Abstract
Objectives
To identify disease-related spatial covariance patterns of grey matter volume as an aid in the classification of Parkinson’s disease (PD).
Methods
Seventy structural covariance networks (SCNs) based on grey matter volume covariance patterns were defined using independent component analysis with T1-weighted structural MRI scans (discovery sample, 70 PD patients and 70 healthy controls). An image-based classifier was constructed from SCNs using a multiple logistic regression analysis with a leave-one-out cross-validation-based feature selection scheme. A validation sample (26 PD patients and 26 healthy controls) was further collected to evaluate the generalization ability of the constructed classifier.
Results
In the discovery sample, 13 SCNs, including the cerebellum, anterior temporal poles, parahippocampal gyrus, parietal operculum, occipital lobes, supramarginal gyri, superior parietal lobes, paracingulate gyri and precentral gyri, had higher classification performance for PD. In the validation sample, the classifier had moderate generalization ability, with a mean sensitivity of 81%, specificity of 69% and overall accuracy of 75%. Furthermore, certain individual SCNs were also associated with disease severity.
Conclusions
Although not applicable for routine care at present, our results provide empirical evidence that disease-specific, large-scale structural networks can provide a foundation for the further improvement of diagnostic MRI in movement disorders.
Key Points
• Disease-specific, large-scale SCNs can be identified from structural MRI.
• A new network-based framework for PD classification is proposed.
• An SCN-based classifier had moderate generalization ability in PD classification.
• The selected SCNs provide valuable functional information regarding PD patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Parkinson’s disease (PD) is a common neurodegenerative disorder characterized by bradykinesia, rigidity and resting tremor [1]. In addition, its many non-motor symptoms highlight how PD should be considered a multi-systemic brain disease [2, 3]. Although substantia nigra degeneration is a hallmark of PD, magnetic resonance imaging (MRI) volumetric studies have also shown extensive extra-substantia nigra pathology [4]. Neocortical changes, such as atrophy and impairment in the executive attention network, are related to the freezing of gait [5], while increased cortical thickness, particularly in the limbic region, has been demonstrated to worsen the inhibition of compulsive behaviours in PD [6]. Furthermore, bilateral temporal cortex thinning has been found to be predictive of the early exacerbation of cognitive impairment in PD [7]. These findings raise the question of whether PD originates from vulnerable anatomical regions and further consequent multiple cortico-cortical network alterations that contribute to the clinical phenotype.
Grey matter volume (GMV) covariation was recently proposed as a surrogate method for assessing large-scale structural networks between different cerebral regions [8, 9]. The concept of large-scale structural covariance networks (SCNs) was used to describe the inter-individual differences in regional brain structure covariation with other brain structures across the population. This approach can reflect shared variation in grey matter morphology [10] and provide a quantitative way to investigate the features of cortical organization [11] Voxel-based morphometry studies have previously explored widespread grey matter damage and have been shown to be capable of differentiating between different forms of parkinsonism [12,13,14]. However, the analysis of inter-regional dependencies from SCNs could potentially supplement information obtained from common analyses that consider each anatomical region separately [15]. In addition, alterations in disease-specific, spatially restricted networks have been proven to be associated with the development of disease [16,17,18], suggesting that the SCN approach could be a powerful tool for identifying at-risk individuals and aiding in early differential diagnosis in patients with neurodegenerative diseases.
Recently, the loss of integrity of SCNs in PD has been demonstrated through the comparison of PD patients with healthy individuals [19], and several particular SCNs have also been correlated with various cognitive impairments, such as visuospatial deficits, and loss of letter verbal fluency [20] . The PD-specific spatial atrophy pattern is compatible with a trans-neuronal spreading and can happen even in the early stage of the disease [21]. These results further suggest a network-degenerative mechanism in PD. However, particular investigations focus on large-scale SCNs in PD patients and their utility in terms of disease classification is still limited in number.
In the present study, we aim to evaluate the feasibility of using SCN analysis for the discrimination of PD patients from healthy controls at an individual level by utilizing a discovery and validation sample study design. To this end, first, we extracted SCNs using a data-driven multivariate independent component analysis (ICA) with anatomical MRI scans. In order to identify a set of SCNs with high classification ability, we constructed an SCN-based classifier with a feature selection framework from the discovery sample, and then tested the generalization ability in the validation sample. Finally, to determine the clinical significance of the derived SCNs, we performed post hoc statistical analyses of selected SCNs between study groups and further evaluated their relationship with disease severity.
Materials and methods
Participants in the discovery and validation samples
All the participants or their guardians provided written informed consent prior to participation in the study. Seventy PD patients in the discovery sample and 26 PD patients in the validation sample with no previous history of neurological or psychiatric illnesses or psychotropic medication usage were prospectively enrolled in the neurology department. Patients were included if they had idiopathic PD diagnosed according to the Parkinson’s Disease Society criteria [22] by an experienced neurologist. The disease severity and functional status of each patient were evaluated with the unified Parkinson’s disease rating scale (UPDRS) [23], the modified Hoehn and Yahr stages (HY stage) [24] and the Schwab and England activities of daily living scale (SE-ADL) [25] in the “OFF” state (i.e. 12 h after the withdrawal of medication).
For comparison, 70 healthy controls in the discovery sample and another 26 healthy controls in the validation sample with no medical history of neurologic diseases or psychiatric illnesses, alcohol/substance abuse or head injury were also recruited.
Image acquisition and analytical framework
Whole-brain T1W scans were performed on a 3-T MRI scanner (Signa, General Electric Healthcare, Milwaukee, WI, USA) with an eight-channel phased-array head coil using an axial 3D inversion-recovery prepared fast-spoiled gradient-recalled echo pulse sequence and the following parameters: repetition time/echo time/inversion time = 9.5/3.9/450 ms; flip angle = 15°; number of excitations = 1; field of view = 24 cm; and matrix size = 512 × 512 × 110; yielding size = 0.47 × 0.47 × 1.3 mm3 (without inter-slice gap and interpolation). The acquisition time for the T1W scan was 5 min and 38 s. A systematic overview of the analytical framework is shown in Fig. 1, and the processing steps are summarized below.
Systematic overview of the method used in this study. a A conventional VBM preprocessing pipeline was used to generate the MNI space GMV images. b The set of spatially organized large-scale SCNs and corresponding loading parameters were estimated using spatial ICA. c Spatial regression analysis with the SCN maps was applied to the GMV images to calculate the network integrity indices of each subject. d FS-MLR and an LOOCV-based feature selection scheme were used to construct the classifier based on the network integrity indices of the large-scale SCNs from the discovery sample. e To assess the generalization ability of the classifier, we used an additional validation sample to evaluate the classification accuracy of the constructed classifier. f Multiple regression analyses were also conducted to provide the additional network-symptom information. DARTEL diffeomorphic anatomical registration exponentiated lie algebra, FS-MLR forward stepwise binary multiple logistic regression, GMV grey matter volume, ICA independent component analysis, LOOCV leave-one-out cross-validation, ROC receiver operating characteristic, MNI Montreal Neurological Institute, SCN structural covariance network, VBM voxel-based morphometry
Estimation of voxel-wise grey matter volume
To generate the voxel-wise standard space GMV images, T1W scans were first preprocessed using the voxel-based morphometry (VBM) approach [26]. The pipeline used for the VBM analysis followed that of our previous studies [9, 14, 27] (for details of the voxel-wise GMV estimation, see the eMethods in the Supplemental Material).
Structural covariance network identification
Multivariate spatial ICA, a data-driven method, was used to extract SCNs for all the participants in the discovery sample. This analysis characterized the common inter-subject GMV covariations without setting a priori regions of interests. We used the multivariate exploratory linear optimized decomposition into independent components (MELODIC; FSL v5.0.9; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/) tool to decompose the GMV data set into a set of spatially distinct components and corresponding loading parameters that represents the relative integrity of the SCN for each participant. We performed the ICA decomposition with 70 components (pre-specified approach) to make it consistent with previous high-order SCN and intrinsic functional network studies [28,29,30]. All 70 of these SCN maps can be downloaded and reused via a NeuroVault permanent link (https://neurovault.org/collections/3112/)
Calculation of network integrity indices
The ICA approach provides 70 SCNs for the validation data set. In order to construct the disease classifier based on the degree of network integrity level in the validation sample, an additional spatial regression analysis was conducted to obtain the network integrity indices of these participants. The network integrity indices were calculated using the four-dimensional data set of GMV images in a spatial regression against the 70 unthresholded SCN maps with the single general linear model. The FSL command-line tool “fsl_glm” was used for this calculation [31]. This analytical procedure provided the network integrity index of each corresponding SCN for each participant in the discovery sample (i.e. the beta weights of the spatial regression analysis) [18]. A larger network integrity index indicated a stronger expression of the corresponding SCN in that participant and vice versa. Subsequent SCN-based disease classifier construction and related statistical analyses (post hoc group comparisons of large-scale SCNs and multiple linear correlation analyses with clinical evaluations) were performed on these network integrity indices for the validation sample.
Statistical analysis
Demographic and group comparison of global brain tissue volume
Differences between the study groups were investigated using the two-sample Student t test (age) and analysis of covariance (ANCOVA) for continuous variables, and using the chi-square test for categorical variables (sex). The threshold of statistical significance was set at Bonferroni corrected p value less than 0.05 (SPSS for Windows, Version 17.0, SPSS, Chicago, IL, USA).
Construction of SCN-based classifier of the discovery sample
To construct the SCN-based PD classifier, a forward stepwise binary multiple logistic regression (FS-MLR, inclusion criteria of p < 0.05) was used with 70 network integrity indices as candidate predictors, diagnosis as the dependent variable, and age and gender as nuisance variables. To construct the classifier unbiasedly, the leave-one-out cross-validation (LOOCV)-based feature selection scheme was used to identify the subset of SCNs that best discriminated the PD patients from the healthy controls (for details regarding the classifier construction, see the eMethods in the Supplemental Material) [14]. The model parameters of the final classifier with selected SCNs were estimated from the whole discovery sample. The optimal cut-off probability value of the final classifier was determined by the highest sum of sensitivity and specificity squared using receiver operator characteristic (ROC) analysis [32]. Classification accuracy, specificity, sensitivity, positive/negative predictive value and the Nagelkerke R2 test were used as metrics to evaluate the efficacy of the classifier [33, 34]
Evaluation of constructed classifier using additional validation sample
To simulate real-world application, an additional validation sample was also acquired. The same VBM preprocessing pipeline was also applied for the validation sample. To obtain the network integrity index of the corresponding SCNs in the validation sample, we used the same spatial regression analysis with the 70 unthresholded SCN maps which were obtained from the discovery sample. This analytical pipeline provided us with the set of network integrity indices of each individual in the validation sample. Finally, the constructed classifier was applied to this validation sample to evaluate the generalization ability of the classifier. We also performed a binomial test of statistical significance of the classification accuracy in the validation sample compared to chance-level performance.
Clinical significance of selected SCNs
To assist in interpreting the clinical significance of highly predictive SCNs, the ANCOVA model with age and sex as nuisance variables was used to investigate the network integrity changes of the selected SCNs between study groups. To further investigate the possible network-symptom association in PD patients, multiple linear regression analyses were conducted using the network integrity indices of highly predictive SCNs as predictors, the clinical evaluations/illness duration as dependent variables, and age and gender as nuisance variables. The statistical threshold was set at uncorrected p < 0.05.
Results
Demographics and clinical characteristics
The demographic and clinical data of the participants are shown in Table 1. Sex and age did not differ between the healthy controls and PD patients in both the discovery sample and the validation sample. The mean modified HY stage of all the patients with PD was 2.11, indicating a relatively early stage of the disease, and there were no significant differences among all the disease evaluation scorings. Compared to the healthy controls, the PD patients had a smaller total GMV and larger cerebrospinal fluid volume. Among the PD patients, there were no demographic differences between the two samples. In addition, there were no significant differences in any of the demographic variables or global brain volume of the healthy controls in the discovery and validation samples.
Construction of classifier with highly predictive SCNs
By using FS-MLR with the LOOCV-based feature selection scheme to identify the SCNs with high classification ability, we found that the selected frequency of each SCN exhibited a polarized distribution (Supplementary Fig. 1). Thirteen SCNs (frequency > 99%) were selected for the final classifier (namely SCNs of the cerebellum [IC1, IC18], anterior temporal pole [IC8], hippocampus [IC13], parietal operculum [IC23], occipital [IC37, 43, 48, 53], supramarginal gyrus [IC40, 44], paracingulate gyrus [IC49] and supplementary motor area (SMA) [IC58]; Fig. 2 and Supplementary Table 1). The classifier distinguished PD with 89% accuracy, 90% sensitivity, 87% specificity, 88% positive predictive value and 90% negative predictive value in the discovery sample (Table 2). Furthermore, the goodness of fit for the constructed classifier was 0.73 (Nagelkerke R2). All of these 13 SCN maps with highly predictive ability can be downloaded and reused via a NeuroVault permanent link.
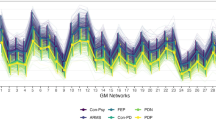
Voxel-wise spatial patterns of the 13 structural covariance networks with high classification ability. This figure displays the spatial location of each SCN and corresponding network integrity index in the two study groups from the discovery sample. To visualize the spatial locations of the SCNs, the spatial maps of each SCN were converted to a spatial mixture model Z-statistic map with thresholded |Z| > 4, and these maps were further overlaid onto an anatomical template in MNI space. The anatomical locations of the SCNs were identified using the Harvard–Oxford cortical and subcortical structural atlases included in the FSL package. The colour bar indicates the corresponding Z value of each SCN. The violin plots show the network integrity index of each SCN in the healthy controls (HC, blue) and the patients with Parkinson’s disease (PD, red) (mean ± SD). The asterisks indicate the statistical differences between the two groups after adjusting for sex and age effects (* indicates a p value less than uncorrected p value 0.05, ** indicates a corrected p value less than 0.05 after FDR correction). All the 13 SCNs can be downloaded and reused via a NeuroVault permanent link. FDR false discovery rate, a.u. arbitrary units, IC independent component, MNI Montreal Neurological Institute, SCN structural covariance network
Generalization performance in the validation sample
In the validation sample, the constructed classifier had moderate generalization ability, with a sensitivity of 81%, specificity of 69% and overall accuracy of 75% (Table 2). The binominal test demonstrated that the classification accuracy in the unseen validation sample was statistically different from chance-level performance (p < 0.001)
Between-group comparisons of selected SCNs
Group differences were found in nine of these 13 SCNs. Compared to the healthy controls, the network integrity indices were lower in the cerebellum (IC1, IC18), anterior temporal pole (IC8), hippocampus (IC13), parietal operculum (IC23) and supramarginal gyrus (IC44) in the PD patients. In contrast, the PD patients exhibited higher network integrity indices in the occipital lobe (IC43, 53) and SMA (IC58). Meanwhile, no significant between-group differences were found in the other four networks (IC37, 40, 48, 49) (Fig. 2 and Supplementary Table 1).
Relationship between clinical evaluations and selected SCNs
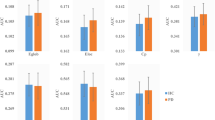
We found that the lower network integrity indices of the selected SCNs were associated with higher disease severities of PD as indicated by the UPDRS I (mentation), UPDRS II (daily activities), UPDRS III (motor function) and general evaluation (UPDRS total, modified HY stage and SE-ASL) scores (Fig. 3). The following network integrity indices of SCNs predicted the UPDRS I score (IC48), UPDRS II score (IC8, IC13, IC18, and IC48), UPDRS III score (IC13, IC18, and IC48), UPDRS total score (IC13, IC18, and IC48), modified HY stage (IC13 and IC48) and SE-ASL (IC13 and IC48). In addition, there were no statistically significant associations between the network integrity indices of selected SCNs and illness duration.
Multiple linear regression analysis of clinical evaluations and network integrity indices of large-scale structural covariance networks in patients with Parkinson’s disease. The relationships between clinical evaluations and the network integrity indices of highly predictive SCNs were revealed by multiple linear regression anlaysis. The standardized beta coefficients are listed in the figure. The grids in black indicate that the beta coefficient is statistically insignificant (uncorrected p > 0.05); the light grey and white grids indicate that the uncorrected p value is less than 0.05 and 0.01, respectively. IC independent component, H&Y Hoehn and Yahr, SCN structural covariance network, SE-ADL Schwab and England activities of daily living scale, UPDRS unified Parkinson’s disease rating scale
Discussion
Using T1W MRI scans with data-driven multivariate ICA analysis, we first extracted the potential whole-brain, large-scale SCNs and further identified a set of SCNs with high classification ability for PD patients. The constructed classifier also demonstrated moderate generalization ability in an unseen validation sample and was associated with several clinical evaluations in individual subjects. Our results showed the value of SCNs as potential network-based image markers for PD classification and disease severity evaluation.
Voxel-based morphometry analysis has been shown to differentiate different forms of parkinsonism with 74–83% sensitivity and 79–94% specificity [12,13,14]. The SCNs have previously been used for exploring the underlying disease pathophysiology. However, the use of this approach for the classification in PD is limited and makes further inter-study comparisons difficult. In one previous study, the accuracy of using SCNs to distinguish between Alzheimer’s patients/controls, Alzheimer’s patients/patients with mild cognitive impairment (MCI) and patients with MCI/controls was 87%, 80% and 86%, respectively [35]. In another study, the combination of CSF biomarkers and ApoE ε4 status with SCNs also achieved excellent diagnostic accuracy for MCI conversion [17]. Resting state functional MRI with a novel covariance of projection approach which integrates multiple network information also achieved acceptable classification accuracy in patients with PD [36]. Similar to a previous study using a multiple-networks approach, our classification model achieved moderate discriminability by using multiple SCNs. The highly predictive SCNs in the classifier, including the basal ganglia, limbic regions, the cerebellum, and other cerebral regions, were consistent with those identified in another functional network study using linear discriminant analysis to evaluate the network regional efficiency [37]. The classification ability of SCNs is comparable to that of voxel-based analysis and the inter-regional dependencies analysis in which supplementary information is obtained from common analyses that consider each anatomical region separately [15].
In addition to such uses of grey matter information, the classification feasibility of different MRI modalities or combined multimodal imaging data has also been studied in PD and exhibited better performance. Nonetheless, the question of which modality or modalities are most useful in a clinical context remains unresolved. The different subjects, sample sizes, image modalities and image analyses of previous studies might affect the classification ability in those studies. For example, one previous study used diffusion-weighted MRI and found that the mean kurtosis of the ipsilateral substantia nigra had good diagnostic accuracy (sensitivity 0.92 and specificity 0.87) in differentiating between PD patients and controls, with that accuracy being better than that of tensor-derived indexes [38]. A recent meta-analysis also demonstrated that the visual assessment of dorsolateral nigral hyperintensity in iron-sensitive MRI provides excellent diagnostic accuracy for distinguishing between PD patients and controls [39]. Furthermore, single basal ganglia network connectivity can also help to differentiate PD patients from controls with 85% accuracy [40]. For all studies, using only the core feature usually yielded higher performance than using the whole brain, such as was used in this SCN-based study. However, those imaging biomarkers with good classification performance are usually a trait marker of the given disease, reflecting a constitutional fault and not the resulting clinical symptoms, and the changes in the biomarker may be profound enough to produce a floor effect where gradation of severity is lost. A multiple-SCNs approach, in contrast, includes the possibility that cortical pathology is predictive of PD if the component regions are more carefully specified a priori.
In the present study, 13 of 70 SCNs were selected to classify PD effectively, including the cerebellum, anterior temporal pole, hippocampus, parietal operculum, supramarginal gyrus, paracingulate gyrus, occipital and SMA. Both higher and lower network integrity indices were identified in PD patients. A higher network integrity index indicates that the spatial pattern of the corresponding SCN is more strongly weighted in the data for that individual. It might reflect malfunction related to disease pathology or compensation that is not efficient enough to counterbalance clinical impairment, while a lower network integrity index might indicate volume atrophy or structural damage. In terms of movement deficits, the lower network integrity indices of the cerebellar SCN in PD suggest declined cerebellar and parietal opercular network integrity in PD and may be related to akinesia/rigidity, tremor, gait disturbance, dyskinesia and the freezing of gait [41, 42]. The lower network integrity indices of the supramarginal gyrus, paracingulate gyrus, anterior temporal lobe and hippocampus might reflect the ventral attentional network disconnection [43], declarative memory impairment [44] and visual hallucinations [45] in PD.
We also found a significantly higher network integrity index in the SMA in PD. The SMA is strongly embedded in motor circuits through its connections with the primary motor cortex, premotor cortex and cingulate cortex [46]. Mal-modulation with increased functional connectivity in the SMA and decreased activity in the inferior frontal cortex [47] can be associated with levodopa-induced dyskinesia in PD. Our aforementioned result thus corresponded to the pathological involvement of the SMA in PD [9, 48] and also might reflect the phenotype of PD.
Actually, brain functions are distributed across different brain areas, and any complex behaviour might require interactions between different brain systems. Therefore, SCNs mainly focused on regional interactions that reveal an information network system in the brain might be useful for clinical phenotype evaluation. Our correlation analyses further demonstrated that those selected diagnostic networks were associated with the common disease severity evaluation scores, such as UPDRS I, II, III and total scores, HY stage and SE-ASL. We found that motor evaluation in PD, like UPDRS III, could be predicted by integrity of hippocampus, cerebellum and occipital network; daily life activities evaluation in UPDRS II can be predicted by anterior temporal lobe, hippocampus, cerebellum and occipital network; and general disease severity evaluation, such as UPDRS total score, modified HY stage and SE-ASL, can also be evaluated by hippocampus, cerebellum and occipital network. The current study suggests that human large-scale SCNs may not only provide the differentiation information for PD diagnosis but also be associated with individual clinical evaluations. The network integrity of SCNs in limbic system, cerebellum and occipital network, which also reflects movement disorder, executive deficits [49] and visual disturbances [45], can become important PD imaging diagnostic makers. Furthermore, an MLR model with SCN analysis might be an alternative method to time-consuming and interviewer-dependent scoring systems to facilitate classification and prognosis evaluation of individual PD. However, further validation is necessary in the future.
Until now, the use of ICA-based SCNs to evaluate PD has been limited. PD-specific atrophy patterns have been identified in the hippocampus, temporal lobes, fronto-parietal regions and the midbrain/cerebellum, and were further correlated with cognitive functions in previous studies [19, 20]. Most of our results were consistent with the findings in those studies. In addition, PD-specific spatial atrophy can mimic intrinsic functional networks derived from healthy participants [19] and highlight the value of SCNs in the evaluation of the network-degenerative mechanism in PD [21]. However, because of the different patient groups enrolled and the different imaging analyses conducted in previous studies, the integrity of some brain networks, such as the occipital lobe, parietal lobe and cerebellum, differed across studies.
Interpretations of the present findings should be viewed with caution. First, it is possible that we included cases that were more “typical” and age- and sex-matched in the present study. Therefore, disease-related changes may be more evident in this study’s sample than in the “real-world” population. Second, it is still unknown whether large-scale SCNs can differentiate between idiopathic PD and “atypical parkinsonism.” In addition, their ability to differentially diagnose PD at an early stage and PD at an advanced stage is not known. Third, the mean education level of the two groups in the present study was different. However, a further supplementary analysis adjusting for education level revealed comparable classification performance in the diagnosis of PD. Last but not least, factors such as the number of ICs, sample size and parameter setting of structural MRI acquisition could also lead to variations in model construction. Future studies with multi-scanners/sites design should be conducted to further confirm our findings.
In conclusion, using structural MRI scans with a data-driven multivariate ICA approach can define the spatial components showing interregional covariant areas relevant to PD pathophysiology. Our results identify an objective set of SCNs to predict PD and can guide future studies aiming to automatically classify PD from a system-level perspective. Application of the ICA-based SCN technique will increase the applicability of structural MRI scans for the identification of potential imaging markers for PD.
Abbreviations
- ANCOVA:
-
Analysis of covariance
- CSF:
-
Cerebrospinal fluid
- DARTEL:
-
Diffeomorphic anatomical registration exponentiated lie algebra
- GMV:
-
Grey matter volume
- HY stage:
-
Hoehn and Yahr stages
- ICA:
-
Independent component analysis
- LOOCV:
-
Leave-one-out cross-validation
- MNI:
-
Montreal Neurological Institute
- MELODIC:
-
Multivariate exploratory linear optimized decomposition into independent components
- PD:
-
Parkinson’s disease
- ROC:
-
Receiver operator characteristic
- SE-ADL:
-
Schwab and England activities of daily living scale
- SCNs:
-
Structural covariance networks
- UPDRS:
-
Unified Parkinson’s disease rating scale
- VBM:
-
Voxel-based morphometry
- WM:
-
White matter
References
Sveinbjornsdottir S (2016) The clinical symptoms of Parkinson's disease. J Neurochem 139:318–324
Chaudhuri KR, Healy DG, Schapira AH, National Institute for Clinical Excellence (2006) Non-motor symptoms of Parkinson's disease: diagnosis and management. Lancet Neurol 5:235–245
Luo C, Song W, Chen Q et al (2014) Reduced functional connectivity in early-stage drug-naive Parkinson's disease: a resting-state fMRI study. Neurobiol Aging 35:431–441
Pan PL, Song W, Shang HF (2012) Voxel-wise meta-analysis of gray matter abnormalities in idiopathic Parkinson's disease. Eur J Neurol 19:199–206
Tessitore A, Amboni M, Cirillo G et al (2012) Regional gray matter atrophy in patients with Parkinson disease and freezing of gait. AJNR Am J Neuroradiol 33:1804–1809
Tessitore A, Santangelo G, De Micco R et al (2016) Cortical thickness changes in patients with Parkinson's disease and impulse control disorders. Parkinsonism Relat Disord 24:119–125
Mak E, Su L, Williams GB et al (2015) Baseline and longitudinal grey matter changes in newly diagnosed Parkinson's disease: ICICLE-PD study. Brain 138:2974–2986
Mechelli A, Friston KJ, Frackowiak RS, Price CJ (2005) Structural covariance in the human cortex. J Neurosci 25:8303–8310
Chou KH, Lin WC, Lee PL et al (2015) Structural covariance networks of striatum subdivision in patients with Parkinson's disease. Hum Brain Mapp 36:1567–1584
Alexander-Bloch A, Giedd JN, Bullmore E (2013) Imaging structural co-variance between human brain regions. Nat Rev Neurosci 14:322–336
Pagani M, Bifone A, Gozzi A (2016) Structural covariance networks in the mouse brain. NeuroImage 129:55–63
Paviour DC, Price SL, Stevens JM, Lees AJ, Fox NC (2005) Quantitative MRI measurement of superior cerebellar peduncle in progressive supranuclear palsy. Neurology 64:675–679
Marquand AF, Filippone M, Ashburner J et al (2013) Automated, high accuracy classification of Parkinsonian disorders: a pattern recognition approach. PLoS One 8:e69237
Lin WC, Chou KH, Lee PL et al (2017) Parkinson's disease: diagnostic utility of volumetric imaging. Neuroradiology 59:367–377
Fornito A, Zalesky A, Breakspear M (2015) The connectomics of brain disorders. Nat Rev Neurosci 16:159–172
Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD (2009) Neurodegenerative diseases target large-scale human brain networks. Neuron 62:42–52
Willette AA, Calhoun VD, Egan JM, Kapogiannis D, Alzheimers Disease Neuroimaging Initiative (2014) Prognostic classification of mild cognitive impairment and Alzheimer's disease: MRI independent component analysis. Psychiatry Res 224:81–88
Hafkemeijer A, Moller C, Dopper EG et al (2016) Differences in structural covariance brain networks between behavioral variant frontotemporal dementia and Alzheimer's disease. Hum Brain Mapp 37:978–988
Zeighami Y, Ulla M, Iturria-Medina Y et al (2015) Network structure of brain atrophy in de novo Parkinson's disease. elife. https://doi.org/10.7554/eLife.08440
Rektorova I, Biundo R, Marecek R, Weis L, Aarsland D, Antonini A (2014) Grey matter changes in cognitively impaired Parkinson's disease patients. PLoS One 9:e85595
Li X, Xing Y, Schwarz ST, Auer DP (2017) Limbic grey matter changes in early Parkinson's disease. Hum Brain Mapp. https://doi.org/10.1002/hbm.23610
Postuma RB, Berg D, Stern M et al (2015) MDS clinical diagnostic criteria for Parkinson's disease. Mov Disord 30:1591–1601
Gasser T, Bressman S, Durr A, Higgins J, Klockgether T, Myers RH (2003) State of the art review: molecular diagnosis of inherited movement disorders. Movement Disorders Society task force on molecular diagnosis. Mov Disord 18:3–18
Goetz CG, Poewe W, Rascol O et al (2004) Movement Disorder Society Task Force report on the Hoehn and Yahr staging scale: status and recommendations. Mov Disord 19:1020–1028
Schwab RS, Engeland A (1969) Projection technique for evaluating surgery in Parkinson's disease. Livingstone, Edinburgh
Ashburner J, Friston KJ (2000) Voxel-based morphometry–the methods. NeuroImage 11:805–821
Yang FC, Chou KH, Fuh JL et al (2013) Altered gray matter volume in the frontal pain modulation network in patients with cluster headache. Pain 154:801–807
Douaud G, Groves AR, Tamnes CK et al (2014) A common brain network links development, aging, and vulnerability to disease. Proc Natl Acad Sci U S A 111:17648–17653
Segall JM, Allen EA, Jung RE et al (2012) Correspondence between structure and function in the human brain at rest. Front Neuroinform 6:10
Smith SM, Fox PT, Miller KL et al (2009) Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci U S A 106:13040–13045
Filippini N, MacIntosh BJ, Hough MG et al (2009) Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc Natl Acad Sci U S A 106:7209–7214
Perkins NJ, Schisterman EF (2006) The inconsistency of "optimal" cutpoints obtained using two criteria based on the receiver operating characteristic curve. Am J Epidemiol 163:670–675
van Stralen KJ, Stel VS, Reitsma JB, Dekker FW, Zoccali C, Jager KJ (2009) Diagnostic methods I: sensitivity, specificity, and other measures of accuracy. Kidney Int 75:1257–1263
Nagelkerke NJD (1991) A note on a general definition of the coefficient of determination. Biometrika 78:691–692
Ortiz A, Munilla J, Alvarez-Illan I, Gorriz JM, Ramirez J, Alzheimer's Disease Neuroimaging Initiative (2015) Exploratory graphical models of functional and structural connectivity patterns for Alzheimer's Disease diagnosis. Front Comput Neurosci 9:132
Ng B, Varoquaux G, Poline JB, Thirion B, Greicius MD, Poston KL (2017) Distinct alterations in Parkinson's medication-state and disease-state connectivity. Neuroimage Clin 16:575–585
Zhang D, Liu X, Chen J, Liu B (2014) Distinguishing patients with Parkinson's disease subtypes from normal controls based on functional network regional efficiencies. PLoS One 9:e115131
Wang JJ, Lin WY, Lu CS et al (2011) Parkinson disease: diagnostic utility of diffusion kurtosis imaging. Radiology 261:210–217
Mahlknecht P, Krismer F, Poewe W, Seppi K (2017) Meta-analysis of dorsolateral nigral hyperintensity on magnetic resonance imaging as a marker for Parkinson's disease. Mov Disord 32:619–623
Szewczyk-Krolikowski K, Menke RA, Rolinski M et al (2014) Functional connectivity in the basal ganglia network differentiates PD patients from controls. Neurology 83:208–214
Wu T, Hallett M (2013) The cerebellum in Parkinson's disease. Brain 136:696–709
Lenka A, Naduthota RM, Jha M et al (2016) Freezing of gait in Parkinson's disease is associated with altered functional brain connectivity. Parkinsonism Relat Disord 24:100–106
Leh SE, Petrides M, Strafella AP (2010) The neural circuitry of executive functions in healthy subjects and Parkinson's disease. Neuropsychopharmacology 35:70–85
Carlesimo GA, Piras F, Assogna F, Pontieri FE, Caltagirone C, Spalletta G (2012) Hippocampal abnormalities and memory deficits in Parkinson disease: a multimodal imaging study. Neurology 78:1939–1945
Cardoso EF, Fregni F, Maia FM et al (2010) Abnormal visual activation in Parkinson's disease patients. Mov Disord 25:1590–1596
Luppino G, Matelli M, Camarda R, Rizzolatti G (1993) Corticocortical connections of area F3 (SMA-proper) and area F6 (pre-SMA) in the macaque monkey. J Comp Neurol 338:114–140
Cerasa A, Pugliese P, Messina D et al (2012) Prefrontal alterations in Parkinson's disease with levodopa-induced dyskinesia during fMRI motor task. Mov Disord 27:364–371
Sawada M, Imamura K, Nagatsu T (2006) Role of cytokines in inflammatory process in Parkinson's disease. J Neural Transm Suppl: 373–381
Monchi O, Petrides M, Mejia-Constain B, Strafella AP (2007) Cortical activity in Parkinson's disease during executive processing depends on striatal involvement. Brain 130:233–244
Funding
This work was supported by funds from the National Science Council (MOST 103-2314-B-182A-010 -MY3 to W-C Lin, MOST 104-2221-E-010-013- to C-P Lin, and MOST 104-2314-B-182A-053 to H-L Chen) and Chang Gung Memorial Hospital (CMRPG891511 and CMRPG8B0831 to W-C Lin, and CMRPG8E0621 to M-H Chen). The authors would like to thank the patients affected by PD and their families for their involvement and altruism. The authors declare no competing financial interests.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Wei-Che Lin in Kaohsiung Chang Gung Memorial Hospital.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
We declare that all human and animal studies have been approved by the Institutional Review Board of Chang Gung Memorial Hospital and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. We declare that all patients gave informed consent prior to inclusion in this study.
Methodology
• prospective
• diagnostic or prognostic study
• performed at one institution
Electronic supplementary material
ESM 1
(DOCX 270 kb)
Rights and permissions
About this article
Cite this article
Lee, PL., Chou, KH., Lu, CH. et al. Extraction of large-scale structural covariance networks from grey matter volume for Parkinson’s disease classification. Eur Radiol 28, 3296–3305 (2018). https://doi.org/10.1007/s00330-018-5342-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-018-5342-1