Abstract
Objective
To establish a radiomics nomogram based on dynamic contrast-enhanced (DCE) MR images to preoperatively differentiate combined hepatocellular-cholangiocarcinoma (cHCC-CC) from mass-forming intrahepatic cholangiocarcinoma (IMCC).
Methods
A total of 151 training cohort patients (45 cHCC-CC and 106 IMCC) and 65 validation cohort patients (19 cHCC-CC and 46 IMCC) were enrolled. Findings of clinical characteristics and MR features were analyzed. Radiomics features were extracted from the DCE-MR images. A radiomics signature was built based on radiomics features by the least absolute shrinkage and selection operator algorithm. Univariate and multivariate analyses were used to identify the significant clinicoradiological variables and construct a clinical model. The radiomics signature and significant clinicoradiological variables were then incorporated into the radiomics nomogram by multivariate logistic regression analysis. Performance of the radiomics nomogram, radiomics signature, and clinical model was assessed by receiver operating characteristic and area under the curve (AUC) was compared.
Results
Eleven radiomics features were selected to develop the radiomics signature. The radiomics nomogram integrating the alpha fetoprotein, background liver disease (cirrhosis or chronic hepatitis), and radiomics signature showed favorable calibration and discrimination performance with an AUC value of 0.945 in training cohort and 0.897 in validation cohort. The AUCs for the radiomics signature and clinical model were 0.848 and 0.856 in training cohort and 0.792 and 0.809 in validation cohort, respectively. The radiomics nomogram outperformed both the radiomics signature and clinical model alone (p < 0.05).
Conclusion
The radiomics nomogram based on DCE-MRI may provide an effective and noninvasive tool to differentiate cHCC-CC from IMCC, which could help guide treatment strategies.
Key Points
• The radiomics signature based on dynamic contrast-enhanced magnetic resonance imaging is useful to preoperatively differentiate cHCC-CC from IMCC.
• The radiomics nomogram showed the best performance in both training and validation cohorts for differentiating cHCC-CC from IMCC.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Combined hepatocellular-cholangiocarcinoma (cHCC-CC) is believed to arise from hepatic progenitor cells with the expression of both biliary and hepatocellular markers [1, 2], which accounts for 0.4–14.2% of primary liver malignancies [2, 3]. In contrast, intrahepatic cholangiocarcinoma (ICC) is the second most common primary liver malignancy after hepatocellular carcinoma (HCC), which arises from intrahepatic biliary epithelium [4, 5].
CHCC-CC is gaining increasing attention clinically and pathologically, due to its unique biology, histopathology, and clinical behavior, despite being rare. Surgery remains the mainstay of treatment for the majority of patients with resectable primary liver malignancies [3]. Previous studies reported that the survival rates after resection of cHCC-CC appeared to be worse than those of HCC and similar to those of ICC [2, 6]. In addition, Machairas et al reported that long-term results of liver transplantation in the setting of cHCC-CC were associated with fairly unfavorable overall outcomes compared to HCC yet were improved compared to ICC [7]. Therefore, the ability to diagnose cHCC-CC accurately before surgery is of great value, and it may play a critical role in further treatment guidance.
In current clinical practice, computed tomography (CT) and magnetic resonance imaging (MRI) have been widely used for detection and characterization of liver tumors [8, 9]. However, it is still extremely difficult to preoperatively diagnose cHCC-CC by imaging studies, due to the complex imaging features of the two entities and the rarity of this tumor [3]. The imaging characteristics of cHCC-CC include not only features typical of HCC, but also those of ICC [3, 10]. Imaging misdiagnosis has been reported in almost two-thirds of patients with cHCC-CC [11]. Liver-specific MR contrast agents such as gadoxetic acid are increasingly used in the evaluation of focal liver lesions because of their dual actions, namely as extracellular contrast agent and hepatobiliary agent [12, 13]. Hwang et al reported that gadoxetic acid-enhanced MRI was valuable for differentiation between cHCC-CC and mass-forming ICC [14]. But its value is mainly shown in the diagnosis of HCC-predominant cHCC-CC [14]. Although using needle biopsy is a minimally invasive and a rapid method of pathologic evaluation of hepatic masses, its clinical application is limited due to increasing the potential risk of needle tract seeding and hematogenous dissemination [15]. Thus, the best choice is still to diagnose cHCC-CC by as noninvasive a technique as possible.
Radiomics can provide important information on the entire underlying intra-tumor heterogeneity and cancer phenotype by extracting numerous quantitative features from radiologic images [16]. Previous studies have demonstrated that radiomics has favorable abilities to help characterize lesions, assess tumors, and evaluate patients’ responses to treatment in the liver [17,18,19,20]. Mokrane et al reported that radiomics could be used to noninvasively diagnose HCC in cirrhotic patients with indeterminate liver nodules [18]. Li et al built a new radiomics model for the differential diagnosis of HCC and hepatic metastasis of rectal cancer [19]. Therefore, it is conceivable that radiomics on MRI has potential to help distinguish between cHCC-CC and ICC.
This study focused on the differentiation of cHCC-CC from mass-forming intrahepatic cholangiocarcinoma (IMCC), because previous studies reported that imaging features of cHCC-CC appeared to more closely resemble ICC rather than HCC [21,22,23]. Thus, the aim of our study was to develop a radiomics nomogram based on dynamic contrast-enhanced (DCE) MR images for preoperative differentiation of cHCC-CC from IMCC.
Materials and methods
This retrospective study was approved by the institutional review board and the requirement for informed consent was waived.
Patients
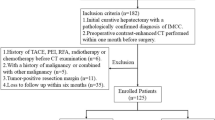
This study was conducted in patients with pathologically proven cHCC-CC or IMCC at surgery between June 2015 and September 2020 at the Zhongshan Hospital, Shanghai. The inclusion criteria were as follows: (1) preoperative liver dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) in the Zhongshan Hospital, Shanghai, within 2 weeks before surgery; (2) patients with single cHCC-CC or single IMCC; (3) lesion size ≥ 1cm; (4) no history of previous treatment for liver lesion; (5) available MR imaging including optimal image quality for our analysis.
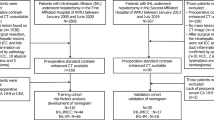
The final cohort consisted of 216 patients (64 cHCC-CC and 152 IMCC) and was randomly divided into a training cohort (45 cHCC-CC and 106 IMCC) and a validation cohort (19 cHCC-CC and 46 IMCC) with a proportion of 7:3. Flowchart of patients’ collection and scheme for analysis is shown in Fig. 1.
Clinical data and pathology
The clinical data including demographic, preoperative tumor markers (AFP, CEA, CA19-9), background liver disease (cirrhosis or chronic hepatitis), and pathologic information were obtained from medical records.
MRI data acquisition
MRI examinations were performed with a 1.5-T (uMR 560; United Imaging Healthcare) scanner. The conventional MR protocol included transverse breath-hold T2-weighted with fat-suppression turbo spin echo sequence (2693/85.58 repetition time/echo time (TR/TE), 201 × 288 matrix, 6 mm section thickness, 380 mm × 360 mm field of view (FOV)) and transverse T1-weighted breath-hold in-phase and out-of-phase gradient echo sequence (115.8/4.4 (in-phase), 2.2 (out-of-phase) TR/TE, 230 × 288 matrix, 6 mm section thickness, 380 mm × 390 mm FOV). A breath-hold single-shot echo-planar diffusion-weighted imaging (DWI) was performed with b values of 0, 500 mm2/s (2807/75.7 TR/TE, 115 × 128 matrix, 6 mm section thickness, 380 mm × 300 mm FOV). For dynamic MRI, the following images were obtained using a breath-hold 3D T1-weighted with fat-suppression fast spoiled gradient echo sequence (4.43/2.2 TR/TE, 192 × 256 matrix, 3 mm section thickness, 400 mm × 280 mm FOV): unenhanced phase, arterial phase (AP, 20–35 s), portal phase (PP, 70–90 s), and delayed phase (DP, 180 s). A total dose of 0.2 mL/kg gadopentetate dimeglumine (Gd-DTPA) was administered intravenously using a power injector at a rate of 2 mL/s, followed by a 20-mL saline flush.
Radiologic evaluation
Two abdominal radiologists (Y.Z. and G.F.Z. with 9 and 16 years of experience, respectively) retrospectively reviewed the MR images together on PACS, who were blinded to pathological outcomes and clinical findings. If disagreements occurred, a third senior radiologist (P.J.X. with 27 years of experience) would resolve their differences. MR features of each lesion were evaluated as follows: (1) tumor margin (well-defined or irregular); (2) enhancement pattern on the arterial phase were registered as follows: (a) rim enhancement: hyperenhancement limited to the periphery of the lesion, involving ≤ 25% of its area; (b) partial enhancement: hyperenhancement involving 25–75% of the lesion; (c) global enhancement: hyperenhancement involving > 75% of the lesion; (3) target sign, defined as peripheral diffusion restriction with central isointensity/hypointensity on DWI image; (4) dynamic enhancement pattern: (A) progressive: the tumor progressively enhanced over time, centripetal enhancement was contained; (B) persistent: the intensity or range of enhancement remained unchanged on all three phases; (C) wash in with wash out: arterial hyperenhancement followed by hypoenhancement on portal or delayed phases; (D) degressive (wash in without wash out): decreasing hyperintensity over time with no contrast agent wash out on portal or delayed phases; (5) biliary dilation; (6) hepatic capsule retraction; (7) tumor size: defined as maximum tumor diameter on transverse MR images on the delayed phase.
Radiomics analysis of MR Images
Workflow
The workflow of a typical radiomics process in our study included tumor segmentation, feature extraction, feature selection, and model construction and evaluation (Fig. 2).
Tumor segmentation and radiomics feature extraction
Three-dimensional manual segmentation was performed by reader 1 (Y.Z. with 9 years of work experience) using ITK-SNAP v.3.6.0 from UPenn (www.itksnap.org). Volumes of interests (VOIs) were manually drawn along the visible borders of tumor on the dynamic arterial, portal, and delayed phase images. In total, 788 radiomics features were extracted from each VOI using an in-house software written in Python (Pyradiomics version 2.12; https://pyradiomics.readthedocs.io/en/2.1.2/), giving a total of 2364 features for each patient (tumors in the arterial, portal, and delayed phases). The radiomics features extracted included shape (n = 14), first-order (n = 18), textural features (n = 68), and wavelet features (n = 688). Information on the feature extraction is detailed in Supplemental Table 1.
To explore the reproducibility of radiomics features, a cohort of 40 lesions were randomly chosen and two radiologists (reader 1; reader 2, G.F.Z. with 16 years of work experience) independently performed repeat segmentation. Then, the intra- and interclass correlation coefficients (ICC) were calculated, and values > 0.8 were considered almost perfect agreement.
Radiomics feature selection and signature construction
All the data of radiomics features were transformed into z-score normalization to reduce the bias caused by different index dimensions. Then, feature selection was carried out in the following three steps. First, features with greater ICC than 0.8 were kept in for further analysis [24]. Second, Pearson’s correction coefficients were calculated to examine redundant and collinear features, and features with mutual correlation coefficients > 0.9 were removed [25]. Third, the least absolute shrinkage and selection operator (LASSO) method with 10-fold cross-validation was applied to select most optimal features and construct a radiomics signature [25, 26]. The radiomics signature was calculated as the Radscore for quantification via a linear combination of selected features with their corresponding weights [25, 26].
Clinical model construction
The univariate analysis was used to assess the difference of clinicoradiological characteristics between cHCC-CC and IMCC in the training cohort. Then, variables with p < 0.05 in univariate analysis were applied to a multivariate logistic regression analysis to elucidate the independent factors. Meanwhile, the clinical model was built on the basis of these independent factors.
Radiomics nomogram model construction
The novel radiomics nomogram model incorporating independent clinicoradiological factors and radiomics signature (Radscore) was constructed by using multivariable logistic regression analysis in the training set. Backward stepwise selection was employed with the Akaike information criterion (AIC) as the stopping rule. The model with the minimum AIC score generated the final combination. Calibration curves of the nomogram were plotted to assess the consistency between prediction and observation, accompanied by the Hosmer-Lemeshow test.
Performance and validation of models
The discrimination performance of the above three models was evaluated by the receiver operating characteristic curve (ROC) and area under the ROC curve (AUC) was compared using the Delong test. The corresponding sensitivity, specificity, and accuracy were also calculated. Internal validation of models was performed using an independent validation.
Statistical analysis
The chi-square test or Fisher exact test was used for categorical variables. Continuous variables were compared by using the Mann-Whitney U test or Student t test, when appropriate. All statistical analyses were performed using SPSS (version 21.0; IBM) and R software (Version 3.6.1). A two-sided p value < 0.05 was considered statistically significant.
Results
Comparisons of demographics, clinical parameters, and radiologic features are summarized in Table 1. No statistical difference in the ratio of cHCC-CC (45/151 vs 19/65, p = 0.933) was observed between training and validation cohorts. Univariate analysis indicated that age, sex, AFP, and background liver disease were significantly different between cHCC-CC and IMCC in training cohort (p < 0.05). At the multivariate analysis, only AFP and background liver disease were independent factors to differentiate cHCC-CC from IMCC (Table 2). The AUCs of clinical model constructed with the two independent factors were 0.856 (95% CI: 0.790, 0.908) in the training cohort and 0.809 (95% CI: 0.693, 0.896) in the validation cohort (Table 3). The formula for clinical model is described in Table 4.
Feature selection and radiomics signature construction
Among 2364 extracted radiomics features, 2166 features showed high stability (Supplemental Fig. 1). Of these features, 478 features were identified as independent after Pearson’s correlation analysis. The further analysis of LASSO regression was used to select 11 features to derive a radiomics signature in the training cohort (Fig. 3). The formula for the radiomics signature (Radscore) is described in Table 4. The details of the 11 selected features are presented in Supplemental Table 2. In general, cHCC-CC showed a significantly higher Radscore than IMCC in the training cohort (−0.386 ± 0.593 vs −1.151 ± 0.483, p < 0.001), and then was confirmed in the validation cohort (−0.554 ± 0.424 vs −1.075 ± 0.571, p = 0.001) (Table 1, Supplemental Fig. 2). The radiomics signature yields an AUC of 0.848 (95% CI: 0.780, 0.901) in the training cohort and 0.792 (95% CI: 0.673, 0.883) in the validation cohort (Table 3).
The least absolute shrinkage and selection operator (LASSO) regression for radiomics features selection and signature construction. a In the LASSO model, the penalization parameter λ selection used 10-fold cross-validation as the minimum criteria. The log (λ) (x-axis) was plotted against the partial likelihood deviance (y-axis). The minimum criteria and the 1−SE criteria were used to draw dotted vertical lines. As a result, an optimal λ of 0.064, with log (λ) = −2.745 was chosen (minimum criteria). b LASSO coefficient profiles of the radiomics features. Tenfold cross-validation in the log (λ) sequence was used to draw the vertical line at the value selected; also indicated are 11 features with nonzero coefficients
Radiomics nomogram model construction
The final model integrating independent clinicoradiological factors and radiomics signature obtained the best performance for differentiation of cHCC-CC from IMCC with an AUC of 0.945 (95% CI: 0.896, 0.975) in the training cohort and 0.897 (95% CI: 0.796, 0.959) in the validation cohort (Table 3). The formula for radiomics nomogram model is described in Table 4. The model is presented as the nomogram in Fig. 4a. Favorable calibrations of the nomogram were obtained in both the training and validation cohorts (Fig. 4b, c). Hosmer-Lemeshow test yields a p value of 0.455 and 0.373, respectively.
The radiomics nomogram incorporating the AFP level, background liver disease, and radiomics signature (Radscore) (a). Good calibration of the radiomics nomogram is shown in the training (b) and in the validation (c) cohorts. Diagnostic performance of the the clinical model, radiomics signature, and radiomics nomogram was assessed and compared through ROC curves in both the training (d) and validation (e) cohorts
Diagnostic performance comparisons by ROC between three different models in the training and validation cohorts are given in Fig. 4d and e. The radiomics nomogram model outperformed either the radiomics signature or clinical model in the training cohort (AUC: 0.945 vs 0.848, 0.856; p = 0.001, < 0.001), and then was confirmed in the validation cohort (AUC: 0.897 vs 0.792, 0.809; p = 0.045, 0.037). However, the Delong test illustrated that there was no significant difference between the radiomics signature and clinical model in both training (p = 0.857) and validation (p = 0.842) cohorts.
Discussion
In this work, we sought to establish a novel radiomics nomogram incorporating AFP, background liver disease, and the developed radiomics signature of DCE-MRI for preoperative differentiation of cHCC-CC from IMCC. The combined model achieved a satisfactory discrimination performance and showed a favorable reliability.
As far as we know, only few studies have been reported in the literature including radiomics-based methods for diagnosis of cHCC-CC [27, 28]. Zhang et al built a new CT-based radiomics model to differentiate cHCC-CC from ICC preoperatively, with an AUC of 0.942 [27]. They also found that radiomics features extracted from the peritumoral area had the potential for differentiation of cHCC-CC from IMCC; however, no statistical significant difference could be observed compared with radiomics features inside the tumor [27]. A study by Liu et al showed that radiomics features extracted from gadoxetic acid-enhanced MR images demonstrated the best performance for differentiation of cHCC-CC from non-cHCC-CC with the highest AUC of 0.770, while CT was of limited value [28]. The weakness of their study is lack of separate training and validation sets, and it did not contain relevant clinical factors. According to previous reports, contrast-enhanced imaging is often used in radiomics analyses of tumors and is beneficial to help highlight vascularity and spatial heterogeneity [28, 29]. Liu et al’s study demonstrated that radiomics analysis in non-contrast MRI sequences appeared to have less value compared with contrast MRI sequences to differentiate cHCC-CC from other primary liver tumors [28]. This may reflect that essential differences in enhancement patterns between cHCC-CC and non-cHCC-CC tumors were helpful to make a correct diagnosis. Therefore, we tried to construct a novel radiomics nomogram of good reliability and investigate the potential value of MRI radiomics analysis to differentiate cHCC-CC from IMCC before surgery.
The radiomics signature consisting of 11 radiomics features in our study was able to predict the classification of cHCC-CC vs IMCC with acceptable performance in the training (AUC of 0.848) and validation (AUC of 0.792) cohorts. Consistent with previous studies, shape and wavelet features were included in our radiomics signature. Shape features were often extracted to provide a quantitative description of the physical appearance of the tumors, surface irregularity, and eccentricity, which could be useful for differential diagnosis of tumors [30,31,32]. Yap et al’s study indicated that shape metrics alone could achieve high prediction performance and hold high variable importance in the combined shape and texture radiomics model to discriminate benign from malignant renal masses [31]. Cuocolo et al found that the radiomics shape features derived from MRI index lesion could be potential imaging biomarkers for detecting clinically significant prostate cancer (csPCa) [32]. Wavelet features are extracted from the images transformed by wavelet filter (which can decompose special patterns hidden in mass of data) [33]. Some previous studies have suggested that wavelet features may better explore tumor biology and heterogeneity [34,35,36]. Liang et al reported that wavelet features were of great value to predict early recurrence of ICC after partial hepatectomy [34]. A study by Zhou et al showed that a radiomics signature built via 7 wavelet features extracted from MR imaging could predict MVI of IMCC [35]. We assumed that differences of these wavelet features in cHCC-ICC and IMCC might be related to the differences in their histopathological tumor composition (such as tumor cellularity, fibrosis, necrosis, and occasionally mucin). However, it is still challenging to figure out the association between a single radiomics feature and complex tumor biological processes [37].
In line with previous studies, AFP and background liver disease were independent predictors for cHCC-CC, which suggested that patients of cHCC-CC are much more likely to have higher AFP level or background liver disease than those of IMCC [10, 27, 38]. In clinical practice, these predictors can be easily detected and incorporated into a nomogram for differential diagnosis of cHCC-CC and IMCC. In addition, we discovered that cHCC-CC was more often in male and in people under the age of 60, compared with IMCC; our results were in accordance with earlier studies [38, 39]. For radiologic features, we found that enhancement pattern on the arterial phase, dynamic enhancement pattern, target sign, biliary dilation, and hepatic capsule retraction were significantly different between cHCC-CC and IMCC in training cohort. These findings were partly close to previous studies [14, 22]. However, these radiologic features were not independent factors after multivariate analysis and were excluded in our nomogram.
Finally, we constructed a radiomics nomogram in combination of clinical factors (serum AFP level and background liver disease) and the radiomics signature. ROC analyses revealed that the nomogram provided the best performance for differentiation of cHCC-CC from IMCC, which outperformed either the radiomics signature or clinical model in both training set and validation cohort. Meanwhile, there were no statistically significant differences in diagnosis performance between clinical model and the radiomics signature in both training set and validation cohort. These findings were partly close to a study by Zhang et al, where a CT-based radiomics nomogram for differentiation of cHCC-CC from ICC reached the highest AUC than other single single-factor models [27].
Limitation
We do acknowledge several limitations in this study. First, because of retrospective nature of this study, it is more susceptible to potential selection bias. Second, the sample size of patient cohorts, especially the validation cohort of 19 patients with cHCC-CC, was relatively small. Third, our results were from a single-center study and further prospective multicenter studies are required to obtain high-level evidence for clinical application. Fourth, we established the models based on patients with only single lesion; thus, the results cannot be extended to patients with multiple lesions. Last, we did not investigate the value of radiomics for prognosis prediction in cHCC-CC and IMCC, as the follow-up data for patients in the study is still being collected. It will be interesting and useful to explore this issue in further research.
Conclusion
To summarize, this study presents a radiomics nomogram that incorporates both the radiomics signature and clinical factors, and can be a noninvasive and promising tool for preoperative differentiation of cHCC-CC from IMCC.
Abbreviations
- ADC:
-
Apparent diffusion coefficient
- AFP:
-
Alpha fetoprotein
- AP:
-
Arterial phase
- AUC:
-
Area under curve
- CA19-9:
-
Cancer antigen 19-9
- CHCC-CC:
-
Combined hepatocellular-cholangiocarcinoma
- CT:
-
Computed tomography
- DP:
-
Delayed phase
- HCC:
-
Hepatocellular carcinoma
- ICC:
-
Intrahepatic cholangiocarcinoma
- IMCC:
-
Mass-forming intrahepatic cholangiocarcinoma
- LASSO:
-
Least absolute shrinkage and selection operator
- MRI:
-
Magnetic resonance imaging
- PP:
-
Portal phase
- ROC:
-
Receiver operating characteristic curve
References
Ogasawara S, Akiba J, Nakayama M, Nakashima O, Torimura T, Yano H (2015) Epithelial cell adhesion molecule-positive human hepatic neoplastic cells: development of combined hepatocellular-cholangiocarcinoma in mice. J Gastroenterol Hepatol 30:413–420
BeaufrèeaufrrCalderaro J, Paradis V (2021) Combined hepatocellular-cholangiocarcinoma: An update. J Hepatol 74:1212–1224
Weber SM, Ribero D, O'Reilly EM, Kokudo N, Miyazaki M, Pawlik TM (2015) Intrahepatic cholangiocarcinoma: expert consensus statement. HPB (Oxford) 17:669–680
Bergquist A, von Seth E (2015) Epidemiology of cholangiocarcinoma. Best Pract Res Clin Gastroenterol 29:221–232
Sempoux C, Jibara G, Ward SC et al (2011) Intrahepatic cholangiocarcinoma: new insights in pathology. Semin Liver Dis 31:49–60
Tang Y, Wang L, Teng F, Zhang T, Zhao Y, Chen Z (2021) The clinical characteristics and prognostic factors of combined hepatocellular carcinoma and cholangiocarcinoma, hepatocellular carcinoma and intrahepatic cholangiocarcinoma after surgical resection: a propensity score matching analysis. Int J Med Sci 18:187–198
Machairas N, Stamopoulos P, Kostakis ID et al (2019) Mixed hepatocellular cholangiocarcinoma: a review of long-term outcomes following liver transplantation. Transplant Proc 51:437–439
Hori M, Murakami T, Kim T, Tomoda K, Nakamura H (2004) CT scan and MRI in the differentiation of liver tumors. Dig Dis 22:39–55
Donato H, França M, Candelária I, Caseiro-Alves F (2017) Liver MRI: from basic protocol to advanced techniques. Eur J Radiol 93:30–39
Wang Y, Yang Q, Li S, Luo R, Mao S, Shen J (2019) Imaging features of combined hepatocellular and cholangiocarcinoma compared with those of hepatocellular carcinoma and intrahepatic cholangiocellular carcinoma in a Chinese population. Clin Radiol 74:407.e401–407.e410
Nishie A, Yoshimitsu K, Asayama Y et al (2005) Detection of combined hepatocellular and cholangiocarcinomas on enhanced CT: comparison with histologic findings. AJR Am J Roentgenol 184:1157–1162
Thian YL, Riddell AM, Koh DM (2013) Liver-specific agents for contrast-enhanced MRI: role in oncological imaging. Cancer Imaging 13:567–579
Reimer P, Schneider G, Schima W (2004) Hepatobiliary contrast agents for contrast-enhanced MRI of the liver: properties, clinical development and applications. Eur Radiol 14:559–578
Hwang J, Kim YK, Park MJ et al (2012) Differentiating combined hepatocellular and cholangiocarcinoma from mass-forming intrahepatic cholangiocarcinoma using gadoxetic acid-enhanced MRI. J Magn Reson Imaging 36:881–889
Wee A (2011) Fine needle aspiration biopsy of hepatocellular carcinoma and hepatocellular nodular lesions: role, controversies and approach to diagnosis. Cytopathology 22:287–305
Yip SS, Aerts HJ (2016) Applications and limitations of radiomics. Phys Med Biol 61:R150–R166
Wei J, Jiang H, Gu D et al (2020) Radiomics in liver diseases: current progress and future opportunities. Liver Int 40:2050–2063
Mokrane FZ, Lu L, Vavasseur A et al (2020) Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol 30:558–570
Li J, Xue F, Xu X, Wang Q, Zhang X (2020) Dynamic contrast-enhanced MRI differentiates hepatocellular carcinoma from hepatic metastasis of rectal cancer by extracting pharmacokinetic parameters and radiomic features. Exp Ther Med 20:3643–3652
Wu J, Liu A, Cui J, Chen A, Song Q, Xie L (2019) Radiomics-based classification of hepatocellular carcinoma and hepatic haemangioma on precontrast magnetic resonance images. BMC Med Imaging 19:23
Potretzke TA, Tan BR, Doyle MB, Brunt EM, Heiken JP, Fowler KJ (2016) Imaging features of biphenotypic primary liver carcinoma (hepatocholangiocarcinoma) and the potential to mimic hepatocellular carcinoma: LI-RADS analysis of CT and MRI features in 61 cases. AJR Am J Roentgenol 207:25–31
Sammon J, Fischer S, Menezes R et al (2018) MRI features of combined hepatocellular-cholangiocarcinoma versus mass forming intrahepatic cholangiocarcinoma. Cancer Imaging 18:8
Fowler KJ, Sheybani A, Parker RA 3rd et al (2013) Combined hepatocellular and cholangiocarcinoma (biphenotypic) tumors: imaging features and diagnostic accuracy of contrast-enhanced CT and MRI. AJR Am J Roentgenol 201:332–339
Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33:159–174
Zhou HF, Han YQ, Lu J et al (2019) Radiomics facilitates candidate selection for irradiation stents among patients with unresectable pancreatic cancer. Front Oncol 9:973
Tibshirani R (2011) Regression shrinkage and selection via the lasso: a retrospective. J R Stat Soc Series B Stat Methodol 73:273–282
Zhang J, Huang Z, Cao L et al (2020) Differentiation combined hepatocellular and cholangiocarcinoma from intrahepatic cholangiocarcinoma based on radiomics machine learning. Ann Transl Med 8:119
Liu X, Khalvati F, Namdar K et al (2021) Can machine learning radiomics provide pre-operative differentiation of combined hepatocellular cholangiocarcinoma from hepatocellular carcinoma and cholangiocarcinoma to inform optimal treatment planning? Eur Radiol 31:244–255
Shur JD, Doran SJ, Kumar S et al (2021) Radiomics in oncology: a practical guide. Radiographics 41:1717–1732
Limkin EJ, Reuzé S, Carré A (2019) The complexity of tumor shape, spiculatedness, correlates with tumor radiomic shape features. Sci Rep 9:4329
Yap FY, Varghese BA, Cen SY et al (2021) Shape and texture-based radiomics signature on CT effectively discriminates benign from malignant renal masses. Eur Radiol 31:1011–1021
Cuocolo R, Stanzione A, Ponsiglione A et al (2019) Clinically significant prostate cancer detection on MRI: a radiomic shape features study. Eur J Radiol 116:144–149
Mechee MS, Hussain ZM, Salman ZI (2021) Wavelet theory: applications of the wavelet. Wavelet Theory
Liang W, Xu L, Yang P et al (2018) Novel nomogram for preoperative prediction of early recurrence in intrahepatic cholangiocarcinoma. Front Oncol 8:360
Zhou Y, Zhou G, Zhang J, Xu C, Wang X, Xu P (2021) Radiomics signature on dynamic contrast-enhanced MR images: a potential imaging biomarker for prediction of microvascular invasion in mass-forming intrahepatic cholangiocarcinoma. Eur Radiol 31:6846–6855
Kim TY, Cho NH, Jeong GB, Bengtsson E, Choi HK (2014) 3D texture analysis in renal cell carcinoma tissue image grading. Comput Math Methods Med 2014:536217
Tang TY, Li X, Zhang Q et al (2020) Development of a novel multiparametric MRI radiomic nomogram for preoperative evaluation of early recurrence in resectable pancreatic cancer. J Magn Reson Imaging 52:231–245
Yin X, Zhang BH, Qiu SJ et al (2012) Combined hepatocellular carcinoma and cholangiocarcinoma: clinical features, treatment modalities, and prognosis. Ann Surg Oncol 19:2869–2876
Kim SH, Park YN, Lim JH, Choi GH, Choi JS, Kim KS (2014) Characteristics of combined hepatocelluar-cholangiocarcinoma and comparison with intrahepatic cholangiocarcinoma. Eur J Surg Oncol 40:976–981
Funding
This study was supported by grants from the Shanghai Municipal Key Clinical Specialty (shslczdzk03202).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Pengju Xu.
Conflict of interest
The authors declare no competing intrests.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
•performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 166 kb)
Rights and permissions
About this article
Cite this article
Zhou, Y., Zhou, G., Zhang, J. et al. DCE-MRI based radiomics nomogram for preoperatively differentiating combined hepatocellular-cholangiocarcinoma from mass-forming intrahepatic cholangiocarcinoma. Eur Radiol 32, 5004–5015 (2022). https://doi.org/10.1007/s00330-022-08548-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08548-2