Abstract
Objectives
To develop and validate a radiomics nomogram for preoperative prediction of microvascular invasion (MVI) in patients with hepatocellular carcinoma (HCC).
Methods
The study included 157 patients with histologically confirmed HCC with or without MVI, and 110 patients were allocated to the training dataset and 47 to the validation dataset. Baseline clinical factor (CF) data were collected from our medical records, and radiomics features were extracted from the artery phase (AP), portal venous phase (PVP) and delay phase (DP) of preoperatively acquired CT in all patients. Radiomics analysis included tumour segmentation, feature extraction, model construction and model evaluation. A final nomogram for predicting MVI of HCC was established. Nomogram performance was assessed via both calibration and discrimination statistics.
Results
Five AP features, seven PVP features and nine DP features were effective for MVI prediction in HCC radiomics signatures. PVP radiomics signatures exhibited better performance than AP and DP radiomics signatures in the validation datasets, with the AUC 0.793. In the clinical model, age, maximum tumour diameter, alpha-fetoprotein and hepatitis B antigen were effective predictors. The final nomogram integrated the PVP radiomics signature and four CFs. Good calibration was achieved for the nomogram in both the training and validated datasets, with respective C-indexes of 0.827 and 0.820. Decision curve analysis suggested that the proposed nomogram was clinically useful, with a corresponding net benefit of 0.357.
Conclusions
The above-described radiomics nomogram can preoperatively predict MVI in patients with HCC and may constitute a usefully clinical tool to guide subsequent personalised treatment.
Key Points
• No previously reported study has utilised radiomics nomograms to preoperatively predict the MVI of HCC using 3D contrast-enhanced CT imaging.
• The combined radiomics clinical factor (CF) nomogram for predicting MVI achieved superior performance than either the radiomics signature or the CF nomogram alone.
• Nomograms combing PVP radiomics and CF may be useful as an imaging marker for predicting MVI of HCC preoperatively and could guide personalised treatment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is very common worldwide and a major cause of mortality [1, 2]. Surgical resection is one of the potentially curative treatments available for HCC, but the recurrence rate remains high [3, 4]. The 5-year postoperative recurrence rate of HCC is approximately 70% in cases with liver resection (LR) and 25% in cases with liver transplantation [4]. One of the key factors is microvascular invasion (MVI). MVI is defined as microscopic tumour invasion in smaller intrahepatic vessels, including microvessels of the portal venous vein or hepatic artery and small lymphatic vessels [5]. Several studies have reported that MVI is a validated, independent predictor of early recurrence and poor survival after LR [6,7,8]. In addition, an accurate preoperative estimation of MVI presence may help surgeons to choose appropriate surgical procedures for patients [9]. If LR is considered for patients with a high risk of MVI, a procedure incorporating a wide resection margin may be preferable [10]. Thus, it is important that the presence of MVI can be evaluated preoperatively. However, MVI is currently only diagnosed after surgical resection, via histopathologic evaluation. It is currently difficult to diagnose MVI via preoperative examinations such as computed tomography (CT), magnetic resonance (MR) imaging, serum markers and preoperative biopsy [10,11,12,13]. Thus, there is an urgent need for a quantitative means of predicting MVI preoperatively.
Recently, radiomics has become an evolving topic in conjunction with hypotheses that medical imaging may provide crucial information pertaining to tumour pathophysiology [14, 15]. Some studies have shown that radiomics features may potentially be useful as diagnostic or prognostic imaging markers for tumour lesion detection, subtype classification and therapeutic response assessment [14, 16,17,18,19,20,21,22]. And, Lei et al [9] showed that the nomogram, including diameter, number, status of capsule, boundary, location and typical dynamic pattern of CT imaging, achieved an optimal preoperative prediction of MVI in HBV-related HCC. However, no previously reported study has utilised radiomics nomograms to preoperatively predict the MVI of HCC using CT imaging. Moreover, our study was a direct extension of Lei et al’s work [9].
The aim of the current study was to develop and validate a CT-based radiomics signature to predict MVI in HCC preoperatively with a graphical nomogram that is user-friendly for clinicians, to assist in the determination of individual therapeutic strategies for patients with HCC.
Materials and methods
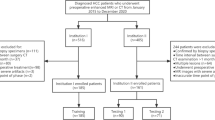
Patients
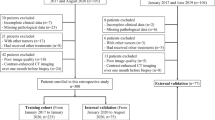
Ethical approval was obtained for this retrospective study, and the requirement for informed consent was waived. The entire cohort dataset was acquired from the January 2012 to December 2016 records of the institutional picture archiving and communication system (PACS; Toshiba), which was used to identify patients who had histologically confirmed HCC with MVI (MVI+) or without MVI (MVI−). Two pathologists with over 5 years of working experience independently reviewed all postoperative specimens histologically and assessed the presence or absence of MVI. All patients underwent contrast-enhanced CT (CECT) scans before LR. Figure 1 shows the patient recruitment process and the inclusion and exclusion criteria. A total of 157 patients conformed to the criteria, 134 men and 23 women (median age 57 years, age range 34–64 years), and were divided into two datasets at a ratio of 2:1 according to the time of surgery. The training dataset included 110 patients (93 men and 17 women, median age 55 years, age range 46–64 years), and 47 patients were allocated to the time-independent validation dataset (41 men and 6 women, median age 56 years, age range 34–64 years) (Table 1).
Baseline clinical-pathologic data were collected from our PACS medical records. Clinical factors (CFs) included age, gender, alpha-fetoprotein (AFP) level, tumour location (left, right or caudate lobe), hepatitis B surface antigen (HBsAg) status (positive or negative) and postoperative pathologic differentiation (well, moderate or poor). The WHO histologic grade system was used to determine the pathologic grade of hepatocellular carcinoma. Laboratory analysis consisted of routine blood tests performed within 1 week before surgery.
CT technology and segmentation
All CT examinations were performed with 64-row spiral CT scanners (Optima CT660 and Discovery 750 HD, GE Healthcare). A 1.2–1.5 mL/kg body weight bolus of iohexol (Omnipaque, GE Healthcare Co., Ltd.) was injected intravenously at a flow rate of 2.5 mL/s, followed by a 20-mL saline flush. Arterial phase (AP), portal venous phase (PVP) and delay phase (DP) were obtained at 35 s, 65 s and 120 s, respectively, after intravenous injection of contrast. The scanning parameters were 120 kV, 280 mAs, 0.8 s rotation time, 5 mm slice thickness and a 5-mm interval. The protocol requirements used for CECT imaging met the criteria recommended by the LI-RADS guideline [23].
Workflow
Radiomics extracts high-throughput quantitative imaging features to perform subsequent data analysis related to target clinical outcome. The workflow of a typical radiomics process consists of four steps: tumour segmentation, feature extraction, model construction and model evaluation.
Tumour segmentation
Three-dimensional manual segmentation was performed by a radiologist with work experience of 8 years, using ITK-SNAP software (http://www.radiantviewer.com). Regions of interest (ROIs) were drawn on all AP, PVP and DP images slice-by-slice for each patient, slightly along the visible borders of the lesion to include the entire lesion’s volume approximated. In the case of the blurred edges, we drew the maximum range of the lesion. The final segmentation results were validated by a senior radiologist with 15 years of work experience.
Feature extraction and selection
A set of 647 radiomics features that reflected the heterogeneity of the tumour was extracted from the AP, PVP and DP images for each patient.
The extracted radiomics features could be divided into two kinds: non-textural features and textural features. Non-textural features included shape and size features and intensity features. Shape and size features captured the direct-viewing characteristics of the lesion, and intensity features depicted the characteristics of the histogram of the lesion. Textural features were extracted based on four textural matrixes: grey level co-occurrence matrix (GLCM), grey level run-length matrix (GLRLM), grey level size zone matrix (GLSZM) and neighborhood grey-tone difference matrix (NGTDM). All the feature extraction was implemented using Matlab 2014a (MathWorks). We applied a wavelet filter onto the original image dataset in order to extract high-dimensional features from different frequency scales. We finally obtained nine image datasets including the original image dataset and eight filtered image datasets in different frequencies. A detailed description of feature definition and extraction is provided in Supplementary Appendix 1. All feature extraction was implemented using Matlab (version 2014a) (MathWorks).
To verify the robustness of the radiomics features, we randomly selected 20 patients for test and re-test analysis and multiclinician segmentation. The intraclass correlation coefficient (ICC) and concordance correlation coefficient (CCC) were calculated to determine the stability of the features. Features with an ICC and CCC lower than 0.75 were excluded from the final feature dataset (Supplement Fig. 1). For AP, 618 out of 647 features were retained; for PVP, 621 out of 647 features; and for DP, 563 out of 647 features.
We used the least absolute shrinkage and selection operator (LASSO) method to reduce the redundancy and dimensionality of the features in the training dataset. We chose the optimal feature dataset with the least cross-validation binominal deviance. Non-zero coefficients were defined as the weight for each selected feature, which indicated the correlation between the feature and MVI. The LASSO model was implemented using the glmnet package [24].
Model construction
Three models were constructed to preoperatively predict MVI status: a respective radiomics model, a clinical model and a combined model. The radiomics signature was calculated using support vector machine (SVM) with the LASSO-selected features as the input factors. The formula of the radiomics signature was as follows:
where b is the intercept, N is the support vector number, svi is the ith support vector, Ci is the coefficient of the ith support vector and x is the new data consisting of the values of the selected feature.
For the construction of the clinical model, we performed multivariable logistic regression analysis of clinical parameters including age, sex, maximum tumour diameter (MTD), cirrhosis, AFP, HBsAg, pathologic grade and location. Backward step-wise variable selection was implemented with the Akaike information criterion. The clinical model was constructed by integrating the final selected clinical predictors using logistic regression modelling.
The combined models incorporated the radiomics signature and the related clinical predictors together with a logistic regression model, which predicted MVI status by synthesising both the radiomics and clinical characteristics.
Model evaluation
Receiver operating characteristic (ROC) curve analysis was utilised to illustrate the diagnostic performance of the three models constructed. Delong validation was used to compare the areas under the curve (AUCs) in different models and determine whether they differed significantly. We also constructed a nomogram for the effective prediction model to provide a more direct way for the clinician to assess the possibility of MVI. Calibration curves were adopted to analyse the diagnostic performance of the nomogram in both the training and the validation datasets. Decision curve analysis was conducted to determine the clinical usefulness of the nomogram by quantifying the net benefits at different threshold probabilities in the entire cohort.
Statistical analysis
All statistical analysis was performed using the PASW Statistics 18.0.0 software package (SPSS). Categorical variables were expressed as numbers or percentages, and continuous variables were expressed as mean ± SD, or median. The two-sample t test was used to determine whether the values of the demographic variables differed significantly between the training and validation groups. Two-sided p values < 0.05 were considered statistically significant.
Results
Clinical characteristics
The clinical characteristics of the training and validation datasets are summarised in Table 1. There were no significant differences in age, gender, tumour location, MTD, AFP, cirrhosis, HBsAg or histologic grade between the training and validation cohorts (p = 0.124–0.873).
Based on the results of histopathology, the patients were classified into two groups: the MVI+ group and the MVI− group. There were no significant differences between the MVI+ group and the MVI− group in the training and validation datasets in terms of gender, tumour location, cirrhosis, HBsAg or pathologic grade. There were significant differences in age and MTD between the two groups in the training and validation datasets (p < 0.05). AFP differed significantly in the training dataset, but this was not confirmed in the validation dataset (Table 1).
Clinical model construction
Multivariable analysis showed that age (odds ratio (OR) 0.94; 95% confidence interval (CI) 0.90–0.98), MTD (OR 1.37; 95% CI 1.07–1.77), AFP (OR 1.52; 95% CI 0.88–2.63) and HBsAg (OR 0.43; 95% CI 0.14–1.31) were effective factors for clinical model construction.
Radiomics signature calculation
We performed LASSO modelling on AP, PVP and DP feature datasets in order to investigate the effectiveness of MVI discrimination via CECT (Supplement Fig. 2). For AP, five features were selected for radiomics signature construction. For the tri-phasic CECT image, five, seven and nine features were ultimately selected as putatively effective features for AP, PVP and DP radiomics signature construction, respectively. Details of the selected features are shown in Table 2.
Performance of the six proposed models
To evaluate the diagnostic performance of the developed model, we included a time-independent validation dataset with 47 patients. The PVP radiomics model exhibited better predictive performance with regard to MVI in the validation datasets, with the AUC of 0.793 compared with the AP radiomics model (AUC 0.684) and DP radiomics model (AUC 0.490) (Fig. 2). Separate bar charts of the PVP radiomics signatures are shown in Supplement Fig. 3, and box plots are shown in Fig. 3. The clinical model did not perform well in terms of discrimination, with AUCs of 0.734 in the training dataset and 0.761 in the validation dataset. After combining the PVP radiomics signature with the effective CF, the predictive performance improved significantly, with AUCs of 0.835 in the training dataset and 0.801 in the validation dataset. For AP and DP, the performance of the combined model showed a significant improvement in the training dataset than the single clinical model (AUC 0.703 and 0.798 vs. 0.734); however, the result was inverse in the validation dataset than with the clinical model (AUC 0.684 and 0.490 vs. 0.761). The performance of the PVP radiomics signature combining with the effective CF was better than that of the tri-phasic radiomics signature combining with the effective CF in the validation dataset (AUC 0.801 vs. 0.680) (Table 3, Fig. 2). Comparison was made between the best performed PVP plus CF model and the single CF model. The Delong test manifested a significant difference in the training dataset with a p value of 0.005. In the validation dataset, the p value was 0.281, but it was a trend that when adding PVP signature, the performance of the validation dataset was better than the single CF model with higher AUC.
Nomogram construction and validation
As the combined model incorporating the PVP radiomics signature and the effective CF had the best predictive performance, we built a nomogram for the graphical representation of predictive outcome (Fig. 4). Good calibration was observed with both the training and the validated datasets, with respective C-indexes of 0.827 and 0.820 (Fig. 5). The Hosmer-Lemeshow test yielded no significant difference between the predictive calibration curve and the ideal curve for MVI prediction with both the training and the validation datasets (p = 0.371 and 0.094, respectively). In the current study, the threshold probability of the decision curve was 3% and the corresponding net benefit was 0.357 (Fig. 6).
Decision curve analysis for the combined nomogram in the validation dataset. The y-axis represents the net benefit, and the x-axis represents the threshold probability. In our study, the threshold probability of the decision curve is 4% and the corresponding net benefit is 0.357. It indicates that the nomogram improves the benefit compared with the measures that treat all patients and treat none patient when threshold probability is > 4%
Discussion
We developed and validated a radiomics signature-based nomogram for preoperative and individualised prediction of MVI in patients with HCC. In previous studies, some researchers [9, 25] have analysed subjective imaging characteristics determined by radiologists and combined clinical-pathologic or gene expression factors to preoperatively predict the MVI of HCC. However, in the current study, radiomics analysis was applied to quantitatively extract CECT imaging features to assess subtle textural variation within tumour lesions, which may contain holistic information related to tumour physiology or microenvironment. Furthermore, we developed an easy-to-use nomogram integrating both proposed PVP radiomics signature and CF to facilitate the preoperative individualised prediction of MVI. The predictive calibration curves of the training and validation datasets demonstrated agreement with the ideal curve. Decision curve analysis showed that the radiomics nomogram was clinically useful in the current study.
Previous studies incorporating radiomics nomograms have yielded many results pertaining to the predictive values of tumour diagnoses and therapy effectiveness [26,27,28,29]. There is also growing interest in multimarker analysis [26,27,28,29]. In the current study, the combined radiomics-CF nomogram for predicting MVI was superior to both the radiomics signature and the CF nomogram alone, with a higher C-index and better calibration. Furthermore, the results implied that the combined PVP radiomics-CF nomogram was robust with regard to the prediction of MVI in HCC. The results of the study emphasised the great importance of the radiomics signature developed for MVI prediction according to the weights of the nomogram.
In the current study, PVP radiomics signature performed better than AP, DP or any kind of the combined radiomics signatures (Table 3). In a previous study, Banerjee et al [25] reported that radiogenomic venous invasion (RVI), as a non-invasive radiogenomic marker, could accurately and preoperatively predict MVI in HCC. In Banerjee et al’s [25] study, all feature scoring was also based exclusively on PVP images. But, what differs between Banerjee’s work and the current study is that our CECT-based PVP radiomics features were more objective than the PVP radiological characteristics assessed by doctors [9, 30, 31]. Furthermore, the predictive accuracy of our proposed model was higher than that of previously reported studies [9, 30,31,32]. In addition, according to the pathological definition of MVI [5], it is mainly detected in the small branches of the portal vein and may also be found in the small branches of the hepatic artery and/or within the small lymphatic vessels of the liver [33]. This suggests another hypothetical explanation as to why the PVP radiomics model reflected MVI better than the AP or DP radiomics model. In addition, the PVP radiomics signature was simpler than the other eight radiomics signature. This approach could be easier to operate in the clinical practice.
The proposed radiomics features are categorised into non-textural and textural features, based on statistical methods [34]. The final predictive model demonstrated that the non-textural radiomics features ‘fos_skewness’ and ‘fos_minimum’ of the PVP were significantly related to MVI. The fos_skewness measures the asymmetry of the distribution of values about the mean grey value, and the fos_minimum reflects the intensity of tumour region. The results showed that the fos_skewness was positively and the fos_minimum was negatively associated with the occurrence of MVI. Other significant radiomics features of the PVP in the current study were textural features. The textural radiomics parameters could not be identified via visual inspection, but these reflected heterogeneity in the tumour. Tumour heterogeneity may be difficult to identify and quantify via traditional imaging tools, the subjective assessment of images or random sampling biopsy [35], but which were approved to be significant with cancer pathophysiology. Although radiomics is not a new tool, it may be a useful imaging marker that improves the assessment and quantification of tumour spatial heterogeneity [36]. However, the radiomics features are extracted and calculated by the computer. Explaining the associations between the radiomics features—especially the higher-order radiomics features—and pathological manifestations is very challenging [37]. On the one hand, the pathophysiologic processes involve multiple interacting components; on the other hand, the maximised information obtained from computer-based radiologic image analysis is far beyond that which is attainable via visual inspection.
In the current study, we developed a clinical model incorporating preoperative age, MTD, HBsAg and AFP. The clinical model exhibited good predictive efficiency for the MVI of HCC, especially when combined with a PVP radiomics model. Furthermore, preoperative AFP and MTD were positively correlated with MVI. As MVI is a common event in advanced HCC [33], an estimate during early T-stage HCC has specific clinical significance. In addition, Lei et al [9] reported that large tumour diameter was one of the preoperative factors associated with MVI. Thus, we selected early T-stage tumours with MTDs less than 6 cm. However, MTD was still one of the key factors in the preoperative estimation of MVI. Some previous studies have reported that AFP levels were significantly higher in patients with MVI [9, 38]. It has also been reported that AFP levels were positively associated with tumour size [31, 39]. In addition, preoperative age and HBsAg were correlated with the MVI of HCC. Fundamental research has revealed that the HBV-initiated tumourigenic process can play an important role in the development of MVI in HCC [40]. Recently, Lei et al [9] and Wei et al [41] reported that high HBV infection and active HBV replication were associated with the development of MVI in HCC patients. The results of the current study are consistent with those findings. The age of the MVI+ group was significantly lower than that of the MVI− group in both the training and the validation datasets. The predictive value of age with regard to MVI of HCC remains unknown and requires further investigation.
The limitations of the current study include the relatively small sample size, the fact that it was entirely retrospective and thus needs be validated via prospective studies and the lack of multicentre validation. MVI grade was also not taken into account in the MVI+ group, and the tumour signs of CECT were not analysed. One hundred twenty seconds after the contrast injection may be too late as a delay phase.
In conclusion, the radiomics signature identified may be useful as an imaging marker for predicting MVI of HCC preoperatively. Nomograms combining PVP radiomics and CF may prove useful as a tool to guide personalised treatment, although this would require further external validation prior to widespread application in clinical practice.
Abbreviations
- AFP:
-
Alpha-fetoprotein
- AP:
-
Arterial phase
- AUC:
-
Area under the curve
- CECT:
-
Contrast-enhanced computed tomography
- CF:
-
Clinical factor
- CI:
-
Confidence interval
- CT:
-
Computed tomography
- DP:
-
Delay phase
- HBsAg:
-
Hepatitis B surface antigen
- HCC:
-
Hepatocellular carcinoma
- ICC:
-
Intraclass correlation coefficient
- LASSO:
-
Least absolute shrinkage and selection operator
- LR:
-
Liver resection
- MR:
-
Magnetic resonance
- MTD:
-
Maximum tumour diameter
- MVI:
-
Microvascular invasion
- OR:
-
Odds ratio
- PACS:
-
Picture archiving and communication system
- PVP:
-
Portal venous phase
- ROI:
-
Region of interest
- RVI:
-
Radiogenomic venous invasion
- SVM:
-
Support vector machine
References
Ferlay J, Soerjomataram I, Dikshit R et al (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136:E359–E386
Chen W, Zheng R, Baade PD et al (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66:115–132
Forner A, Llovet JM, Bruix J (2012) Hepatocellular carcinoma. Lancet 379:1245–1255
Bruix J, Gores GJ, Mazzaferro V (2014) Hepatocellular carcinoma: clinical frontiers and perspectives. Gut 63:844–855
Gouw AS, Balabaud C, Kusano H, Todo S, Ichida T, Kojiro M (2011) Markers for microvascular invasion in hepatocellular carcinoma: where do we stand? Liver Transpl 17(Suppl 2):S72–S80
Clavien PA, Lesurtel M, Bossuyt PM, Gores GJ, Langer B, Perrier A (2012) Recommendations for liver transplantation for hepatocellular carcinoma: an international consensus conference report. Lancet Oncol 13:e11–e22
Huang C, Zhu XD, Ji Y et al (2017) Microvascular invasion has limited clinical values in hepatocellular carcinoma patients at Barcelona Clinic Liver Cancer (BCLC) stages 0 or B. BMC Cancer 17:58
Shim JH, Jun MJ, Han S et al (2015) Prognostic nomograms for prediction of recurrence and survival after curative liver resection for hepatocellular carcinoma. Ann Surg 261:939–946
Lei Z, Li J, Wu D et al (2016) Nomogram for preoperative estimation of microvascular invasion risk in hepatitis B virus-related hepatocellular carcinoma within the Milan criteria. JAMA Surg 151:356–363
Hirokawa F, Hayashi M, Miyamoto Y et al (2014) Outcomes and predictors of microvascular invasion of solitary hepatocellular carcinoma. Hepatol Res 44:846–853
Choi YS, Rhee H, Choi JY et al (2013) Histological characteristics of small hepatocellular carcinomas showing atypical enhancement patterns on gadoxetic acid-enhanced MR imaging. J Magn Reson Imaging 37:1384–1391
Sterling RK, Wright EC, Morgan TR et al (2012) Frequency of elevated hepatocellular carcinoma (HCC) biomarkers in patients with advanced hepatitis C. Am J Gastroenterol 107:64–74
Shim JH, Han S, Lee YJ et al (2013) Half-life of serum alpha-fetoprotein: an early prognostic index of recurrence and survival after hepatic resection for hepatocellular carcinoma. Ann Surg 257:708–717
Aerts HJ, Velazquez ER, Leijenaar RT et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006
Choi ER, Lee HY, Jeong JY et al (2016) Quantitative image variables reflect the intratumoral pathologic heterogeneity of lung adenocarcinoma. Oncotarget 7:67302–67313
Braman NM, Etesami M, Prasanna P et al (2017) Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res 19:57
Kuo MD, Jamshidi N (2014) Behind the numbers: decoding molecular phenotypes with radiogenomics—guiding principles and technical considerations. Radiology 270:320–325
Mazurowski MA (2015) Radiogenomics: what it is and why it is important. J Am Coll Radiol 12:862–866
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Zhou Y, He L, Huang Y et al (2017) CT-based radiomics signature: a potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdom Radiol (NY) 42:1695–1704
Yamamoto S, Korn RL, Oklu R et al (2014) ALK molecular phenotype in non-small cell lung cancer: CT radiogenomic characterization. Radiology 272:568–576
Kickingereder P, Burth S, Wick A et al (2016) Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology 280:880–889
Elsayes KM, Hooker JC, Agrons MM et al (2017) 2017 version of LI-RADS for CT and MR imaging: an update. Radiographics 37:1994–2017
Friedman J, Hastie T, Tibshirani R (2010) Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33:1–22
Banerjee S, Wang DS, Kim HJ et al (2015) A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology 62:792–800
Huang Y, Liu Z, He L et al (2016) Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung Cancer. Radiology 281:947–957
Huang YQ, Liang CH, He L et al (2016) Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 34:2157–2164
Zhang B, Tian J, Dong D et al (2017) Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin Cancer Res 23:4259–4269
Zhou M, Leung A, Echegaray S et al (2018) Non-small cell lung cancer radiogenomics map identifies relationships between molecular and imaging phenotypes with prognostic implications. Radiology 286:307–315
Renzulli M, Brocchi S, Cucchetti A et al (2016) Can current preoperative imaging be used to detect microvascular invasion of hepatocellular carcinoma? Radiology 279:432–442
Chou CT, Chen RC, Lin WC, Ko CJ, Chen CB, Chen YL (2014) Prediction of microvascular invasion of hepatocellular carcinoma: preoperative CT and histopathologic correlation. AJR Am J Roentgenol 203:W253–W259
Bakr S, Echegaray S, Shah R et al (2017) Noninvasive radiomics signature based on quantitative analysis of computed tomography images as a surrogate for microvascular invasion in hepatocellular carcinoma: a pilot study. J Med Imaging (Bellingham) 4:041303
Rodríguez-Perálvarez M, Luong TV, Andreana L, Meyer T, Dhillon AP, Burroughs AK (2013) A systematic review of microvascular invasion in hepatocellular carcinoma: diagnostic and prognostic variability. Ann Surg Oncol 20:325–339
Lubner MG, Malecki K, Kloke J, Ganeshan B, Pickhardt PJ (2017) Texture analysis of the liver at MDCT for assessing hepatic fibrosis. Abdom Radiol (NY) 42:2069–2078
Davnall F, Yip CS, Ljungqvist G et al (2012) Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging 3:573–589
Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ (2017) CT texture analysis: definitions, applications, biologic correlates, and challenges. Radiographics 37:1483–1503
Tran B, Dancey JE, Kamel-Reid S et al (2012) Cancer genomics: technology, discovery, and translation. J Clin Oncol 30:647–660
Liu C, Xiao GQ, Yan LN et al (2013) Value of alpha-fetoprotein in association with clinicopathological features of hepatocellular carcinoma. World J Gastroenterol 19:1811–1819
Du M, Chen L, Zhao J et al (2014) Microvascular invasion (MVI) is a poorer prognostic predictor for small hepatocellular carcinoma. BMC Cancer 14:38
Chen L, Zhang Q, Chang W, Du Y, Zhang H, Cao G (2012) Viral and host inflammation-related factors that can predict the prognosis of hepatocellular carcinoma. Eur J Cancer 48:1977–1987
Wei X, Li N, Li S et al (2017) Hepatitis B virus infection and active replication promote the formation of vascular invasion in hepatocellular carcinoma. BMC Cancer 17:304
Funding
This study has received funding from the CAMS Innovation Fund for Medical Sciences (CIFMS) (2016-I2M-1-001), PUMC Youth Fund (2017320010), Chinese Academy of Medical Sciences (CAMS) Research Fund (ZZ2016B01), National Natural Science Foundation of China (81227901, 61231004) and National Key R&D Program of China (2017YFA0205200, 2017YFC1309100).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Jie Tian.
Conflict of interest
The authors declare that they have no conflict of interest.
Statistics and biometry
Jingwei Wei kindly provided statistical advice for this manuscript.
One of the authors (Dongsheng Gu) has significant statistical expertise.
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the institutional review board.
Ethical approval
Institutional review board approval was obtained.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 7154 kb)
Rights and permissions
About this article
Cite this article
Ma, X., Wei, J., Gu, D. et al. Preoperative radiomics nomogram for microvascular invasion prediction in hepatocellular carcinoma using contrast-enhanced CT. Eur Radiol 29, 3595–3605 (2019). https://doi.org/10.1007/s00330-018-5985-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-018-5985-y