Abstract
Genetic and environmental factors interact throughout life and give rise to individual differences, i.e., individuality. The diversifying effect of environmental factors is counteracted by genetic mechanisms to yield persistence of specific features (robustness). Here, we compared robustness between cohorts of isogenic mice of eight different commonly used strains by analyzing to what extent environmental variation contributed to individuality in each of the eight genotypes, using a previously published dataset. Behavior was assessed in the home-cage, providing control over environmental factors, to reveal within-strain variability in numerous spontaneous behaviors. Indeed, despite standardization and in line with previous studies, substantial variability among mice of the same inbred strain was observed. Strikingly, across a multidimensional set of 115 behavioral parameters, several strains consistently ranked high in within-strain variability (DBA/2J, 129S1/Sv A/J and NOD/LtJ), whereas other strains ranked low (C57BL/6J and BALB/c). Strain rankings of within-strain variability in behavior were confirmed in an independent, previously published behavioral dataset using conventional behavioral tests administered to different mice from the same breeding colonies. Together, these show that genetically inbred mouse strains consistently differ in phenotypic robustness against environmental variation, suggesting that genetic factors contribute to variation in robustness.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
There is a keen interest in society as well as in fundamental and medical research in factors that make us unique as individuals. Generally, these factors are divided into genetic factors (nature) and environmental factors (nurture), which through complex interactions shape the behavioral characteristics of an individual. Accordingly, in the complete absence of variation-contributing genetic factors, such as in the case of monozygotic (genetic identical) twins, or inbred (isogenic) mice, any difference in behavior should result from environmental factors. Nonetheless, in experimental studies employing isogenic (genetic identical) mice, in which environmental conditions are highly standardized to reduce environmental variation, behavioral differences between isogenic mice are observed (Gärtner 1990; Crabbe et al. 1999; Freund et al. 2013). A number of factors may contribute to this individuality in behavior in mice, including uncontrollable environmental factors such as intrauterine position (Lathe 2004). Ultimately, such environmental differences may lead to the profound epigenetic differences (e.g., DNA methylation levels) that have been observed between genetically identical individuals (Kaminsky et al. 2009; van Dongen et al. 2012). Interestingly, there is clear evidence for molecular mechanisms that buffer environmental variation. This phenomenon is known as phenotypic robustness in model organisms (Queitsch et al. 2012), and is broadly defined as the ability of a given genetic background to produce a constant phenotype, despite environmental variability (Debat and David 2001). For instance, experimental evidence in plants and flies shows that impairing HSP90 function decreases buffering of environmental variation, thereby leading to increased phenotypic variation (Queitsch et al. 2002; Sangster et al. 2008). The aim of the present study was to investigate, in mice, whether genetic variation contributes to phenotypic robustness.
The extent of variation among genetically identical individuals is the net result of factors that promote individuality and those that buffer against variation. In the present study, we kept isogenic animals under strictly controlled environmental conditions (e.g., living environment, nutrition). Nonetheless, some environmental factors are uncontrollable [e.g., intrauterine position (Lathe 2004)], and we used this environmental variation and perturbation to study robustness. Importantly, these environmental factors are of similar nature, and are uncontrollable in a similar fashion in different strains of mice. Thus, behavioral differences between strains, in response to these uncontrollable environmental factors, should arise largely from strain differences in the capacity to buffer. To test this buffering capacity (robustness), we compared within-strain variability in behavior between eight genetically different laboratory isogenic inbred strains of mice.
To quantify variability in behavioral phenotypes of sufficient numbers of isogenic mice, systematic, accurate, and unbiased measurements are crucial (Bendesky and Bargmann 2011). Here, we relied on studying behavioral phenotypes in an automated home-cage environment (Loos et al. 2014), in which behavior is automatically tracked for multiple days in a row. Such a home-cage provides a continuous living and testing environment without human interference to reduce environmental factors contributing to within-strain variation. Genetic contribution to phenotypic robustness would then become manifest as differences in within-strain variability between isogenic lines with respect to the many behavioral parameters that automated home-cage testing yield.
Materials and methods
Mice
Mice were obtained from Jackson Laboratory and bred in the facilities of the Neuro-Bsik consortium (VU University Amsterdam, The Netherlands or Harlan Laboratories, Horst, The Netherlands; 129S1/SvImJ n = 59, A/J n = 43, BALB/cJ n = 44, C3H/HeJ n = 29, C57BL/6J n = 107, DBA/2J n = 39, FVB/NJ n = 36, NOD/ShiLtJ n = 37). 8 to 12-week-old male mice were singly housed on sawdust in standard Makrolon type II cages enriched with cardboard nesting material for at least 1 week prior to experiments, with water and food ad libitum (7:00/19:00 lights on/off; providing an abrupt phase transition). Experiments were carried out in accordance with the European Communities Council Directive of 24 November 1986 (86/609/EEC), and with approval of the local animal care and use committee of the VU University.
Automated home-cage observation and data analyses
Observation was performed in a home-cage environment (PhenoTyper model 3000, Noldus Information Technology, Wageningen, The Netherlands), described in detail previously (Maroteaux et al. 2012). Mice were housed for seven consecutive days, of which the first 3 days were used to analyze spontaneous behavior. Mice were introduced in the cage in the second half of the subjective light phase (14:00–16:00 h), and video tracking started at the onset of the first subjective dark phase (19:00 h). The cages (L = 30 × W = 30 × H = 35 cm) were made of transparent Perspex walls with an opaque Perspex floor covered with bedding based on cellulose. A feeding station and a water bottle were attached on to two adjacent walls. A triangular-shaped shelter compartment (height: 10 cm; non-transparent material) with two entrances was fixed in the corner of the opposite two walls. The top unit of each cage contained an array of infrared LEDs and an infrared-sensitive video camera used for video tracking. The X–Y coordinates of the center of gravity (COG) of mice sampled at a resolution of 15 coordinates per second were acquired and smoothed (Hen et al. 2004) using EthoVision software (EthoVision HTP 2.1.2.0, based on EthoVision XT 4.1, Noldus Information Technology, Wageningen, The Netherlands). In occasions where mice were not detected by the video tracking system, due to processor overload (missing frames) or in particular areas of the cage where image contrast was low (not found frames; particularly when climbing into the feeding station), linear interpolation (interpolated frames) between last observed position and newly detected position was implemented. Resulting digital tracks were processed to generate behavioral parameters using AHCODA™ analysis software (Sylics, Synaptologics BV, Amsterdam, The Netherlands), as described in detail previously (Loos et al. 2014).
Statistical analyses
Before statistical analyses, parameters were log10 transformed in case this decreased the positive skew of the distribution (see Table S1 for parameters and the applied transformation). Estimates of the genetic effect size (narrow sense heritability) were calculated as described by Hegmann and Possidente (Hegmann and Possidente 1981) using a custom function (Microsoft Excel) as reported previously (Loos et al. 2009), which takes the differences in the number of animals per group into account when estimating the within and between-strain variance (Lynch and Walsh 1998). Data were checked for skewness, and if skewness could be improved by log10 transformations, this was executed. Outlier data points were defined when exceeding the threshold of ±5 times the standard deviation of the entire dataset (i.e., not per strain), and outlying values were replaced by this threshold. Principal component analysis (PCA) was performed on data of individual mice, and after subtraction of strain means, missing data were replaced my strain means (i.e., 0 after subtraction); principal components were retained if the Eigenvalue was larger than 1, which is typically used as threshold (Kaiser’s criterion; (Kaiser 1960)). Subject’s scores on PC were estimated using regression. ANOVA and PCA were performed with SPSS version 20.0 (IBM, Armonk, New York, USA).
Results
Systematic analysis of within-strain variability
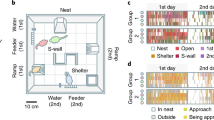
Our analysis yielded qualitatively different move, arrest, and shelter segments, of which an example is shown in Fig. 1a. The methods and between-strain differences in behavior of 11 strains have been described previously (Loos et al. 2014). In the present study, however, we investigated within-strain differences in behavior of 8 of those strains for which a large number of individuals was measured (between 29 and 107 individual mice per strain), which is required for an accurate assessment of within-strain variability. In short, mouse activity was studied with respect to time windows ranging from activity bouts in the subminute range (Fig. 1b), habituation effects across multiple days (Fig. 1c), diurnal 12/12 h dark/light rhythms (Fig. 1d), to the hours surrounding the phase. Together, the analysis provided 115 parameters to study within-strain variability in behavior. Given that several parameters were related in time or location in the cage, we examined to what extent each of the 115 parameters contributed to insight into within-strain variability. Hereto, we performed principal component (PC) analysis with Varimax rotation on the data of all individual mice (394) belonging to eight strains, after subtraction of their respective strain mean, to specifically focus on within-strain variability. PC analysis identified 22 orthogonal PCs of within-strain variability across the entire dataset. However, the principal components identified in the pooled data of mice of all strains may not be truly independent in each strain. In other words, the variance of some behaviors could be highly correlated in some strains but not in others and pooling them might mask those differences. To explore the independence of the parameters within each strain, we repeated PC analyses within each of the 8 strains. In all strains, PC analysis identified 20 or more PCs, showing that this dataset of 115 parameters is truly multidimensional and, therefore, suited to investigate overall phenotypic robustness.
Segmentation of sheltering behavior and activity into elements, studied at four different time scales. a A representative track of approximately 17 min of a C57BL/6J mouse, obtained during the dark phase of the first night in the cage (7:34:22–7:52:00), dissected into elements by individually determined thresholds. b Active elements of behavior occur in bouts, interchanged with periods of inactivity during long arrest or long shelter segments. c At a multi-day timescale, activity level changes due to habituation effects. d At a timescale of 12 h, there is an effect of the dark/light phase. e At the timescale of hours, especially around phase transitions, activity shows strong increases and/or decreases. For illustrative purposes, panels c–e contain reference data of C57BL/6J mice that were previously published (Loos et al. 2014)
Strain comparison of within-strain variability in home-cage behavior
To study strain differences in phenotypic robustness, we compared the within-strain standard deviation. The coefficient of variation (standard deviation divided by the mean) was unsuitable as metric for within-strain variability, since the coefficient of variation will approach infinity when the mean is close to zero (e.g., as in the case of measures of habituation, which are inferred from the slope of the activity during the first 3 days). The standard deviations were plotted as z-scores in a heatmap (Fig. 2), representing the high (red) or low (blue) variability of a strain compared to the average variability of all eight strains. Although all strains displayed both relatively high and low variability on particular parameters in comparison to other strains, some strains stood out in terms of the relatively low number of highly variable parameters (C57BL/6J for instance) or a high number of highly variable parameters (for instance 129S1/Sv). To quantify differences between strains in overall phenotypic robustness, i.e., variability in many behavioral parameters, the average variability of the eight strains was calculated (Fig. 3a). Indeed, this quantification indicated that C57BL/6J and BALB/c mice showed the lowest variability across these 115 parameters in contrast to highly variable strains such as 129S1/Sv and A/J. Since larger means might be associated with larger standard deviations, we investigated whether there was a relation between the z-scores of the standard deviations (as used in Figs. 2, 3) and the z-scores of the means, for each parameter in the dataset. There was no consistent correlation between the z-scores of the means and the z-score of the standard deviations, indicating that the metric of variability in this study (i.e., the standard deviation) was not confounded by differences in the mean performance of the strains (Fig S1).
Strain comparison of within-strain variability across 115 behavioral parameters. For each behavioral parameter (rows), the standard deviation within each strain (columns) was calculated. The standard deviations are plotted as z-scores, representing the high (red) or low (blue) deviation of a strain compared to the average standard deviation of the eight strains for that parameter. The dendrogram on the left side of the figure visualizes the result of average linkage clustering of the parameters with Euclidian distance as metric, illustrating the dimensionality of the dataset
Strain comparison of overall phenotypic robustness. Phenotypic robustness was inferred from the average within-strain variability for a given strain, calculated from a collection of behavioral parameters. Before calculating the average, within-strain variability was standardized (z-scores). a Jittered scatterplot of the average variability of each individual mouse in the experiment, as obtained from 115 parameters in the home-cage (Loos et al. 2014). The average variability per strain is indicated by the horizontal lines. b Bar plot of the average variability per strain, as obtained from the 22 PCs extracted from the 115 parameters. c Bar plot of the average variability per strain, obtained from previously published behavioral parameters obtained in a battery of conventional behavioral tests (Loos et al. 2009). The error bars represent the standard error of the mean
As PCA had indicated, the 115 parameters were not independent but reflect 22 orthogonal components. Thus, the average variability of a strain, as depicted in Fig. 3a, represents the average of 22 independent assessments of within-strain variability in a given strain. Thus, to statistically test for strain differences in variability, while accommodating the dependency among parameters, the average within-strain variability on the 22 PCs was statically evaluated. Indeed, ANOVA showed that the strain difference in variability in the 22 orthogonal components was highly significant [F(7168) = 4.33, P < 0.001; Fig. 3b].
The group size varied considerably between the eight inbred strains in the study, and the accuracy of the estimation of a standard deviation is higher with increasing sample sizes. Although the lower standard deviation in the largest group (i.e., C57BL/6J) cannot readily be explained by the larger group size, we nevertheless investigated the effect of group size on the observed variability by randomly drawing smaller samples for all strains, i.e., samples the size of the smallest group (C3H/HeJ; n = 29) as well as a typical size for behavioral experiments (n = 12), and calculated the average variability based on these smaller samples (Fig. S2, intermediate and light gray shaded bars). The pattern of strain differences in variability did not change; again C57BL/6J and BALB/c mice showed the lowest variability, while strains such as 129S1/Sv and A/J displayed the highest variability.
Furthermore, to check whether variability between mice could have been due to measurement accuracy, we correlated measures of video tracking accuracy of each strain with within-strain variability. None of these measures correlated significantly with variability (missing frames: Spearman’s ρ = .12, P = .78; not found frames: Spearman’s ρ = .33, P = .42; interpolated frames: Spearman’s ρ = .10, P = .82), indicating no evidence for methodological sources of strain differences in variability.
Next, we sought to confirm our strain ranking of variability with an independent second behavioral dataset. We re-analyzed 29 previously published behavioral parameters obtained in a battery of conventional behavioral tests of activity/anxiety in a panel of inbred strains [(Loos et al. 2009); n = 12 per strain], using five strains that were also tested in the current study. In line with the home-cage data in the current study, a significant strain difference in variability was detected [F(4140) = 9.61, P < 0.001], and again, the average variability of C57BL/6J was low, whereas the A/J showed the highest variability.
In conclusion, we found consistent differences in within-strain variability among inbred strains, suggesting a genetic contribution to overall robustness of behavioral phenotypes in mice.
Discussion
The longitudinal automated home-cage data used in the current study allowed us to study a wide range of behaviors in individual mice under highly standardized conditions over prolonged periods of time (72 h). Consistent differences in within-strain variability between eight inbred strains were observed across many behavioral phenotypes, showing that distinct genetic backgrounds differ in robustness, i.e., their capacity to produce a constant behavioral phenotype.
Within-strain variability, i.e., variation between highly isogenic individuals, has been observed in behavioral experiments in general (Gärtner 1990; Crabbe et al. 1999; Freund et al. 2013), and has been attributed to a number of sources. First, despite careful inbreeding, genetic variation can still exist in these highly inbred mice. However, all strains used here have been subjected to decades of inbreeding by the same breeder (Jackson Laboratories) that implemented a genetic stability program (Taft et al. 2006), virtually eliminating the possibility for systematic differences in the amount of residual genetic variation between the lines in the experiment. Nonetheless, at present, it is unknown whether different strains may differ in the rate of, e.g., de novo mutations and copy number variation (Egan et al. 2007; Watkins-Chow and Pavan 2008). Thus, differences in within-strain variability may be due to strain differences in the fidelity of the DNA replication machinery and/or resistance to transposon-based mutations.
Second, despite standardized breeding and housing in the same facility, uncontrollable environmental influences, such as intrauterine position of the embryo and feeding hierarchy in newborns, cause within-strain variability (Lathe 2004). The majority of these phenomena can be expected to be present in all strains tested in this study and, therefore, provide no direct explanation for strain differences in within-strain variability.
Third, stochastic events in molecular and cellular processes contribute to the emergence of phenotypic variation between isogenic individuals. Experiments in unicellular organisms have for instance shown that events stochastically affecting chromatin structure create so-called intangible phenotypic variation (Pujadas and Feinberg 2012), for review (Whitelaw et al. 2010b)). Especially those stochastic events that occur during a critical developmental phase may non-linearly affect the phenotype of an organism (Molenaar et al. 1993). The effect of stochastic events promoting variability, and mechanisms contributing to robustness cannot be distinguished in the present study and may, to a certain extent, be two sides of the same token.
All three sources of within-strain variation mentioned above may contribute to initial variation between individual isogenic mice, and set the stage for a unique developmental trajectory of each individual (Lynch and Kemp 2013). As recently shown in a longitudinal (3 months) home-cage observation of exploratory behavior of inbred mice, these individual trajectories increasingly diverge over time (Freund et al. 2013). Importantly, the present study shows that the eight common inbred strains differed in their sensitivity to these three sources of variation; some strains are more robust than others.
Of note, strain differences in maternal care (Carlier et al. 1982) are known to affect behavior of the offspring at later developmental stages; however, these factors would systematically impact on all offspring of a particular strain, and contribute to the plethora of strain differences in home-cage behavior (Loos et al. 2014), but would not specifically explain observed differences in phenotypic variation within strains.
There is evidence for mechanisms that buffer these three sources of within-strain variation. In plants and flies, there is clear evidence for buffering by the HSP90 pathway. Reduced HSP90 function impairs buffering of genetic and environmental variation, thereby increasing the penetrance of genetic variants to change the phenotype (Rutherford and Lindquist 1998; Queitsch et al. 2002; Sangster et al. 2008). In mice, reduced levels of two modifiers of epigenetic gene silencing (Dnmt3a and Trim28) increase within-strain variation gene expression and behavior (Whitelaw et al. 2010a). Thus, genetic differences between common inbred strains of mice in molecular chaperones or epigenetic modifiers might underlie the differences in phenotypic robustness described in the current study. The observation of differences in variability among strains suggests that some strains have superior mechanisms to buffer diversifying factors, and maintain a constant phenotype (i.e., C57BL/6J and BALB/c), whereas others show reduced phenotypic robustness (i.e., DBA/2J, 129S1/Sv A/J,and NOD/LtJ). A study on variation in gene expression has shown that segregating mouse lines are instrumental to identify loci underlying within-strain variability (Fraser and Schadt 2010). In fact, a recent study using different Drosophila inbred lines confirmed the idea that different genotypes vary dramatically in their propensity for variability, and that loci affecting variability could be mapped (Ayroles et al. 2015). We anticipate that systematic automated behavioral analysis panels of inbred or mutant mice in the home-cage could provide the throughput, multidimensionality and sensitivity for genetic dissection of phenotypic robustness in mice as well.
In conclusion, the present study in a panel of inbred mouse strains showed that, in addition to the previously described strain differences the response to environmental factors (G × E interaction), mouse strains also differ in phenotypic robustness. This may have implication for the choice of mouse strain in future behavioral genetics studies, in which within-strain variability is of great concern. Moreover, this conclusion could only be reached by measuring the behavior of multiple genetically identical individuals of the same genetic background, which is not possible in humans. Thus, these findings in mice might be exemplary for our thinking of how the human genome, by influencing phenotypic robustness, renders each individual more or less prone to subtle environmental perturbations.
References
Ayroles JF, Buchanan SM, O’Leary C et al (2015) Behavioral idiosyncrasy reveals genetic control of phenotypic variability. Proc Natl Acad Sci 112:201503830. doi:10.1073/pnas.1503830112
Bendesky A, Bargmann CI (2011) Genetic contributions to behavioural diversity at the gene-environment interface. Nat Rev Genet 12:809–820. doi:10.1038/nrg3065
Carlier M, Roubertoux P, Cohen-Salmon C (1982) Differences in patterns of pup care in Mus musculus domesticus l-Comparisons between eleven inbred strains. Behav Neural Biol 35:205–210
Crabbe JC, Wahlsten D, Dudek BC (1999) Genetics of mouse behavior: interactions with laboratory environment. Science 284:1670–1672
Debat V, David P (2001) Mapping phenotypes: canalization, plasticity and developmental stability. Trends Ecol Evol 16:555–561. doi:10.1016/S0169-5347(01)02266-2
Egan CM, Sridhar S, Wigler M, Hall IM (2007) Recurrent DNA copy number variation in the laboratory mouse. Nat Genet 39:1384–1389. doi:10.1038/ng.2007.19
Fraser HB, Schadt EE (2010) The quantitative genetics of phenotypic robustness. PLoS ONE 5:e8635. doi:10.1371/journal.pone.0008635
Freund J, Brandmaier AM, Lewejohann L et al (2013) Emergence of individuality in genetically identical mice. Science 340:756–759. doi:10.1126/science.1235294
Gärtner K (1990) A third component causing random variability beside environment and genotype. A reason for the limited success of a 30 year long effort to standardize laboratory animals? Lab Anim 24:71–77
Hegmann JP, Possidente B (1981) Estimating genetic correlations from inbred strains. Behav Genet 11:103–114
Hen I, Sakov A, Kafkafi N et al (2004) The dynamics of spatial behavior: how can robust smoothing techniques help? J Neurosci Methods 133:161–172. doi:10.1016/j.jneumeth.2003.10.013
Kaiser HF (1960) The application of electronic computers to factor analysis. Educ Psychol Meas 20:141–151
Kaminsky ZA, Tang T, Wang S-C et al (2009) DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet 41:240–245. doi:10.1038/ng.286
Lathe R (2004) The individuality of mice. Genes Brain Behav 3:317–327. doi:10.1111/j.1601-183X.2004.00083.x
Loos M, van der Sluis S, Bochdanovits Z et al (2009) Activity and impulsive action are controlled by different genetic and environmental factors. Genes Brain Behav 8:817–828. doi:10.1111/j.1601-183X.2009.00528.x
Loos M, Koopmans B, Aarts E et al (2014) Sheltering behavior and locomotor activity in 11 genetically diverse common inbred mouse strains using home-cage monitoring. PLoS ONE 9:e108563. doi:10.1371/journal.pone.0108563
Lynch KE, Kemp DJ (2013) Nature-via-nurture and unravelling causality in evolutionary genetics. Trends Ecol Evol. doi:10.1016/j.tree.2013.10.005
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer Associates, Sunderland
Maroteaux G, Loos M, van der Sluis S et al (2012) High-throughput phenotyping of avoidance learning in mice discriminates different genotypes and identifies a novel gene. Genes Brain Behav 11:772–784. doi:10.1111/j.1601-183X.2012.00820.x
Molenaar PC, Boomsma DI, Dolan CV (1993) A third source of developmental differences. Behav Genet 23:519–524
Pujadas E, Feinberg AP (2012) Regulated noise in the epigenetic landscape of development and disease. Cell 148:1123–1131. doi:10.1016/j.cell.2012.02.045
Queitsch C, Sangster TA, Lindquist S (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417:618–624. doi:10.1038/nature749
Queitsch C, Carlson KD, Girirajan S (2012) Lessons from model organisms: phenotypic robustness and missing heritability in complex disease. PLoS Genet 8:e1003041. doi:10.1371/journal.pgen.1003041
Rutherford SL, Lindquist S (1998) Hsp90 as a capacitor for morphological evolution. Nature 396:336–342. doi:10.1038/24550
Sangster TA, Salathia N, Undurraga S et al (2008) HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proc Natl Acad Sci U S A 105:2963–2968. doi:10.1073/pnas.0712200105
Taft RA, Davisson M, Wiles MV (2006) Know thy mouse. Trends Genet 22:649–653. doi:10.1016/j.tig.2006.09.010
Van Dongen J, Slagboom PE, Draisma HHM et al (2012) The continuing value of twin studies in the omics era. Nat Rev Genet 13:640–653. doi:10.1038/nrg3243
Watkins-Chow DE, Pavan WJ (2008) Genomic copy number and expression variation within the C57BL/6J inbred mouse strain. Genome Res 18:60–66. doi:10.1101/gr.6927808
Whitelaw NC, Chong S, Morgan DK et al (2010a) Reduced levels of two modifiers of epigenetic gene silencing, Dnmt3a and Trim28, cause increased phenotypic noise. Genome Biol 11:R111. doi:10.1186/gb-2010-11-11-r111
Whitelaw NC, Chong S, Whitelaw E (2010b) Tuning into noise: epigenetics and intangible variation. Dev Cell 19:649–650. doi:10.1016/j.devcel.2010.11.001
Acknowledgments
We thank Rolinka van der Loo for operating the PhenoTyper systems and Ruud Wijnands for assistance, Noldus Information Technology for supplying software free of charge and hardware at cost price, and Ben Loke, Cecilia Herrera, Raymond de Heer, and Willem van der Veer for development of hardware, software, and test scripts. This work was supported by Agentschap NL (NeuroBSIK Mouse Phenomics Consortium, BSIK03053), the Netherlands Organization for Scientific Research (NWO/MaGW: VIDI-452-12-014 to S.v.d.S). The authors declare no conflict of interest. M.L. and B.K. are full time employees of Sylics (Synaptologics BV), a private, VU University spin-off company that offers mouse phenotyping services using AHCODA™. A.B.S. and M.V. participate in a holding that owns Sylics shares and have received consulting fees from Sylics.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Maarten Loos, Bastijn Koopmans, Matthijs Verhage, and August B. Smit have been contributed equally to this work.
Collaborators: Neuro-BSIK Mouse Phenomics Consortium are Brussaard AB, Borst JGG, Elgersma Y, Galjart N, van der Horst GT, Levelt CN, Pennartz CM, Smit AB, Spruijt BM, Verhage M, de Zeeuw CI.
Data will be available on the Mouse Phenome Database (MPD) of Jackson laboratories, as well as on our own publically available website public.sylics.com.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Loos, M., Koopmans, B., Aarts, E. et al. Within-strain variation in behavior differs consistently between common inbred strains of mice. Mamm Genome 26, 348–354 (2015). https://doi.org/10.1007/s00335-015-9578-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-015-9578-7