Abstract
Objective
To compare the image quality and diagnostic performance between standard turbo spin-echo MRI and accelerated MRI with deep learning (DL)–based image reconstruction for degenerative lumbar spine diseases.
Materials and methods
Fifty patients who underwent both the standard and accelerated lumbar MRIs at a 1.5-T scanner for degenerative lumbar spine diseases were prospectively enrolled. DL reconstruction algorithm generated coarse (DL_coarse) and fine (DL_fine) images from the accelerated protocol. Image quality was quantitatively assessed in terms of signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR) and qualitatively assessed using five-point visual scoring systems. The sensitivity and specificity of four radiologists for the diagnosis of degenerative diseases in both protocols were compared.
Results
The accelerated protocol reduced the average MRI acquisition time by 32.3% as compared to the standard protocol. As compared with standard images, DL_coarse and DL_fine showed significantly higher SNRs on T1-weighted images (T1WI; both p < .001) and T2-weighted images (T2WI; p = .002 and p < 0.001), higher CNRs on T1WI (both p < 0.001), and similar CNRs on T2WI (p = .49 and p = .27). The average radiologist assessment of overall image quality for DL_coarse and DL_fine was higher on sagittal T1WI (p = .04 and p < .001) and axial T2WI (p = .006 and p = .01) and similar on sagittal T2WI (p = .90 and p = .91). Both DL_coarse and DL_fine had better image quality of cauda equina and paraspinal muscles on axial T2WI (both p = .04 for cauda equina; p = .008 and p = .002 for paraspinal muscles). Differences in sensitivity and specificity for the detection of central canal stenosis and neural foraminal stenosis between standard and DL-reconstructed images were all statistically nonsignificant (p ≥ 0.05).
Conclusion
DL-based protocol reduced MRI acquisition time without degrading image quality and diagnostic performance of readers for degenerative lumbar spine diseases.
Clinical relevance statement
The deep learning (DL)–based reconstruction algorithm may be used to further accelerate spine MRI imaging to reduce patient discomfort and increase the cost efficiency of spine MRI imaging.
Key Points
• By using deep learning (DL)–based reconstruction algorithm in combination with the accelerated MRI protocol, the average acquisition time was reduced by 32.3% as compared with the standard protocol.
• DL-reconstructed images had similar or better quantitative/qualitative overall image quality and similar or better image quality for the delineation of most individual anatomical structures.
• The average radiologist’s sensitivity and specificity for the detection of major degenerative lumbar spine diseases, including central canal stenosis, neural foraminal stenosis, and disc herniation, on standard and DL-reconstructed images, were similar.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Degenerative lumbar spine disease, the most common cause of chronic lower back pain and sciatica, encompasses a variety of conditions such as disc degeneration, lumbar central canal stenosis, annular fissure, and spondylolisthesis [1,2,3]. Patients with suspected degenerative lumbar spine diseases are initially managed with conservative treatment but may require further tests including a lumbar spine MRI if symptoms persist despite treatment [4, 5]. In these patients, a spine MRI is used to rule out other possible underlying conditions, assess the severity of degenerative spine diseases, and plan further management [1, 4, 6].
A typical noncontrast lumbar spine MRI protocol usually consists of spin-echo-based sagittal T1-weighted images (T1WI), sagittal T2-weighted images (T2WI), and axial T2-WIs and takes approximately 20 min on a high field strength machine [6, 7]. Due to its long scan time, a limited number of patients can undergo MRIs per given time, leading to decreased productivity per MRI scanner and increased cost of lumbar spine MRI [8]. Consequently, reducing lumbar spine MRI acquisition time while maintaining noninferior diagnostic performance can benefit patients by reducing discomfort associated with long MRI acquisition time and increasing cost efficiency by increasing productivity per MRI scanner [6, 8].
Compressed sensing in combination with parallel imaging is currently being utilized to increase MRI scan speed by reducing the amount of acquired data [9, 10]. Parallel imaging uses preestimated coil sensitivities to reconstruct images from multiple k-space data sampled from multichannel coils, while compressed sensing exploits data redundancy to reduce the sampling rate during image reconstruction [10, 11]. The major drawbacks of these methods are long computation time for iterative reconstruction, image blurring, and undersampling artifacts resulting from balancing data consistency with data sparsity [12, 13].
Recently, deep learning (DL)–based models such as U-net and variational networks have been proposed that can be used in conjunction with or as an alternative to preexisting methods for MRI scan acceleration [9, 14,15,16]. These models learn prior image representations from large amounts of data during training and infer missing information in undersampled k-space during inference [17]. DL-based image reconstruction methods may achieve higher quality for reconstructing images than traditional methods and can be used in real time due to reduced computational complexity during inference [9, 17].
In this study, we propose an accelerated lumbar spine MRI protocol utilizing a DL-based reconstruction algorithm for highly accelerated spin-echo data acquisition. We aimed to compare the image quality and diagnostic performance of readers for degenerative lumbar spine diseases between standard turbo spin-echo (TSE) MRI and accelerated MRI with DL-based image reconstruction.
Methods
This study was approved by the institutional review board of Seoul National University Hospital (IRB No.2103-174-1207). Written informed consent was obtained from all study participants.
Study population
Patients with chronic lower back pain or radiculopathy who visited Seoul National University Hospital (SNUH) from February 2022 to May 2022 were consecutively enrolled. The inclusion criteria were as follows: the patients (a) were aged ≥ 18, (b) had chronic lower back pain or radiculopathy, and (c) required lumbar spine MRI for further evaluation due to failed response to conservative treatment. Exclusion criteria were as follows: patients who had (a) any contraindication to MRI, including cardiac pacemaker implanted state or claustrophobia, (b) incomplete MR images, (c) MR images of suboptimal image quality, and (d) did not consent to participate.
Lumbar spine MRI examinations
The study participants underwent MRI examinations at 1.5-T scanners (Philips Ingenia and Siemens Avanto). All participants underwent both standard and accelerated lumbar spine MRI acquisition protocols, including sagittal TSE T1WI, sagittal TSE T2WI, and axial TSE T2WI. Imaging parameters of the sequences used in this study are detailed in Supplemental Table S1. Vendor-specific routine reconstruction and DL-based reconstruction algorithms were applied to raw data acquired with standard and accelerated protocols, respectively.
DL-based reconstruction algorithm
This study utilized commercially available DL-based MR image reconstruction software (SwiftMR v2.0.1.0, AIRS Medical). The algorithm is a variant of U-net, comprising 18 convolutional blocks, 4 max-pooling layers, 4 upsampling layers, 4 feature concatenations, and 3 convolutional layers incorporated in a cascading manner, with each layer enforcing data consistency (Fig. 1) [15]. Unlike its previous version, the algorithm used in this study operates only in the image domain, where undersampled DICOM images are used as input for the reconstruction of output DICOM images [9].
The model was trained and internally validated with 31865 series and 3540 series of MRIs, respectively. MRIs used for algorithm development were serially collected from multiple hospitals in South Korea for predefined time period and were mutually exclusive from the MRIs collected for this study. Images with specific findings were not intentionally included nor excluded from the training and internal validation sets. IRB approval and research agreement were obtained at each individual hospital prior to MRI data collection, and the data were anonymized before being used for reconstruction algorithm development.
Loss function was defined as the structural similarity index (SSIM) between the input and the label image, and the model was optimized with Adam over 20 epochs using a batch size of 4 at a learning rate of 10–3, decaying to 10–4 [18]. The network was trained using four NVIDIA Tesla V100 GPUs with 32 GB memory (NVIDIA Corporation). The DL algorithm generated coarse (DL_coarse) and fine (DL_fine) images based on the amount of denoising applied during training (1/1.42 and 1/1.52).
Quantitative image assessment
The image quality of DL-reconstructed images was quantitatively assessed using the signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR). SNR and CNR were computed according to the following formulas [19]: SNR = SIL1/2 disc/N, CNR =|(SIL1/2 disc − SIL1 bone marrow)|/N, where N is the noise defined as the standard deviation of the background signal intensity outside the patient.
Qualitative image assessment
Four radiologists (with 4, 4, 10, and 10 years of experience in radiology) participated as independent and blinded test readers. Prior to the actual test session, readers engaged in a training session where they evaluated three sample MRIs containing the same sequences as the test MRIs to increase their understanding of the test objective. The readers reviewed each patient’s MRI three times for a total of 150 spine MRIs in three separate sessions, with at least four weeks of time interval between each review [20, 21]. During each session, the MRIs of 50 patients, containing a mixture of standard, DL_coarse, and DL_fine images, were presented in a randomized crossover manner.
The readers were first instructed to qualitatively assess the image quality of the MRIs. For eight anatomical structures of the lumbar spine MRI (bone marrow, endplates, discs, cerebrospinal fluid, cauda equina, facet joints, neural foramina, and paraspinal muscles), each reader recorded the image quality of the anatomical structures on selected sequences using a 5-point scale (1: not visible or not distinguishable, 2: barely visible, 3: adequately visible, 4: good visibility, and 5: excellent visibility) [8, 22]. Bone marrow, endplates, and lumbar discs were assessed on sagittal T1WI and sagittal T2WI, CSF, facet, and cauda equina on sagittal and axial T2WI, neural foramina on sagittal T1WI, and paraspinal muscles on axial T2WI. The readers then evaluated the overall image impression on a 5-point scale (1: not acceptable or no diagnostic value, 2: very limited diagnostic value, 3: acceptable for most diagnoses, 4: good for the majority of diagnoses, 5: optimal) and the presence of artifacts on a 4-point scale (1: massive artifacts, 2: significant artifacts, 3: minimal artifacts, 4: no artifacts) [8].
In the second part of the test, the reader’s performance for the diagnosis of degenerative lumbar spine diseases was assessed [23]. The readers were instructed to evaluate the presence of disc abnormality (herniation or bulging), annular fissure, central canal stenosis, neural foraminal stenosis, Schmorl’s nodes, and spinal instability (spondylolisthesis or retrolisthesis) for each intervertebral or vertebral level and additionally assess the severity of central canal stenosis and neural foraminal stenosis (mild, moderate, and severe) [24,25,26,27,28].
Reference standard
Two senior neuroradiologists (S.H.C. and R.E.Y., with 20 and 12 years of experience in radiology, respectively) independently annotated all 50 lumbar spine MRIs for the presence of select degenerative spine diseases using the same grading scheme used in the reader test. To improve the accuracy of the labeling, the senior radiologists referred to all available clinical information during the radiologic evaluation. After resolving disagreements in annotations with a consensual review of the MRIs, the final labels were used as the reference standard.
Statistical analysis
SNR and CNR of DL-reconstructed images and standard images were compared using paired t-test with Bonferroni correction [29]. Overall image quality, individual structure image quality, and the presence of artifacts were compared using the Wilcoxon signed-rank test with Bonferroni correction [29]. The reader sensitivity and specificity for the diagnosis of select degenerative lumbar spine diseases in DL-reconstructed images and standard images were compared with generalized estimated equations. p < 0.05 was considered to be indicative of a significant difference for each statistical analysis. All statistical analyses were performed with R statistical software version 3.6.2 (R Project for Statistical Computing).
Results
Patient and image characteristics
A total of 51 consecutive patients with chronic lower back pain or radiculopathy who required lumbar spine MRI for further evaluation were initially included. One patient was excluded due to consent withdrawal, and the final study population consisted of 50 patients (mean age ± standard deviation, 69.3 years ± 11.1; range, 30–91 years) (Fig. 2). Fourteen (28%) and 36 (72%) patients underwent MRI examination at Avanto 1.5-T MRI scanner and Ingenia 1.5-T MRI scanner, respectively. Patient characteristics are summarized in Table 1.
Of 250 intervertebral levels evaluated for 50 patients, L2/3, L3/4, and L4/5 of one patient and L4/5 of two patients were excluded from the analysis of disc abnormality and annular fissure due to a history of posterior lumbar interbody fusion (PLIF) surgery or interbody cage with posterior instrumentation. Ninety-three percent [227/245], 33% [80/245], 30% [75/250], and 17% [42/250] of intervertebral levels were positive for disc abnormality, annular fissure, central canal stenosis, and spinal instability, respectively. Seventeen percent [85/500] of neural foramina and 38% [114/300] of vertebral levels were positive for neural foraminal stenosis and Schmorl’s nodes, respectively. The distribution of radiologic diagnoses is summarized in Table 1.
Lumbar spine MRI acquisition time
In the standard protocol, sagittal TSE T1WI, sagittal TSE T2WI, and axial TSE T2WI lasted 153.9 s and 166.6 s, 100.6 s and 211.9 s, and 157.2 s and 194.9 s, resulting in overall scan times of 411.74 s and 573.4 s on Philips Ingenia 1.5 T and Siemens Avanto 1.5 T, respectively. In the accelerated protocol, the sequences lasted 102.6 s and 93.2 s, 65.1 s and 165.8 s, and 104.8 s and 137.1 s, resulting in overall scan times of 272.5 s and 396.1 s, respectively. With the accelerated protocol, the total acquisition time was reduced by 33.8% and 30.9%, respectively, for an average reduction of 32.3% compared to the standard protocol.
Quantitative image assessment
SNR of both DL_coarse and DL_fine images were significantly higher on T1WI (p < 0.001 and p < 0.001) and T2WI (p = 0.002 and p < 0.001), as compared with the standard images. CNR of both DL_coarse and DL_fine images were significantly higher on T1WI (p < 0.001 and p < 0.001) but not on T2WI (p = 0.49 and p = 0.27), as compared with the standard images. The SNR and CNR values for standard and DL-reconstructed images are presented in Table 2. Subgroup analysis showed that for Siemens Avanto, SNR and CNR of DL_coarse and DL_fine were significantly higher for both T1WI and T2WI (all p < 0.05), while for Philips Ingenia, CNR of DL_fine was significantly higher on T1WI as compared with standard images (p = 0.021) (Supplemental Table S2).
Qualitative image assessment
The average radiologist assessment of overall image impression for both DL_coarse and DL_fine was higher on sagittal T1WI (4.1 ± 0.7 and 4.2 ± 0.7 vs. 4.0 ± 0.7; p = 0.04 and p < 0.001) and axial T2WI (4.1 ± 0.7 and 4.1 ± 0.8 vs. 4.0 ± 0.7; p = 0.006 and p = 0.01), and similar on sagittal T2WI (4.1 ± 0.7 and 4.1 ± 0.7 vs. 4.1 ± 0.7; p = 0.90 and p = 0.91), as compared with that for standard images (Table 3). The presence of artifacts was similar for both DL_coarse and DL_fine on sagittal T1WI (3.4 ± 0.7 and 3.4 ± 0.7 vs. 3.3 ± 0.7; p = 0.27 and p = 0.41), sagittal T2WI (3.5 ± 0.7 and 3.6 ± 0.7 vs. 3.6 ± 0.7; p = 0.71 and p = 1.00), and axial T2WI (3.5 ± 0.7 and 3.5 ± 0.7 vs. 3.5 ± 0.7; p = 0.97 and p = 0.44), as compared with that for standard images (Table 3).
The average radiologist assessment of the image quality of the endplate on sagittal T1WI, cauda equina on axial T2WI, and paraspinal muscles on axial T2WI were higher for both DL_coarse and DL_fine as compared with that for standard images (p = 0.003 and p = 0.01 for endplate, p = 0.04 and p = 0.04 for cauda equina, and p = 0.008 and p = 0.002 for paraspinal muscles). On the other hand, the image quality of bone marrow on sagittal T2WI was lower for both DL_coarse and DL_fine (p = 0.03 and p = 0.007). The image quality of the neural foramina on sagittal T1 was similar for DL_coarse and higher for DL_fine (p = 0.07 and p = 0.001). The image quality of all other anatomical structures on DL-reconstructed images was graded similarly to that of standard images (p > 0.05). A more detailed comparison of the image quality of individual anatomical structures can be found in Supplemental Table S3.
Reader test for diagnosis of degenerative lumbar spine diseases
The average sensitivity and specificity for the diagnosis of central canal stenosis were 0.81 and 0.88 for DL_coarse, 0.82 and 0.90 for DL_fine, and 0.83 and 0.88 for standard images, and the differences in sensitivity and specificity of DL_coarse and DL_fine vs. standard images were statistically nonsignificant (p = 0.29 and p = 0.85 for sensitivity; p = 0.46 and p = 0.07 for specificity) (Fig. 3, Table 4). Likewise, the average sensitivity and specificity for the diagnosis of neural foraminal stenosis were 0.75 and 0.97 for DL_coarse, 0.70 and 0.97 for DL_fine, and 0.74 and 0.96 for standard images, and the differences in sensitivity and specificity of DL_coarse and DL_fine vs. standard images were statistically nonsignificant (p = 0.51 and p = 0.06 for sensitivity; p = 0.70 and p = 0.68 for specificity) (Fig. 4).
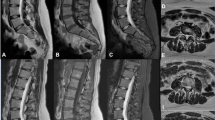
Sagittal T2WI, sagittal T1WI, and axial T2WI of a 57-year-old female who underwent an MRI due to radiculopathy. On the sagittal T2WI, bulging discs at L4/5 and L5/S1 levels are well depicted on DL-reconstructed images as well as on standard images. Similarly, on the sagittal T1WI, moderate neural foraminal stenosis at L5/S1 level is well visualized on both DL-reconstructed and standard images. Of note, on the axial T2WI, nerve roots, and paraspinal muscles appear sharper on DL-reconstructed images than on standard images
The average sensitivities were both higher on DL_coarse and DL_fine (p < 0.001 and p = 0.004), but specificities were both lower (both p < 0.001) for the detection of the annular fissure. The average specificities were both higher (p < 0.001 and p = 0.001) for the detection of Schmorl’s nodes. The average sensitivity was higher on DL_coarse (p = 0.02), and specificity was lower on DL_coarse (p = 0.04) for the detection of spinal instability. The diagnostic performances of DL-reconstructed images and standard images for other degenerative diseases, including the detection of disc abnormalities, were similar (p ≥ 0.05).
Discussion
We investigated the feasibility of using an accelerated MRI protocol with deep learning (DL)-based image reconstruction for imaging degenerative lumbar spine diseases. Our study demonstrated that using a DL-based reconstruction algorithm in combination with an accelerated MRI protocol represents a promising means to reduce scanning time without affecting image quality and reader performance for the diagnosis of major degenerative lumbar spine diseases.
A previous study by Sun S. et al showed that images processed with a DL-based reconstruction algorithm demonstrated significantly higher image quality and lower motion artifact while maintaining similar reader agreement for assessing degenerative lumbar spine diseases compared to those processed with standard reconstruction [30]. Our study further explored the value of the DL-based reconstruction algorithm by showing that MRI images obtained with accelerated protocol and processed with DL-based reconstruction have similar or better image quality, and a similar number of artifacts, despite the reduction in scan time for image acquisition.
Such results suggest that by leveraging pre-learned information about the underlying data distribution in the input image domain, the DL algorithm successfully reconstructs information that may have been lost during sparse data acquisition [31]. In that regard, the proposed DL algorithm for MRI reconstruction is analogous to the DL-based denoising algorithms for lower-dose scanning in CT, which learn to map high-noise images to the corresponding low-noise images while preserving key structural information; these DL-based CT denoising algorithms have already been proven to be effective methods for reducing noise while preserving the natural texture of the images [32,33,34].
During model training, high-resolution images were used to enable the DL algorithm’s image resolution enhancement effect. Such training enables the estimation of truncated high-frequency data in the image domain. This was reflected as both DL-reconstructed images having a superior performance for the characterization of cauda equina and paraspinal muscles on axial T2WI as compared with standard images, possibly due to better delineation of the boundary of nerve roots and increased sharpness of muscle fascicle and fascia resulting from increased resolution.
As the training dataset included MRIs collected from multiple vendors with different scan parameters, we expected the DL reconstruction algorithm to be vendor-neutral. Subgroup analysis comparing SNR and CNR of DL-reconstructed images and standard images for the two MRI scanners used showed that while there is a slight difference in the performance of the DL-based reconstruction algorithm for the two MRI instruments tested, similar SNR and CNR trend is observed. Such results suggest that the DL-based reconstruction algorithm learns representations of diverse noise patterns of the training dataset to produce high-quality images across different MRI scanners and acquisition parameters.
In the reader test for the diagnosis of degenerative lumbar spine diseases, the statistical differences in sensitivity and specificity for the diagnosis of disc abnormality, central canal stenosis, and neural foraminal stenosis between the DL-reconstructed images and standard image were nonsignificant. For other lesions, the sensitivity or specificity was significantly higher or lower in DL_coarse or DL_fine, but the absolute difference did not exceed 0.1. These results suggest that by using the proposed DL-based accelerated MRI protocol, MRI scan time can be reduced without a significant decrease in diagnostic performance for major degenerative lumbar spine diseases and that radiologists can select the degree of denoising for output images based on their preference.
Of note, sensitivity was significantly higher in both DL-reconstructed images, whereas specificity was significantly lower than that of the standard image for the detection of the annular fissure, which is a relatively subtle finding in spine MRI. This suggests that increased lesion visibility is associated with both better detection performance and may also be subject to overinterpretation. To minimize false positive readings, careful evaluation and customization of denoising levels should be conducted prior to the actual deployment of the DL-based protocol. In addition, routine monitoring of the quality of the DL-reconstructed images may minimize erroneous diagnosis.
The strengths of our study can be summarized as follows. First, although a few previous retrospective studies have tested the feasibility of DL-based MRI reconstruction methods for various MRI reconstruction tasks [29,30,31, 35], we are the first to prospectively evaluate the feasibility of the approach for the detection of degenerative lumbar spine diseases. We comprehensively compared the DL-reconstructed images with standard images using both quantitative and qualitative evaluation methods on all sequences included in the lumbar spine MRI protocol, including sagittal TSE T1WI, sagittal TSE T2WI, and axial TSE T2WI. Next, unlike other DL-based methods for MRI acceleration that require k-space data [9], whose information may be difficult to access for some vendors, the DL algorithm proposed in this study operates only in the image domain, so this approach can be generalized across multiple MRI instruments. In addition, pairs of blurred and high-resolution images were used to optimize the parameters of the CNN-based reconstruction algorithm, so we do not expect to introduce unseen structures into the images, as compared with generative adversarial networks that are used for image reconstruction. Yet, because DL-reconstructed images have higher SNRs and higher or similar CNRs, the original artifacts that are present in the images may be more emphasized, particularly for DL_fine images [36]. Finally, we demonstrated that the DL algorithm can reliably produce two types of output, suggesting that DL algorithms can readily be fine-tuned to produce output that fits user preference.
There are several limitations to this study. First, due to the limited number of enrolled patients, we could not test for the noninferiority of sensitivity and specificity for the detection of degenerative lumbar spine diseases. Second, because this is a single-center study, the patient population and the type of MRI scanners and scan parameters were limited, and future studies validating the algorithm in a multicenter setting are warranted. Third, due to the high prevalence of facet arthrosis in the study population, we could not reliably calculate reader specificity, and facet arthrosis was thus excluded from the reader performance test. Finally, in the actual clinical deployment of the protocol, patients may have incidental findings unrelated to degenerative lumbar spine diseases, such as spinal arteriovenous malformation (AVM) or spinal tumor, and the effect of the DL-based protocol on these incidental findings has not been demonstrated.
In conclusion, we have shown that by using a DL-based reconstruction method in combination with an accelerated MRI protocol, MRI acquisition time can be greatly reduced while achieving similar or higher SNR and CNR, similar or higher overall image quality, a similar number of artifacts, and similar reader sensitivity and specificity for the detection of major degenerative lumbar spine diseases. Our results demonstrate the potential of employing the DL reconstruction algorithm for further acceleration of spine MR imaging.
Abbreviations
- CNN:
-
Convolutional neural network
- CNR:
-
Contrast-to-noise ratio
- CSF:
-
Cerebrospinal fluid
- DL:
-
Deep learning
- SNR:
-
Signal-to-noise ratio
- SSIM:
-
Structural similarity index
- T1WI:
-
T1-weighted images
- T2WI:
-
T2-weighted images
- TSE:
-
Turbo spin-echo
References
Ravindra V, Senglaub S, Rattani A et al (2018) Degenerative lumbar spine disease: estimating global incidence and worldwide volume. Global Spine J 8:219256821877076. https://doi.org/10.1177/2192568218770769
Hoy D, Bain C, Williams G et al (2012) A systematic review of the global prevalence of low back pain. Arthritis Rheum 64:2028–2037. https://doi.org/10.1002/art.34347
Lurie J, Tomkins-Lane C (2016) Management of lumbar spinal stenosis. BMJ 352:h6234. https://doi.org/10.1136/bmj.h6234
Balagué F, Mannion AF, Pellisé F, Cedraschi C (2012) Non-specific low back pain. Lancet 379:482–491. https://doi.org/10.1016/S0140-6736(11)60610-7
Humphreys S, Eck J, Hodges S (2002) Neuroimaging in low back pain. Am Fam Physician 65:2299–2306
Sayah A, Jay AK, Toaff JS et al (2016) Effectiveness of a rapid lumbar spine MRI protocol using 3D T2-weighted SPACE imaging versus a standard protocol for evaluation of degenerative changes of the lumbar spine. AJR Am J Roentgenol 207:614–620. https://doi.org/10.2214/AJR.15.15764
Zanchi F, Richard R, Hussami M et al (2020) MRI of non-specific low back pain and/or lumbar radiculopathy: do we need T1 when using a sagittal T2-weighted Dixon sequence? Eur Radiol 30:2583–2593. https://doi.org/10.1007/s00330-019-06626-6
Bratke G, Rau R, Weiss K et al (2019) Accelerated MRI of the lumbar spine using compressed sensing: quality and efficiency. J Magn Reson Imaging 49:e164–e175. https://doi.org/10.1002/jmri.26526
Muckley MJ, Riemenschneider B, Radmanesh A et al (2021) Results of the 2020 fastMRI challenge for machine learning MR image reconstruction. IEEE Trans Med Imaging 40:2306–2317. https://doi.org/10.1109/TMI.2021.3075856
Yoon JH, Nickel MD, Peeters JM, Lee JM (2019) Rapid imaging: recent advances in abdominal MRI for reducing acquisition time and its clinical applications. Korean J Radiol 20:1597–1615
Ye JC (2019) Compressed sensing MRI: a review from signal processing perspective. BMC Biomed Eng 1:8. https://doi.org/10.1186/s42490-019-0006-z
Jaspan O, Fleysher R, Lipton M (2015) Compressed sensing MRI: a review of the clinical literature. Br J Radiol 88:20150487. https://doi.org/10.1259/bjr.20150487
Liang D, Liu B, Wang J, Ying L (2009) Accelerating SENSE using compressed sensing. Magn Reson Med 62:1574–1584. https://doi.org/10.1002/mrm.22161
Sriram A, Zbontar J, Murrell T et al (2020) End-to-End Variational Networks for Accelerated MRI Reconstruction. In: Martel AL, Abolmaesumi P, Stoyanov D et al (eds) Medical image computing and computer assisted intervention – MICCAI 2020. Springer International Publishing, Cham, pp 64–73
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Springer International Publishing, Cham, pp 234–241
Jung W, Kim J, Ko J et al (2022) Highly accelerated 3D MPRAGE using deep neural network–based reconstruction for brain imaging in children and young adults. Eur Radiol 32:5468–5479. https://doi.org/10.1007/s00330-022-08687-6
Zeng G, Guo Y, Zhan J et al (2021) A review on deep learning MRI reconstruction without fully sampled k-space. BMC Med Imaging 21:195. https://doi.org/10.1186/s12880-021-00727-9
Kingma D, Ba J (2015) Adam: a method for stochastic optimization. International Conference on Learning Representations, San Diego, CA, USA. https://arxiv.org/abs/1412.6980
Maeder Y, Dunet V, Richard R et al (2017) Bone marrow metastases: T2-weighted Dixon spin-echo fat images can replace T1-weighted spin-echo images. Radiology 286:948–959. https://doi.org/10.1148/radiol.2017170325
Jang S, Song H, Shin YJ et al (2020) Deep learning–based automatic detection algorithm for reducing overlooked lung cancers on chest radiographs. Radiology 296:652–661. https://doi.org/10.1148/radiol.2020200165
Yoo H, Lee SH, Arru CD et al (2021) AI-based improvement in lung cancer detection on chest radiographs: results of a multi-reader study in NLST dataset. Eur Radiol 31:9664–9674. https://doi.org/10.1007/s00330-021-08074-7
Nölte I, Gerigk L, Brockmann MA et al (2008) MRI of degenerative lumbar spine disease: comparison of non-accelerated and parallel imaging. Neuroradiology 50:403–409. https://doi.org/10.1007/s00234-008-0363-0
Hallinan JTPD, Zhu L, Yang K et al (2021) Deep learning model for automated detection and classification of central canal, lateral recess, and neural foraminal stenosis at lumbar spine MRI. Radiology 300:130–138. https://doi.org/10.1148/radiol.2021204289
Fujiwara A, Tamai K, Yamato M et al (1999) The relationship between facet joint osteoarthritis and disc degeneration of the lumbar spine: an MRI study. Eur Spine J 8:396–401. https://doi.org/10.1007/s005860050193
Lee S, Lee JW, Yeom JS et al (2010) A practical MRI grading system for lumbar foraminal stenosis. AJR Am J Roentgenol 194:1095–1098. https://doi.org/10.2214/AJR.09.2772
Li Y, Fredrickson V, Resnick DK (2015) How should we grade lumbar disc herniation and nerve root compression? A systematic review. Clin Orthop Relat Res 473:1896–1902. https://doi.org/10.1007/s11999-014-3674-y
Ko Y, Lee E, Lee JW et al (2020) Clinical validity of two different grading systems for lumbar central canal stenosis: Schizas and Lee classification systems. PLoS One 15:e0233633
Fardon DF, Williams AL, Dohring EJ et al (2014) Lumbar disc nomenclature: version 2.0: Recommendations of the combined task forces of the North American Spine Society, the American Society of Spine Radiology and the American Society of Neuroradiology. Spine J 14:2525–2545. https://doi.org/10.1016/j.spinee.2014.04.022
Ueda T, Ohno Y, Yamamoto K et al (2022) Deep learning reconstruction of diffusion-weighted MRI improves image quality for prostatic imaging. Radiology 303:373–381. https://doi.org/10.1148/radiol.204097
Sun S, Tan ET, Mintz DN et al (2022) Evaluation of deep learning reconstructed high-resolution 3D lumbar spine MRI. Eur Radiol 32:6167–6177. https://doi.org/10.1007/s00330-022-08708-4
Recht MP, Zbontar J, Sodickson DK et al (2020) Using deep learning to accelerate knee MRI at 3 T: results of an interchangeability study. AJR Am J Roentgenol 215:1421–1429. https://doi.org/10.2214/AJR.20.23313
Yeoh H, Hong SH, Ahn C et al (2021) Deep learning algorithm for simultaneous noise reduction and edge sharpening in low-dose CT images: a pilot study using lumbar spine CT. Korean J Radiol 22:1850–1857
Mohammadinejad P, Mileto A, Yu L et al (2021) CT noise-reduction methods for lower-dose scanning: strengths and weaknesses of iterative reconstruction algorithms and new techniques. Radiographics 41:1493–1508. https://doi.org/10.1148/rg.2021200196
Shan H, Padole A, Homayounieh F et al (2019) Competitive performance of a modularized deep neural network compared to commercial algorithms for low-dose CT image reconstruction. Nat Mach Intell 1:269–276. https://doi.org/10.1038/s42256-019-0057-9
Jung W, Kim EH, Ko J et al (2022) Convolutional neural network-based reconstruction for acceleration of prostate T2 weighted MR imaging: a retro- and prospective study. Br J Radiol 95:20211378. https://doi.org/10.1259/bjr.20211378
Yi X, Walia E, Babyn P (2019) Generative adversarial network in medical imaging: a review. Med Image Anal 58:101552. https://doi.org/10.1016/j.media.2019.101552
Acknowledgements
We thank Yunhee Choi at the Division of Medical Statistics, Medical Research Collaborating Center, Seoul National University Hospital for providing advice on statistical interpretation of the data.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (NRF-2021R1A4A1028713 and NRF-2023R1A2C3003250) and the Korea Medical Device Development Fund grant funded by the Korean government (the Ministry of Science and ICT, South Korea; the Ministry of Trade, Industry and Energy; the Ministry of Health and Welfare, Republic of Korea; and the Ministry of Food and Drug Safety) (project no. 9991007218, KMDF_PR_20200901_0086). This study received technical support and a research grant (grant number 06- 2021–2210) from AIRS Medical Inc.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Roh-Eul Yoo, Seoul National University Hospital.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Roh-Eul Yoo is a Junior Deputy Editor of European Radiology. She has not taken part in the review or selection process of this article.
Statistics and biometry
Yunhee Choi at the Division of Medical Statistics, Medical Research Collaborating Center, Seoul National University Hospital, kindly provided statistical advice for this manuscript.
Informed consent
Written informed consent was obtained from all subjects in this study.
Ethical approval
This study was approved by the institutional review board of Seoul National University Hospital (IRB No.2103-174-1207).
Study subjects or cohorts overlap
None
Methodology
• prospective
• diagnostic study
• performed at one institution
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yoo, H., Yoo, RE., Choi, S.H. et al. Deep learning–based reconstruction for acceleration of lumbar spine MRI: a prospective comparison with standard MRI. Eur Radiol 33, 8656–8668 (2023). https://doi.org/10.1007/s00330-023-09918-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-023-09918-0