensitivity in the abscission zone. Funct Plant Biol 33:539–546
Bunya-aticharta K, Ketsa S, van Doorn WG (2011) Ethylene-sensitive and ethylene-insensitive abscission in Dendrobium: correlation with polygalacturonase activity. Postharvest Biol Technol 60:71–74
Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262:539–544
Chen J-F, Gallie DR (2010) Analysis of the functional conservation of ethylene receptors between maize and Arabidopsis. Plant Mol Biol 74:405–421
Clark DG, Gubrium EK, Barrett JE, Nell TA, Klee HJ (1999) Root formation in ethylene-insensitive plants. Plant Physiol 121:53–59
Debener T, Winkelmann T (2010) Ornamentals. In: Kempken F, Jung C (eds) Genetic modification of plants, Biotechnology in agriculture and forestry. vol 64. Springer, Berlin, pp 369–391
Gamble RL, Qu X, Schaller GE (2002) Mutational analysis of the ethylene receptor ETR1. Role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiol 128:1428–1438
Guo H, Ecker JR (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7:40–49
Hall BP, Shakeel SN, Schaller E (2007) Ethylene receptors: ethylene perception and signal transduction. J Plant Growth Regul 26:118–130
Han BH, Suh EJ, Lee SY, Shin HK, Lim YP (2007) Selection of non-branching lines induced by introducing Ls-like cDNA into Chrysanthemum (Dendranthema x grandiflorum (Ramat.) Kitamura) “Shuho-no-chikara”. Sci Hortic 115:70–75
Hew CS, Clifford PE (1993) Plant growth regulators and the orchid cut-flower industry. Plant Growth Regul 13:231–239
Hou CJ, Yang CH (2009) Functional analysis of FT and TFL1 orthologs from orchid (Oncidium Gower Ramsey) that regulate the vegetative to reproductive transition. Plant Cell Physiol 50:1544–1557
Huang CC, Paull RE (2009) The responses of Oncidium cut flowers to ethylene and 1-MCP. J Taiwan Agric Res 58:1–6
Huang WF, Huang PL, Do YY (2007) Ethylene receptor transcript accumulation patterns during flower senescence in Oncidium ‘Gower Ramsey’ as affected by exogenous ethylene and pollinia cap dislodgment. Postharvest Biol Technol 44:87–94
Immink RGH, Ferrario S, Busscher-Lange J, Kooiker M, Busscher M, Angenent GC (2003) Analysis of the petunia MADS-box transcription factor family. Mol Gen Genom 268:598–606
Kim JH, Botella JR (2004) Etr1-1 gene expression alters regeneration patterns in transgenic lettuce stimulating root formation. Plant Cell Tiss Organ Cult 78:69–73
Langston BJ, Bai S, Jones ML (2005) Increases in DNA fragmentation and induction of a senescence-specific nuclease are delayed during corolla senescence in ethylene. J Exp Bot 56:15–23
Liu Q, Wen C-K (2012) Arabidopsis ETR1 and ERS1 differentially repress the ethylene response in combination with other ethylene receptor genes. Plant Physiol 158:1193–1207
Livak KJ, Schmittgen TD (2001) A quantitative assay for measuring mRNA decapping by splinted ligation reverse transcription polymerase chain reaction: qSL-RT-PCR. RNA 17:535–543
Mapeli AM, De Oliveira LS, Megguer CA, Barbosa JG, Barros RS, Finger FL (2009) Characterisation of respiration, ethylene production, and carbohydrate contents during flower opening in Epidendrum ibaguense. J Hort Sci Biotechnol 84:609–612
Merchante C, Alonso JM, Stepanova AN (2013) Ethylene signaling: simple ligand, complex regulation. Curr Opin Plant Biol 16:554–560
Mibus H, Heckl D, Serek M (2011) Cloning and characterisation of three APETALA1 homologous genes in different flower types of Rosa hybrida L. J Plant Growth Regul 30:272–285
Mondragon-Palomino M, Theißen G (2009) Why are orchid flowers so diverse? Reduction of evolutionary constraints by paralogues of class B floral homeotic genes. Ann Bot 104:583–594
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Patterson SE, Bleecker AB (2004) Ethylene-dependent and—independent processes associated with floral organ abscission in Arabidopsis. Plant Physiol 134:194–203
Raffeiner B, Serek M, Winkelmann T (2009a) Agrobacterium tumefaciens mediated transformation of Oncidium and Odontoglossum orchid species with the ethylene receptor mutant gene etr1-1. Plant Cell Tiss Organ Cult 98:125–134
Raffeiner B, Serek M, Winkelmann T (2009b) 1-Methylcyclopropene inhibits ethylene effects in cut inflorescences and potted plants of Oncidium and Odontoglossum orchid species. Europ J Hortic Sci 74:10–15
Raffeiner B, Serek M, Winkelmann T (2010) Establishment and optimization of an efficient in vitro regeneration system of Oncidium, Wilsonara, Odontocidium and Vuylstekeara. Acta Hortic 878:445–452
Rattanawisalanon C, Ketsa S, van Doorn WG (2003) Effect of aminooxyacetic acid and sugars on the vase life of Dendrobium flowers. Postharvest Biol Technol 29:93–100
Sanikhani M, Mibus H, Stummann BM, Serek M (2008) Kalanchoë blossfeldiana plants expressing Arabidopsis etr1-1 allele show reduced ethylene sensitivity. Plant Cell Rep 27:729–737
Shakeel SN, Wang X, Binder BM, Schaller GE (2013) Mechanisms of signal transduction by ethylene: overlapping and nonoverlapping signalling roles in a receptor family. AoB Plants 5:plt010
Shaw JF, Chen HH, Tsai MF, Kuo CI, Huang LC (2002) Extended flower longevity of Petunia hybrida plants transformed with boers, a mutated ERS gene of Brassica olercea. Mol Breed 9:211–216
Sriskandarajah S, Mibus H, Serek M (2007) Transgenic Campanula carpatica plants with reduced ethylene sensitivity showing specific expression of etr1-1 in flowers and buds. Plant Cell Rep 26:805–813
Sun Y, Christensen B, Liu F, HQ W, Mueller R (2009) Effects of ethylene and 1-MCP (1-methylcyclopropene) on bud and flower drop in mini Phalaenopsis cultivars. Plant Growth Regul 59:83–91
Tanase K, Aida R, Yamaguchi H, Tanikawa N, Nagata M, Onozaki T, Ichimura K (2011) Heterologous expression of a mutated carnation ethylene receptor gene, Dc-ETR1nr, suppresses petal abscission and autocatalytic ethylene production in transgenic Torenia fournieri Lind. J Jpn Soc Hortic Sci 80:113–120
Tsai WC, Hsiao YY, Pan ZJ, Kuoh CS, Chen WH, Chen HH (2008) The role of ethylene in orchid ovule development. Plant Sci 175:98–105
van Doorn WG, Woltering EJ (2008) Physiology and molecular biology of petal senescence. J Exp Bot 59:453–480
Wang H, Stier G, Lin J, Liu G, Zhang Z, Chang Y, Reid MR, Jiang CZ (2013) Transcriptome changes associated with delayed flower senescence on transgenic petunia by inducing expression of etr1-1, a mutant ethylene receptor. PLOS one 8(7):e65800
Wilkinson JQ, Lanahan MB, Clark DG, Bleecker AB, Chang C, Meyerowitz EM, Klee HJ (1997) A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat Biotechnol 15:444–447
Woltering EJ, van Doorn WG (1988) Role of ethylene in senescence of petals—morphological and taxonomical relationships. J Exp Bot 39:1605–1616
Zahn LM, Leebens-Mack J, De Pamphilis CW, Ma H, Theissen G (2005) To B or not to B a flower: the role of DEFICIENS and GLOBOSA orthologs in the evolution of the angiosperms. J Hered 96:225–240
Acknowledgments
The authors thank Ewa Schneider, Bärbel Ernst, Peter Pietrzyk, and Annette Steding for their excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Winkelmann, T., Warwas, M., Raffeiner, B. et al. Improved Postharvest Quality of Inflorescences of fbp1::etr1-1 Transgenic Burrageara ‘Stefan Isler Lava Flow’. J Plant Growth Regul 35, 390–400 (2016). https://doi.org/10.1007/s00344-015-9545-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-015-9545-2