Abstract
Objective
To explore the application of deep learning in patients with primary osteoporosis, and to develop a fully automatic method based on deep convolutional neural network (DCNN) for vertebral body segmentation and bone mineral density (BMD) calculation in CT images.
Materials and methods
A total of 1449 patients were used for experiments and analysis in this retrospective study, who underwent spinal or abdominal CT scans for other indications between March 2018 and May 2020. All data was gathered from three different CT vendors. Among them, 586 cases were used for training, and other 863 cases were used for testing. A fully convolutional neural network, called U-Net, was employed for automated vertebral body segmentation. The manually sketched region of vertebral body was used as the ground truth for comparison. A convolutional neural network, called DenseNet-121, was applied for BMD calculation. The values post-processed by quantitative computed tomography (QCT) were identified as the standards for analysis.
Results
Based on the diversity of CT vendors, all testing cases were split into three testing cohorts: Test set 1 (n = 463), test set 2 (n = 200), and test set 3 (n = 200). Automated segmentation correlated well with manual segmentation regarding four lumbar vertebral bodies (L1–L4): the minimum average dice coefficients for three testing sets were 0.823, 0.786, and 0.782, respectively. For testing sets from different vendors, the average BMDs calculated by automated regression showed high correlation (r > 0.98) and agreement with those derived from QCT.
Conclusions
A deep learning–based method could achieve fully automatic identification of osteoporosis, osteopenia, and normal bone mineral density in CT images.
Key Points
• Deep learning can perform accurate fully automated segmentation of lumbar vertebral body in CT images.
• The average BMDs obtained by deep learning highly correlates with ones derived from QCT.
• The deep learning–based method could be helpful for clinicians in opportunistic osteoporosis screening in spinal or abdominal CT scans.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Osteoporosis is a common and frequently occurring disease in the aging population [1]. About 200 million people suffer from osteoporosis and 89 million fractures occur worldwide every year [2]. Osteoporosis is a disease of bone metabolism that shows a decrease in bone mineral density (BMD) and strength [3], which may lead to low back pain, disc degeneration, or an increased risk of fracture of the vertebral body [4,5,6,7,8,9]. Hence, the early diagnosis of osteoporosis is very important to the progress of disease prevention.
Currently, evaluation methods for osteoporosis consist of the commonly used approaches, such as dual-energy X-ray absorptiometry (DXA), quantitative computed tomography (QCT), and quantitative ultrasound (QUS), and emerging imaging techniques, such as dual-layer spectral CT [10], 1H-MRS [11], and positron emission tomography (PET) [12]. BMD measurement is a reliable and ideal method for early diagnosis of osteoporosis. DXA is a commonly used tool for measuring spinal BMD [13]. BMD measured by DXA is defined as the sum of cortical bone and cancellous bone, considering two-dimensional structures. However, DXA could not eliminate the influence of cortex, hyperosteogeny, and sclerosis on BMD measurement [14], which might underestimate the actual loss of bone mass [15]. QCT is a recognized method for 3D bone density assessment [16]. Several studies [17,18,19,20] have shown that the detection rate of QCT on osteoporosis is significantly higher than that of DXA. But it requires calibration and standardized software, which means complex post-processing. And compared with DXA, the radiation dose of QCT is much higher. Thus, application of QCT as a screening technology is limited so far.
Millions of CT scans covering part or all of spine, are available from patients with other indications, such as urinary and / or digestive diseases, every year. These CT scans can be used for the opportunistic screening of osteoporosis, without additional exposure and substantial costs [21]. Several literature studies had shown that conventional diagnostic CT scans were used to measure BMD by measuring directly the CT values of cancellous bone, leading to correlation coefficients ranging from 0.399 to 0.891 [22]. However, CT value not only depends on internal factors of vertebral body but also on external factors such as equipment, X-ray tube voltage, and CT device [23]. Therefore, the CT values obtained from different devices need to be calibrated, which is why conventional CT scans in diagnosis of osteoporosis are limited.
Deep learning has been increasingly used in medical imaging analysis and even has entered the stage of rapid development [24,25,26,27,28]. In terms of osteoporosis, several works on application of the deep learning technique have existed. Sangwoo et al [29] combined machine learning and deep learning to predict patients with abnormal BMD by incorporating spine X-ray images. Bergman et al [30] presented a deep learning method to compute the DXA BMD and T-Score from standard chest or abdomen CT scans. Pan et al [31] developed a deep learning–based system to automatically measure BMD for opportunistic osteoporosis screening using low-dose chest CT scans obtained for lung cancer screening. However, in this system, only segmenting all vertebral bodies into three classes was used by a 3D CNN model, while isolating and labeling each individual vertebral body was then performed by conventional image processing algorithms. Using BMD values obtained with DXA as reference, Yasaka et al [32] developed a deep learning model to predict the bone mineral density of lumbar vertebrae from unenhanced abdominal CT images. This work only focused on BMD prediction, not provided vertebral body location. To the best of our knowledge, there are no studies regarding the application of deep learning on fully automated location of lumbar vertebral body and calculation of BMD similar to QCT value to date.
In this study, we proposed a deep learning algorithm to locate lumbar vertebral body in CT scans and calculate BMD similar to QCT value accordingly. This work is an exploration on the application of deep learning for diagnosis of osteoporosis, and aims to assess the performance of the automatic method to locate lumbar vertebral body in terms of accuracy as well as to calculate BMD.
Materials and methods
Study population
The retrospective study has been approved by the institutional review board and the ethics committee of the Fifth Affiliated Hospital of Sun Yat-sen University. To the retrospective of the study, the informed consent was waived from all patients, and to protect the patient’s privacy, the data were desensitized before using. We collected the images and data from March 2018 to May 2020 at our hospital. The inclusion criteria were (a) CT examination images including lumbar spine, such as lumbar examination and abdominal examination; and (b) willingness to undergo this clinical study. The exclusion criteria of patient were (a) with an absence of meeting the post-processing requirements for CT examination; (b) with secondary osteoporosis, such as osteoporosis caused by renal failure, diabetes, and hyperparathyroidism; (c) with compression fracture on L1–L4; and (d) with postoperative metal or bone cement implant. In total, 1449 patients fulfilled all criteria were included in this retrospective study. Among them, 586 patients were used as the training cohort for model development, and three independent testing cohorts, comprising 463, 200, and 200 patients, respectively, were later collected to provide evaluation and analysis of the trained models.
CT image acquisition
All CT scans for training and 463 CT scans for testing were obtained by using 128-channel multi-detector CT scanners (uCT 760, United Imaging Healthcare). The remaining 400 CT scans for testing were collected from two different vendors’ CT scanners (n = 200, Somatom Definition Flash unit, Siemens Healthcare; n = 200, Revolution CT scanner, GE Healthcare). All CT parameters were set, in accordance with the “China Health Quantitative CT Big Data Project Research Program” [33], as follows: collimation, 0.625 mm; tube voltage, 120 kVp; tube current, automatic. Reconstruction intervals were 1.0 mm. All CT images were reconstructed to 512 × 512 matrices using iterative reconstruction algorithms available with the vendor’s CT scanners.
QCT image post-processing
All CT images were post-processed by QCT Pro Model 4 (Mindways Software, Inc.). The quality control analysis was used by a unified European Spine Phantom (ESP, NO.145). The central layer of the vertebral body was selected to calculate the average bone density value.
Diagnosis was performed according to the guidelines introduced by the International Society for Clinical Densitometry (ISCD) and American College of Radiology (ACR), in which osteoporosis if BMD values blow 80 mg/cm3; osteopenia, from 80 to 120 mg/cm3; normal, over 120 mg/cm3 [34, 35].
Workflow of method
The proposed method mainly contained two steps: (1) lumbar vertebral body segmentation and (2) BMD calculation. In this work, we used deep learning techniques to automatically perform these procedures. The automated calculation was based on accurate segmentation. Specifically, U-Net was employed to perform segmentation task. After segmentation, lumbar vertebral body had been located and its interior was extracted for BMD calculation. BMD calculation was executed via a regression model. Figure 1 shows overview of our proposed framework.
The first stage: segmentation
Images for segmentation and image annotation
In this study, segmentation model was used for determining the position of the first four lumbar vertebral bodies (L1–L4). Hence, CT transverse slices of each patient were converted to sagittal images. We should perform vertebral body detection to ensure that all sagittal images selected for model training contain the vertebral bodies of interest. Yet for simplicity, we performed vertebral foramen detection in the transverse slices. Specifically, given a transverse slice, a binary image was first gained via using threshold method, of which the threshold value was selected by the distribution of HU values in the training cohort. In our work, the threshold value was 150 HU. After a series of operations, including morphological close operation, NOT operation, background removal, and denoising of 3D, only vertebral foramen region remained in the binary image. Given a series of CT slices from a patient, the abovementioned detection was followed by getting vertebral foramen center of each transverse slice. The mean of vertebral foramen centers of all slices was seen as spine center and was used to select sagittal images. Including the sagittal slice where this mean center was, we finally consecutively selected 10 sagittal images as candidates for segmentation tasks.
To reduce time consumption of manual annotation while ensuring the visibility of all concerned vertebral bodies (L1–L4) in the image, the average image of 20 consecutive sagittal images was used for vertebral body contouring on a research platform provided data annotation, i.e., InferScholar 3.0 (Infervision), by several radiologists. All sagittal images were first delineated separately by 2 radiologists (JW.L, a resident with 1 year of experience, and YS.C, a resident with 3 years of experience, respectively). And then, all annotations were reviewed and modified by YJ.F, a board-certified radiologist with 7 years of experience. Sketched region was used for segmentation ground truth. To make the network directly recognize the four different vertebral bodies, in each segmentation ground truth, the labeled region of L1 was filled with “1” and accordingly L2 with “2,” L3 with “3,” L4 with “4,” and others with “0.”
Image preprocessing
The pre-processing included three steps. First, all sagittal images were processed using a window, whose window level (WL) and window width (WW) respectively was [350, 1000]. And then, all of pixel values in the images were scaled to [0, 1] using min-max normalization method. Finally, all the images were processed to images with 512 × 512 pixels. In the final step, image was cropped around the center if its size was out of range; zero padding if inadequate.
Development of the segmentation model in the training cohort
A 2D U-shaped architecture called U-Net [36], which is favored in medical segmentation, was adopted in our work. The U-Net (shown in Fig. 2) employs an encoder-decoder architecture. The encoder part (the left part) is used for hierarchical feature extraction, and the decoder part (the right part) is employed to merge features for obtaining precise results.
Five-fold cross validation was applied to train and validation segmentation model. And then, a selected model was applied to the testing cohort. More details of U-Net architecture, model training, and model selection are described in the Supplementary Material.
When using, given a sequence of CT slices from a patient, we first convert them to sagittal images and find spine center. Using the sagittal slice, where spine center was, as the second-channel image, we then selected three consecutive sagittal images as a three-channel image to do pre-processing. Given such a processed three-channel image, the trained model will output the probability of each pixel for each class via the softmax function. The final segmentation made by the model will be performed by assigning each pixel to the class with the highest probability.
Evaluation of the segmentation model performance in the independent testing cohort
To evaluate automatic segmentation, the resulting regions of vertebral bodies were compared with the manual ground truth annotations. The segmentation performance was evaluated using (a) the dice similarity coefficient (DSC), a measure of spatial overlap of segmentations and ground truths, which is defined as 2TP/(FP + 2TP + FN), where TP, FP, and FN are the numbers of true-positive, false-positive, and false-negative segmentations, respectively; (b) the positive predictive value (PPV), a method for evaluating the numbers of TP segmentations in all positive predictions, which is defined as TP/(TP + FP); and (c) sensitivity, which evaluates the numbers of TP segmentations in all positive truths and is defined as TP/(TP + FN).
The second stage: regression
Images for regression and image labeling
Images for regression were extracted from transverse slices, and each of them was a three-channel image. To obtain such a three-channel image, we performed two steps as follows: (1) determining the second-channel image. It is noted that, viewing from 3D space, sagittal image is in the Y-Z plane and transverse slice is in the X-Y plane. Hence, using sagittal image, we could attain the transverse slice where the second-channel image was, and the selective range of the second-channel image in the transverse slice. Then, using transverse slice, we could accomplish the second-channel image selection and extraction. Specifically, given a vertebral body from the sagittal image via covering mask, we located its center at position (y, z) via related operations of connected component. In other words, z coordinate of this center corresponded to the position of the required transverse slice, and y coordinate of this center mapped to the corresponding y coordinate of the required transverse slice. Setting the height of image for model regression as “h” and using the abovementioned y coordinate as middle point, we then cropped a narrow strip from the abovementioned transverse slice, of which the size was h × 512 pixels. Using threshold method (same as the method of segmentation section), the binary image of the narrow strip was gained followed by remaining the maximum connected region of this image. This region was in the vertebral body of transverse slice. Setting the width of image for model regression as “w” and using the processed binary image as the mask of the narrow strip, we extracted the needed image, a rectangle region in the narrow strip of which the size was h × w pixels, to be seen as the second-channel image. It was noted that when we set “h” and “w,” we should guarantee the extracted image without cortex. In our study, both “h” and “w” were 32. (2) Extracting the first-channel and the third-channel images from corresponding position of the former and later slice of the abovementioned transverse slice, respectively.
After the abovementioned operations, images extracted from vertebral body interiors, of which the size was 32 × 32 × 3 pixels, were finally used for regression task and the corresponding BMDs derived from QCT were used as its reference standard.
Image preprocessing
All extracted images were pre-processed before feeding into automatic regression. The processing pipeline included (1) gray transformation to [0, 255] using the above window and (2) normalization to [0, 1] using min-max normalization method.
Development of the regression model in the training cohort
The DenseNet-121 [37] was used to calculate BMD values. Its structure mainly contains 4 dense blocks and 3 transition layers. Each dense block is composed of a series of 1 × 1 convolutional layers, 3 × 3 convolutional layers, and skip connections. Each transition layer includes a 1 × 1 convolutional layer for reducing network parameters and a 2 × 2 average pooling operation with stride 2 for downsampling.
Five-fold validation was also applied to train and validation regression model. To speed up network convergence and get better regression performance, model parameters were pre-trained on the ImageNet [38].
More details of model training and model selection are described in the Supplementary Material.
Evaluation of the regression model performance in the independent testing cohort
To assess goodness of fit, the regression performance was evaluated using the coefficient of determination (r2) between automated obtained BMDs and reference standards.
To further evaluate the validity of the regression model, we additionally calculated average BMDs and classified them into three categories by the guidelines for evaluating QCT studies [34]. The number of true-positive, false-positive, true-negative, and false-negative findings of classification performance via average BMDs on testing cohort was also described in a 3 × 3 contingency table representing the confusion matrix.
Statistical analysis and software
Continuous variables are expressed as means ± standard deviations, and categorical variables are represented as frequencies. Categorical variables were compared by using the chi-square test. Paired BMD values were compared with the Wilcoxon signed-rank test, without assuming the underlying distribution. p < 0.05 was considered indicative of a statistically significant difference. Pearson correlation coefficient was used to evaluate reliability of BMD calculation. Bland-Altman analysis was also performed to compare average BMDs. Cohen’s kappa coefficient was used to measure the agreement between automated regression and reference. The level of agreement was interpreted as slight if kappa coefficient was 0 to 0.20; fair, 0.21 to 0.40; moderate, 0.41 to 0.60; substantial, 0.61 to 0.80; and almost perfect, 0.81 to 1.
The software used to build the models based on DCNN was based on an Ubuntu 16.04 operating system with the deep learning toolkit MXNet. The training process was run on an Intel® Core™ i7-5930K CPU 3.50GHz with GeForce GTX TITAN X GPU. The overall neural network implementation, evaluation, and statistical analysis were all performed in the Python2.7 environment.
Results
Patient characteristics
A total of 1449 patients were eligible for the final analysis. The patients were aged from 15 to 98 years (average, 53.8 years). Table 1 lists the clinical and demographic features for the training and testing datasets. According to the guidelines, all patients were divided into three categories: osteoporosis (n = 244, 16.8%), osteopenia (n = 605, 41.8%), and normal (n = 600, 41.4%).
The training and testing datasets demonstrated no statistically significant differences in age characteristics (p > 0.05), whereas statistically different in the distribution of BMD (p < 0.01). There was sex and CT examination position existing no statistically significant differences between training dataset and test set 1 (p > 0.05), whereas showing statistically difference and test set 2, as well as test set 3 (p < 0.001).
The performance of the segmentation model in the independent testing cohort
Table 2 shows the evaluation results of the segmentation model. According to the results, automated deep learning–based segmentation and manual segmentation correlated well regarding four lumbar vertebral bodies. Of three testing cohorts, the DSCs for L1 were 0.823 (95% CI [0.815, 0.831]), 0.786 (95% CI [0.773, 0.799], and 0.789 (95% CI [0.780, 0.802]), respectively; for L2 were 0.825 (95% CI [0.817, 0.833]), 0.793 (95% CI [0.779, 0.807]), and 0.786 (95% CI [0.774, 0.797]), respectively; for L3 were 0.862 (95% CI [0.855, 0.868]), 0.813 (95% CI [0.798, 0.824]), and 0.801 (95% CI [0.791, 0.812]), respectively; for L4 were 0.899 (95% CI [0.895, 0.904]), 0.883 (95% CI [0.874, 0.893]), and 0.782 (95% CI [0.769, 0.797]), respectively.
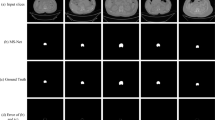
In 69% cases of test set 1, as well as 63.5% of test set 2 and 44% of test set 3, the segmentation model worked well, leading to high DSCs (over 0.90) in at least three of the four lumbar vertebral bodies. Especially in 58% patients of test set 1, as well as 50% of test set 2 and 30% of test set 3, DSCs in all of the four lumbar vertebral bodies were 0.90 or better. Examples of CT images in sagittal view, manual annotations by the radiologists, and automated segmentation results of our method are shown in Fig. 3.
Visual comparison between the segmentation results of our method and manual segmentation. From top to bottom: segmentation results in patients of test sets 1, 2, and 3, respectively. From left to right: CT sagittal images, manual segmentation, and automated segmentation, respectively. Red, green, yellow, and blue colors represent the vertebral bodies of L1, L2, L3, and L4, respectively. The manual and automated segmentation correlate very well that DSCs in all of the four lumbar vertebral bodies were over 0.90
Since the segmentation model in this work is used for locating lumbar vertebral bodies in each image, the segmentation result is effective if four DSCs are all over 0.5. Based on this, approximately 86% cases in test set 1 and 81.5% cases in test set 2, as well as 65.5% cases in test set 3, had good segmentation results.
The performance of the regression model in the independent testing cohort
The performance analysis is represented using part of cases in the three testing cohorts, which had effective segmentation results, i.e., 86% cases (398/463) in test set 1, 81.5% cases (163/200) in test set 2, and 65.5% cases (131/200) in test set 3. The analysis of three whole testing cohorts is described in the Supplementary Material.
BMDs obtained from the regression model were evaluated with reference to ones derived from QCT. The evaluation results of the regression model are illustrated in Table 2.
In this study, only three vertebral bodies of each patient were post-processed by QCT. Thus, the average BMD was defined as the mean of predicted BMD values from this three vertebral bodies. Figure 4 shows that the average BMDs in three testing cohorts were all highly correlated to references (r = 0.992, 0.986, and 0.980, respectively; all p < 0.001). The limit of agreement between the average BMDs obtained by automated regression and the ones calculated by reference was − 11.84 to 13.64 mg/cm3 for test set 1, − 16.8 to 12.6 mg/cm3 for test set 2, and − 8.3 to 29.3 mg/cm3 for test set 3. The Wilcoxon signed-rank test showed that the average BMDs in test set 1 were not significantly different from reference standards (p > 0.3), whereas in test set 2 were underestimated and in test set 3 were overestimated compared with values derived from QCT (p < 0.001).
Moreover, category according to the average BMDs derived from QCT was used as reference standard. There were all strong agreements between prediction and reference standard in three testing cohorts (Cohen’s kappa, 0.888, 0.868, and 0.879, respectively). The classification confusion matrices that report the number of true-positive, false-positive, true-negative, and false-negative results for the average BMDs are shown in Table 3.
Discussion
In this study, we proposed a deep learning–based framework using spinal or abdominal CT scans to segment lumbar vertebral body and automatically calculate BMD value. Specifically, the framework consists of the U-Net trained by sagittal images to tackle segmentation task and a regression network to calculate BMD value. Experimental results demonstrated accuracy and robustness of the proposed framework, and two key findings were further obtained: (1) the model based on DCNN could achieve a good performance for localization and segmentation of L1–L4. (2) The DCNN could efficiently calculate BMD value, and the average BMD value was highly correlated with one derived from QCT.
Lumbar vertebral body localization in sagittal images is the basis for BMD measurement. In this study, we employed U-Net to conduct vertebral body segmentation. The results on three independent testing cohorts showed the average DSCs of four vertebral bodies were all near or over 0.8, indicating automated segmentation was highly correlated with manual annotation. Moreover, in most patients (approximately 80.2% in a total of three testing cohorts), automated segmentation performed well, leading to the DSCs of L1–L4 all over 0.5. In other words, in some cases, automated segmentation did not perform appropriately. There may be several reasons for this: (1) distinguishing vertebral body with similar structure from sagittal image remains a challenging task. (2) We performed semantic segmentation instead of instance segmentation. (3) Even though data were annotated by the domain experts, label noise still could be a limiting factor in developing the model. There may be another reason for the poor segmentation results in test sets 2 and 3 which were obtained from other two CT vendors’ scanners that we trained our model only using CT images from a single CT scanner, leading to lack of data diversity in deep learning.
BMD measurement is a reliable and ideal method for early diagnosis of osteoporosis. DXA is the most widely used technique for diagnosis of osteoporosis and performing serial assessments of BMD, but it is susceptible to abdominal aortic calcification and spinal degeneration [14]. QCT can supplement DXA by geometric and septal bone assessment, and clinically, has gained wide acceptance within the evaluation of osteoporosis. However, QCT is not extensively used in most hospitals because of the need for post-processing equipment and complex post-processing work. We utilized the method based on DCNN to automatically calculate BMDs. For testing sets from different vendors (test set 1, test set 2, and test set 3), the Pearson correlation coefficients of the average BMDs calculated by automated regression were all over 0.98. The results in the independent testing cohorts revealed that strong correlation existed between the average BMD obtained by the automatic method and one derived from QCT. However, the limits of agreements between the average BMDs obtained by automated regression and ones calculated by reference indicated the BMDs obtained by our models had difference in CT scans from different vendors, which were acceptable for the clinician. The main reason for the existence of such differences may be the scarcity of training data diversity, which is essential for the deep learning–based method. In addition, our automatic method only used a simple and brief convolutional neural network to estimate BMD. Thus, the proposed method was more efficient and convenient than post-processing required by QCT.
Patients, who suffered from other indications, may also be accompanied by osteoporosis or osteopenia; thus, their abdominal or spinal CT scans could be used to “opportunistic screening” [16, 25]. However, due to diverse image reconstruction algorithms and various voltage radiating tubes, the CT values for evaluating osteoporosis have been limited [16]. In this work, we also focused on the accuracy of osteoporosis prediction using the BMD values calculated by our method. Underestimating or overestimating BMD occurred mostly in CT scans which were obtained from other different CT vendors, due to lack of training data diversity. However, our experimental results indicated that the proposed method could detect osteoporosis or osteopenia using conventional CT scans, which might contribute to the screening of early osteoporosis and be beneficial to the prevention of osteoporosis.
In summary, our research has several advantages. First, we used BMD values derived from QCT as a reference standard, which was shown to be more accurate in diagnosing osteoporosis compared with that of DXA [17,18,19]. Furthermore, our hospital is one of the participating centers of the China Health Big Data (China Biobank) project, and we regularly calibrate parameters to provide accurate QCT osteoporosis data [33]. Thus, the data used in this study were valid. Second, we employed a single trained regression network to calculate BMD, rather than complex and cumbersome procedures, which might contribute to reducing the workloads of clinicians. Third, our model could predict the risk of osteoporosis through image features extracted from conventional CT scans. So, this method may provide assistance in diagnosing osteoporosis for many “opportunistic screening” without additional costs.
It should be noted that there are also several limitations in our current study. First, the proposed method was established on the basis of data obtained from a single center, and the model was only trained on data obtained with a single CT scanner. This is our preliminary results; studies with training datasets of increasing variability are needed to further validate the robustness and reproducibility of our methods. Even prospective multicenter studies with considerably large datasets are our future works. Second, data from patients with severe scoliosis were not considered in our study. Therefore, the application of our results to populations with this type of disease is limited. Furthermore, the proposed method was not able to automatically exclude vertebral bodies with calcification. The calculated BMD of these vertebral bodies is quite different from the actual situation, which may have a significant effect on clinical diagnosis results.
In conclusion, our study demonstrated that the proposed method based on DCNN could provide accurate segmentation for lumbar vertebral body and automatic calculation of BMD, which had a great potential to be an available tool for clinicians in opportunistic osteoporosis screening.
Abbreviations
- ACR:
-
American College of Radiology
- BMD:
-
Bone mineral density
- DCNN:
-
Deep convolutional neural network
- DSC:
-
Dice similarity coefficient
- DXA:
-
Dual-energy X-ray absorptiometry
- ESP:
-
European spine phantom
- HU :
-
Hounsfield units
- ISCD:
-
International Society for Clinical Densitometry
- PET:
-
Positron emission tomography
- PPV:
-
Positive predictive value
- QCT:
-
Quantitative computed tomography
- QUS:
-
Quantitative ultrasound
- WL:
-
Window level
- WW:
-
Window width
References
Hofbauer LC, Rachner TD (2015) More DATA to guide sequential osteoporosis therapy. Lancet 386:1116–1118. https://doi.org/10.1016/S0140-6736(15)61175-8
Pisani P, Renna MD, Conversano F et al (2016) Major osteoporotic fragility fractures: risk factor updates and societal impact. World J Orthop 7:171–181. https://doi.org/10.5312/wjo.v7.i3.171
Yang J, Pham SM, Crabbe DL (2003) Effects of oestrogen deficiency on rat mandibular and tibial microarchitecture. Dentomaxillofac Radiol 32:247–251. https://doi.org/10.1259/dmfr/12560890
Watanabe M, Sakai D, Yamamoto Y, Sato M, Mochida J (2010) Upper cervical spine injuries: age-specific clinical features. J Orthop Sci 15:485–492. https://doi.org/10.1007/s00776-010-1493-x
Devlin HB, Goldman M (1966) Backache due to osteoporosis in an industrial population. A survey of 481 patients. Ir J Med Sci 6:141–148. https://doi.org/10.1007/bf02943677
Ensrud KE, Blackwell TL, Cawthon PM et al (2016) Degree of trauma differs for major osteoporotic fracture events in older men versus older women. J Bone Miner Res 31:204–207. https://doi.org/10.1002/jbmr.2589
Fechtenbaum J, Etcheto A, Kolta S, Feydy A, Roux C, Briot K (2016) Sagittal balance of the spine in patients with osteoporotic vertebral fractures. Osteoporos Int 27:559–567. https://doi.org/10.1007/s00198-015-3283-y
Yang Z, Griffith JF, Leung PC, Lee R (2009) Effect of osteoporosis on morphology and mobility of the lumbar spine. Spine (Phila Pa 1976) 34:E115–E121. https://doi.org/10.1097/brs.0b013e3181895aca
Lee JJ, Aghdassi E, Cheung AM et al (2012) Ten-year absolute fracture risk and hip bone strength in Canadian women with systemic lupus erythematosus. J Rheumatol 39(7):1378–1384. https://doi.org/10.3899/jrheum.111589
Roski F, Hammel J, Mei K et al (2019) Bone mineral density measurements derived from dual-layer spectral CT enable opportunistic screening for osteoporosis. Eur Radiol 29(11):6355–6363. https://doi.org/10.1007/s00330-019-06263-z
Li GW, Tang GY, Liu Y, Tang RB, Peng YF, Li W (2012) MR spectroscopy and Micro-CT in evaluation of osteoporosis model in rabbits: comparison with histopathology. Eur Radiol 22:923–929. https://doi.org/10.1007/s00330-011-2325-x
Dyke JP, Aaron RK (2010) Noninvasive methods of measuring bone blood perfusion. Ann N Y Acad Sci 1192:95–102. https://doi.org/10.1111/j.1749-6632.2009.05376.x
Ott SM (1991) Methods of determining bone mass. J Bone Miner Res 6:S71–S76. https://doi.org/10.1002/jbmr.5650061416
Rand T, Seidl G, Kainberger F et al (1997) Impact of spinal degenerative changes on the evaluation of bone mineral density with dual energy X-ray absorptiometry (DXA). Calcif Tissue Int 60:430e3. https://doi.org/10.1007/s002239900258
Ito M, Hayashi K, Yamada M, Uetani M, Nakamura T (1993) Relationship of osteophytes to bone mineral density and spinal fracture in men. Radiology 189:497–502. https://doi.org/10.1148/radiology.189.2.8210380
Engelke K (2017) Quantitative computed tomography—current status and new developments. J Clin Densitom 20(3):309–321. https://doi.org/10.1016/j.jocd.2017.06.017
Engelke K, Libanati C, Liu Y et al (2009) Quantitative computed tomography (QCT) of the forearm using general purpose spiral whole-body CT scanners: accuracy, precision and comparison with dual-energy X-ray absorptiometry (DXA). Bone 45(1):110e8. https://doi.org/10.1016/j.bone.2009.03.669
Li N, Li XM, Xu L, Sun WJ, Cheng XG, Tian W (2013) Comparison of QCT and DXA: osteoporosis detection rates in postmenopausal women. Int J Endocrinol 2013:1–5. https://doi.org/10.1155/2013/895474
Xiao-ming X, Na L, Li K et al (2019) Discordance in diagnosis of osteoporosis by quantitative computed tomography and dual-energy X-ray absorptiometry in Chinese elderly men. J Orthop Translat 18:59–64. https://doi.org/10.1016/j.jot.2018.11.003
Löffler MT, Jacob A, Valentinitsch A et al (2019) Improved prediction of incident vertebral fractures using opportunistic QCT compared to DXA. Eur Radiol 29:4980–4989. https://doi.org/10.1007/s00330-019-06018-w
Valentinitsch A, Trebeschi S, Kaesmacher J et al (2019) Opportunistic osteoporosis screening in multi-detector CT images via local classification of textures. Osteoporos Int 30:1275–1285. https://doi.org/10.1007/s00198-019-04910-1
Gausden EB, Nwachukwu BU, Schreiber JJ, Lorich DG, Lane JM (2017) Opportunistic use of CT imaging for osteoporosis screening and bone density assessment. J Bone Joint Surg 99:1580–1590. https://doi.org/10.2106/JBJS.16.00749
Feng-tan L, Dong L, Zhang Y-t (2013) Influence of tube voltage on CT attenuation, radiation dose, and image quality: phantom study. Chin J Radiol 47:458–461. https://doi.org/10.3760/cma.j.issn.1005-1201.2013.05.016
Wolterink JM, Leiner T, de Vos BD, van Hamersvelt RW, Viergever MA, Išgum I (2017) Automatic coronary artery calcium scoring in cardiac CT angiography using paired convolutional neural networks. Med Image Anal 34:123–136. https://doi.org/10.1016/j.media.2016.04.004
Sirinukunwattana K, Ahmed Raza SE, Yee-Wah T, Snead DRJ, Cree IA, Rajpoot NM (2016) Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imaging 35:1196–1206. https://doi.org/10.1109/TMI.2016.2525803
Gulshan V, Peng L, Coram M et al (2016) Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316:2402–2410. https://doi.org/10.1001/jama.2016.17216
Esteva A, Kuprel B, Novoa RA et al (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542:115–118. https://doi.org/10.1038/nature21056
González G, Ash SY et al (2017) Disease staging and prognosis in smokers using deep learning in chest computed tomography. Am J Respir Crit Care Med 197:193–203. https://doi.org/10.1164/rccm.201705-0860OC
Lee S, Choe EK, Kang HY, Yoon JW, Kim HS (2019) The exploration of feature extraction and machine learning for predicting bone density from simple spine X-ray images in a Korean population. Skeletal Radiology pp 1–6. https://doi.org/10.1007/s00256-019-03342-6
Bergman Amitai O, Bar A, Toledano E et al (2017) Computing DEXA score from CT using Deep segmentation networks cascade. Available at: www.zebra-med.com/research-publications/computing-dexa-score-from-ctusing-deep-segmentation-networks-cascade/
Pan Y, Shi D, Wang H et al (2020) Automatic opportunistic osteoporosis screening using low-dose chest computed tomography scans obtained for lung cancer screening. Eur Radiol 30(7):4107–4116. https://doi.org/10.1007/s00330-020-06679-y
Yasaka K, Akai H, Kunimatsu A, Kiryu S, Abe O (2020) Prediction of bone mineral density from computed tomography: application of deep learning with a convolutional neural network. Eur Radiol 30(6):3549–3557. https://doi.org/10.1007/s00330-020-06677-0
Wu Y, Guo Z, Fu X et al (2019) The study protocol for the China Health Big Data (China Biobank) project. Quant Imaging Med Surg 96:1095–1102. https://doi.org/10.21037/qims.2019.06.16
Link TM, Lang TF (2014) Axial QCT: clinical applications and new developments. J Clin Densitom 17:438–448. https://doi.org/10.1016/j.jocd.2014.04.119
American College of Radiology (2018) ACR-SPR-SSR practice parameter for the performance of musculoskeletal quantitative computed tomography (QCT). American College of Radiology, Reston. Available at: https://www.acr.org/-/media/ACR/Files/Practice-Parameters/QCT.pdf
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, pp 234–241. https://doi.org/10.1007/978-3-319-24574-4_28
Iandola F, Moskewicz M, Karayev S, Girshick R, Darrell T, Keutzer K (2014) Densenet: implementing efficient convnet descriptor pyramids. arXiv preprint arXiv:1404.1869
Russakovsky O, Deng J, Su H et al (2015) Imagenet large scale visual recognition challenge. Int J Comput Vision 115:211–252. https://doi.org/10.1007/s11263-015-0816-y
Funding
This study has received funding by Science and Technology Planning Project of Zhuhai (ZH2202200010HJL), by Investigator-Initiated Clinical Trial of The Fifth Affiliated Hospital of Sun Yat-sen University (YNZZ003).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Shaolin Li.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 176 kb)
Rights and permissions
About this article
Cite this article
Fang, Y., Li, W., Chen, X. et al. Opportunistic osteoporosis screening in multi-detector CT images using deep convolutional neural networks. Eur Radiol 31, 1831–1842 (2021). https://doi.org/10.1007/s00330-020-07312-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07312-8